Abstract
The liver is the main organ that clears circulating lipopolysaccharide (LPS), and hepatocytes are a major cell type involved in LPS uptake. Little is known about the mechanisms for LPS internalization in hepatocytes and what signaling pathways are involved. We show here that LPS uptake is initiated after formation of a multi-receptor complex within lipid rafts. We find that essential components for LPS uptake are CD14, TLR4, MD2, and the β2-integrin CD11b/CD18. Activation of p38 MAPK is also essential for the initiation of LPS uptake, and interestingly, we show that this activation is not through TLR4 signaling by MyD88 but through activation of TIRAP via CD11b/CD18. However, TLR4/MD2 remain essential components at the cell surface as part of the LPS receptor complex. We therefore suggest novel roles for TLR4/MD2, CD11b/CD18, TIRAP, and p38 MAPK in LPS uptake by hepatocytes.
The liver is the first major organ downstream of the gut and is responsible for the clearance and initial recognition of the majority of LPS2 (1). Studies using radiolabeled LPS characterized the uptake and processing of LPS within the liver more than 20 years ago (2, 3). LPS is taken up by both parenchymal cells (hepatocytes) and nonparenchymal cells (Kupffer cells, hepatic stellate cells, etc.) in the liver, and it appears in the bile within 15 min of intravenous injection (4). Controversy remains regarding the relative roles of Kupffer cells and hepatocytes in the uptake and clearance of LPS. Kupffer cells rapidly produce a strong cytokine response to LPS (5) and can deacylate LPS using a lipase (6). Another study suggested that some types of LPS are preferentially taken up by Kupffer cells and deacylated, which then allows for more rapid internalization within hepatocytes (7). However other studies using gadolinium chloride to deplete Kupffer cells have determined that hepatocytes also play a major role in LPS clearance (4). Regardless, it is clear that hepatocytes, which form the largest mass of cells in the liver and are constantly exposed to endotoxin from the gut, play an important role in LPS uptake, and innate immune responses to LPS are dependent on this process.
The mechanisms involved in the internalization of LPS remain poorly understood, however. Our laboratory has previously shown that hepatocytes express the cell surface components of the LPS receptor/signaling complex (CD14/TLR4/MD2) (8-10) and that signaling activated by LPS through this receptor complex on hepatocytes leads to the activation of MAPK signaling proteins, translocation of NFκB to the nucleus, and also production and release of acute phase proteins such as soluble CD14 and LPS-binding protein (8, 9). However, the functional role of the LPS recognition complex on hepatocytes remains uncertain. Here we hypothesize that this complex plays a role in the binding and initiation of uptake of LPS into cells.
In this study we have clarified the mechanism of initial uptake of LPS into primary isolated hepatocytes. As expected, we found that CD14, MD2, and TLR4 were required for LPS uptake. Uptake was also dependent on activation of p38 MAPK, but surprisingly this activation was not dependent on TLR4 signaling. TLR4/MD2 was required at the cell surface, but activation of p38 MAPK was dependent on β2-integrin association with the LPS receptor complex within lipid rafts and also on TIRAP (Toll-IL-1 receptor adaptor protein). We therefore show a novel structural role for TLR4/MD2 in the uptake of LPS, as well as a previously unsuspected role for CD11b/CD18 in the activation of p38 MAPK through TIRAP that is essential for LPS uptake.
EXPERIMENTAL PROCEDURES
Reagents—Ultrapure LPS (Escherichia coli 0111:B4) was from List Biological Laboratories, Inc. (Vandell Way, CA). This LPS does not contain a significant amount of contaminating proteins that could stimulate Toll-like receptor 2 (TLR2) nonspecifically. Alexa-488 Fluor™ E. coli and Salmonella minnesota LPS was from Molecular Probes (Carlsbad, CA). All LPS was tested for purity by separation on silver-stained SDS-PAGE gels, and no detectable tumor necrosis factor was produced in TLR4-/- macrophages in response to any of the LPS used. Williams Medium E was from Invitrogen; fetal calf serum was from Hyclone Laboratories (Logan, UT). SB203580, JNK inhibitor, and U0126 were from Calbiochem. Nystatin and filipin III were from Sigma. Rabbit anti-mouse phospho- and total ERK, JNK, and p38 MAPK were from Cell Signaling Technologies (Beverly, MA). Rat anti-mouse CD14 and CD18 were from BD Pharmingen (San Jose, CA). Anti-CD11b and anti-MD2 were from Abcam (Cambridge, MA), rabbit anti-mouse TLR4, TIRAP, and MyD88 (myeloid differentiation factor 88) were from eBioscience (San Diego, CA).
Animals—Experimental protocols were approved by IACUC at the University of Pittsburgh. C57BL/6 (WT) mice, B6. 129S4Itgam (CD11b-/-), Toll-like receptor 4 mutant mice (C3H/HeJ) (11), C3H/HeOuJ, TLR4-/- (C57BL/10ScN) mice, and their control mice C57BL/10, and CD18-deficent (B6.129Itgb2), which were specific pathogen-free, weighed approximately 20 g and were from Jackson Laboratories (Bar Harbor, ME). CD14-/- mice, a kind gift from Dr. Mason Freeman (Massachusetts General Hospital, Boston, MA) were back-crossed at least six times and bred in our facility. MyD88-/- mice were a kind gift from Dr. R Medzhitov (Howard Hughes Medical Institute). These mice and matched control C57BL/6 mice were treated with sulfamethoxazole (40 mg/ml) and trimethoprim (4 mg/ml) orally in drinking water for the first 8 weeks of life. The mice were used 2 weeks after cessation of antibiotic treatment.
Hepatocyte Isolation and Cell Culture—Hepatocytes were isolated from mice by an in situ collagenase (type VI; Sigma) perfusion technique, modified as described previously (12). Hepatocyte purity exceeded 99% by flow cytometric assay, and viability was typically over 95% by trypan blue exclusion. Hepatocytes (150,000 cells/ml) were plated on gelatin-coated culture plates or coverslips precoated with Collagen I (BD Pharmingen) in Williams medium E with 10% calf serum, 15 mm HEPES, 10-6 m insulin, 2 mm l-glutamine, 100 units/ml penicillin, 100 units/ml streptomycin. Hepatocytes were allowed to attach to plates overnight, and prior to treatment the cell culture medium was changed to serum-free medium.
LPS Uptake into Hepatocytes—100 ng/ml Alexa-488 Fluor™ E. coli LPS was added to hepatocytes plated on coverslips for times up to 90 min and then washed twice in PBS before fixation with 2% paraformaldehyde for 15 min and subsequent staining of the nuclei with Hoescht stain for 30 s. LPS uptake was visualized using Olympus Provis fluorescent microscopy and quantified using Metamorph™ imaging software. Fluorescence intensity was linear over the dose range of LPS of 100 ng/ml to 5 μg/ml. Trypan blue quenching was used to quench extracellular fluorescence and reduced fluorescence by up to 15%, suggesting that the majority of the fluorescence recorded is intracellular LPS. As an additional control hepatocytes were pretreated for 30 min with an excess of Polymyxin B. No LPS uptake was seen after binding to Polymyxin B. Additionally uptake of Alexa-488 bound to Albumin (100 ng/ml) was maximal after 30-45 min and was not TLR4-dependent. The results of fluorescent LPS uptake in hepatocytes were verified in two additional ways: immunofluorescence with anti-E. coli LPS antibody (described below) and assessment of uptake of 14C-radiolabeled smooth E. coli LPS (a kind gift from Dr. R. Munford, University of Texas Southwestern). Isolated hepatocytes were treated with 100 ng/ml of 14C-LPS for up to 90 min. The cells were washed three times in PBS and lysed with 1× cell lysis buffer (Cell Signaling Technologies). Radioactivity was measured in cell lysates by scintillation counting and recorded as cpm.
Immunofluorescence—Hepatocytes plated on coverslips were given 100 ng/ml Alexa-488 Fluor E. coli LPS for up to 90 min, fixed, and stained with Hoescht as described above. The cells were then permeabilized with 0.1% Triton X-100, washed in PBS and PBB (0.5% bovine serum albumin in PBS), blocked with 2% bovine serum albumin in PBS for 1 h with further blocking overnight at 4 °C with whole mouse IgG (1:100 dilution). Mouse anti-E. coli LPS antibody (Abcam) or anti-MD2 antibody was added at 1:200 dilution for 1 h at room temperature. Secondary antibody was goat anti-mouse Fab1 fragments (Cy3, 1:1000 dilution). The coverslips were visualized by confocal microscopy.
Preparation of Cell Lysates and Western Blot Analysis—Treated hepatocytes were washed twice in PBS and lysed with 1× cell lysis buffer (Cell Signaling Technologies) containing 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Triton, 2.5 mm sodium pyrophosphate, 1 mm β-glycerolphosphate, 1 mm Na3VO4, 1 μg/ml leupeptin, and 1 μg/ml phenylmethylsulfonyl fluoride on ice for 10 min. Protein content of cell lysates was determined by BCA protein assay (Pierce). For Western blot, equal protein amounts were separated by SDS-PAGE and transferred onto a nitrocellulose membrane followed by immunostaining with optimized dilutions of primary antibody. Horseradish peroxidase-conjugated secondary antibodies were then used in a standard enhanced chemiluminescence reaction according to the manufacturer's instructions (Pierce).
Immunoprecipitation—Whole cell lysate was obtained from isolated hepatocytes using radioimmune precipitation assay buffer (Sigma) with added proteinase inhibitors. To preclear the lysate, 10 μl of protein A/G-Sepharose Plus beads (Santa Cruz) was added to 200 μg of lysate and incubated at 4 °C for 1 h on a rotating wheel before centrifugation at 3000 rpm for 5 min. After preclearance, 2.5 μg of primary antibody (TLR4 or MD2) was added per sample and incubated for a 1-2 h at 4 °C. 20 μl of protein A/G-Sepharose Plus beads/sample were added, and the samples were incubated overnight, rotating at 4 °C. The samples were centrifuged for 5 min at 3000 rpm, and the pellet washed twice with lysis buffer on ice. 30 μl of sample buffer was added per sample, and the samples were boiled for 5 min, followed by centrifugation (5 min/3000 rpm). The samples were then loaded and separated by size on SDS-PAGE gels, with immunoblotting for TIRAP. Appropriate negative and positive controls were used throughout.
Total RNA Isolation and Reverse Transcription-PCR—Total RNA was extracted from whole liver, isolated hepatocytes, and RAW 264.7 macrophages with Qiagen RNeasy kit (Valencia, CA) as per the manufacturer's protocol. After first strand cDNA synthesis, PCR was performed in a model 480 thermocycler (PerkinElmer Life Sciences). PCR conditions for amplifying the cDNA of CD11b were as follows: initial 30 s at 98 °C, then 98 °C for 10 s, 65 °C for 15 s, and 72 °C for 15 s for 35 cycles. The forward primer sequence was 5′-GATGGTGTCGAGCTCTCTGCG-3′. The reverse primer sequence was 5′-TTGTCTCAACTGTGATGGAGCA-3′. PCR products were fractionated by 1.75% agarose gel electrophoresis, and DNA bands were visualized with ethidium bromide. The expected length of the product is 397 bp.
Transfection with Recombinant Adenoviral Vectors—Adenovirus vectors for CD14 were constructed using plasmid vectors generously provided by Regine Landmann (University Hospital, Basel, Switzerland). Recombinant adenoviruses were generated by cotransfection of Ψ5 and pAdlox-CD14 linearized with SwaI into the adenovirus packaging cell line CRE8, which expresses Cre recombinase. Recombinant adenoviruses were propagated in CRE8 cells, purified by ViraBind Adenovirus Purification kit (Cell Biolabs, San Diego, CA) according to manufacturer directions, subjected to subsequent dialysis according to standard protocols, and stored at -80 °C. Adenovirus vectors containing a dominant negative form of p38 MAPK (AdDNp38) were purchased from Cell Biolabs. This construct contains a mutation in the dual phosphorylation site of p38 from TGY to AGF. Virus was amplified using HEK293 cells and purified as above. AdTLR4 and AdmutTLR4 were from the Vector Core Facility at the University of Pittsburgh. Admut-TLR4 has the same point mutation in the TLR4-TIR domain as C3H/HeJ mice (P712H). Adenoviral vectors or control (AdΨ5) were infected into hepatocytes at a multiplicity of infection of 50 for 48 h before cell treatment.
Transient Transfection Assay—For plasmids, plasmid DNA prepared using the Qiagen Maxi Prep™ was transiently transfected into hepatocytes using Lipofectamine 2000™ reagent according to the manufacturer's instructions (Invitrogen) 48 h before experimentation. For siRNA, Smartpool™ siRNA targeting MyD88, TIRAP, or MD2 or a control nontargeting siRNA were purchased from Dharmacon and used at a final concentration of 30 nm siRNA/well. Transfection into hepatocytes was as above 24 h before experimentation.
Recombinant Soluble CD14—A full-length rat CD14 clone was purified from a dZap cDNA library (Stratagene). The clone was truncated to amino acid 16 downstream of the signal sequence and subcloned in-frame into the yeast expression plasmid, YEpFLAG. A six-histidine tag with in-frame stop codon was introduced upstream of the glycosylphosphatidylinositol anchor site at amino acid 338 and transformed into yeast strain BJ3505. Protein expression and purification were then performed as described previously (13).
TIRAP/MyD88 Inhibitory Peptides—Inhibitory peptide sets for TIRAP and MyD88 were purchased from Imgenex (San Diego, CA) together with the control peptide and were used at a final concentration of 100 mm 24 h prior to experimentation. The TIRAP inhibitory protein acts as a decoy and binds to and blocks TIR-TIR interactions. The MyD88 inhibitory peptide interferes with MyD88 homodimer formation. The efficacy of inhibition was determined by decreased MAPK activation.
Lipid Raft Isolation and Analysis—Treated hepatocytes were washed twice with PBS, and cells were pelleted by centrifugation in 1 ml of PBS. Cell pellets were lysed with ice cold 0.5% Brij58 (Sigma) in TKM buffer (6.057 g of Tris, 1.86 g of KCl, 1.02 g of MgCl2, 0.372 g of EDTA for 1 liter of TKM, pH 7.4) with proteinase inhibitors and kept on ice for 30 min. The lysates were separated by density centrifugation in a discontinuous sucrose gradient (40, 36, and 5% sucrose in TKM). The samples were spun at 250,000 × g in a swinging bucket rotor for 16 h at 4 °C. 1-ml fractions were then taken, and those containing lipid rafts were identified by dot-blot Western hybridization on Protran nitrocellulose with horseradish peroxidase-conjugated cholera toxin B (Sigma). The fractions were then separated by SDS-PAGE to identify proteins within each fraction.
Statistical Analysis—The data are presented as the means ± S.E. The experimental results are analyzed for their significance by Student's t test using SigmaStat (Systat software, San Jose, CA). The significance was established at the 95% confidence level (p < 0.05).
RESULTS
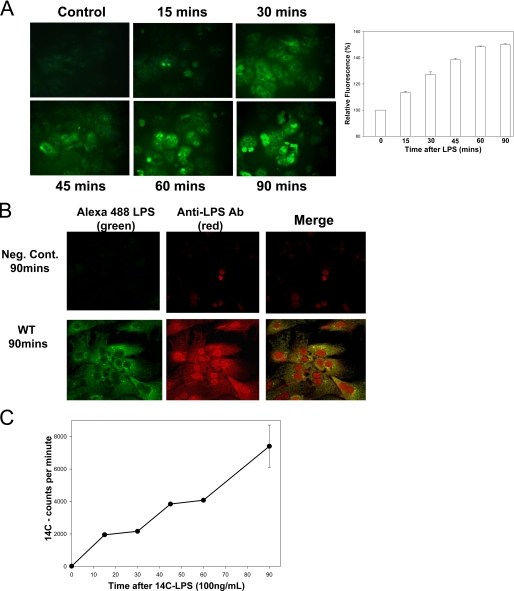
Hepatocytes Take up LPS—The liver is the primary site for the uptake and clearance of circulating LPS (2, 3). To characterize LPS uptake by hepatocytes, we isolated hepatocytes from the livers of C57BL/6 mice and allowed them to adhere overnight to collagen-coated coverslips. The cells were washed, and the medium was changed to serum-free medium so that serum factors able to aid LPS uptake would not interfere with the results. Green fluorescent Alexa-488 Fluor™ E. coli LPS (100 ng/ml) was added for time points up to 90 min before extensive washing to remove excess LPS, followed by fixation in paraformaldehyde. The cells were then visualized by fluorescent microscopy to identify those that had taken up LPS. Hepatocytes were able to take up LPS, with some uptake noted as early as 15 min and maximal uptake between 60 and 90 min (Fig. 1A). The experiments were repeated using a higher concentration of 1 μg/ml LPS to determine whether this altered the dynamics of LPS uptake in hepatocytes. As might be expected there was a higher level of fluorescence of these hepatocytes after LPS uptake, but the uptake occurred over similar time scales (data not shown). In all subsequent experiments we used the lower LPS concentration.
FIGURE 1.
Hepatocytes take up LPS. A, uptake of fluorescent LPS up to 90 min in primary isolated mouse hepatocytes from C57BL/6 (WT) mice. The cells were fixed and visualized by Olympus Provis fluorescent microscopy (×40 magnification) (left panels). Fluorescence was determined relative to base-line background using Metamorph™ software (n = 4/time point) (right panel). B, comparison of LPS uptake in isolated hepatocytes using Alexa Fluor™ E. coli LPS (green) with additional immunostaining with anti-E. coli antibody (red). Negative control (Neg. Cont.) received no fluorescent LPS but was immunostained as above. The merged images show good correlation of green and red fluorescence in the images. C, images representative of three separate experiments. WT (C57BL/6) hepatocytes were given 100 ng/ml radioactive labeled 14C-LPS for time points up to 90 min. The cell lysates were then analyzed for radioactivity by scintillation counting. The error bars show S.E.
We also used two further independent methods to confirm LPS uptake into hepatocytes: immunofluorescence using anti-E. coli LPS antibody and uptake of 14C-radiolableled smooth E. coli LPS (a gift from Dr. R. Munford, University of Texas Southwestern). Similar groups of isolated hepatocytes were treated with Alexa-488 Fluor™ E. coli LPS (100 ng/ml) for up to 90 min and fixed as previously. Immunofluorescence was then performed using anti-E. coli LPS antibody (1:200 dilution) with Cy3-labeled secondary antibodies (1:1000, red) for detection. Both green and red fluorescence were detected separately, and then the images were merged to show colocalization (yellow) (Fig. 1B). There was good matching of green and red fluorescence patterns, confirming internalization of LPS. There was also some staining of the nucleus with the antibody that was not seen with fluorescent LPS. This may represent nonspecific nuclear staining with the antibody or an increased sensitivity of detection of LPS in the nucleus. Uptake of 100 ng/ml 14C-LPS was measured over the same time points in whole cell lysates by scintillation counting. Radioactivity counts in lysates also corresponded with the fluorescent LPS uptake data and the immunofluorescence data (Fig. 1C).
LPS Uptake into Hepatocytes Is CD14- and TLR4-dependent—The requirement of CD14 and TLR4 in the cellular response to LPS is well characterized in macrophages and monocytes (14). CD14 has also been shown to be involved in LPS uptake by murine macrophages (15, 16). We and others have previously shown that hepatocytes express both CD14 and TLR4 (8, 10, 17). TLR4 is part of the LPS receptor complex, although it has not been shown to directly bind LPS (18). LPS binds primarily to other components of the LPS receptor complex at the cell surface, including CD14 and the accessory protein MD2 (19, 20). We therefore wanted to determine whether CD14 and TLR4 were involved in LPS internalization into hepatocytes.
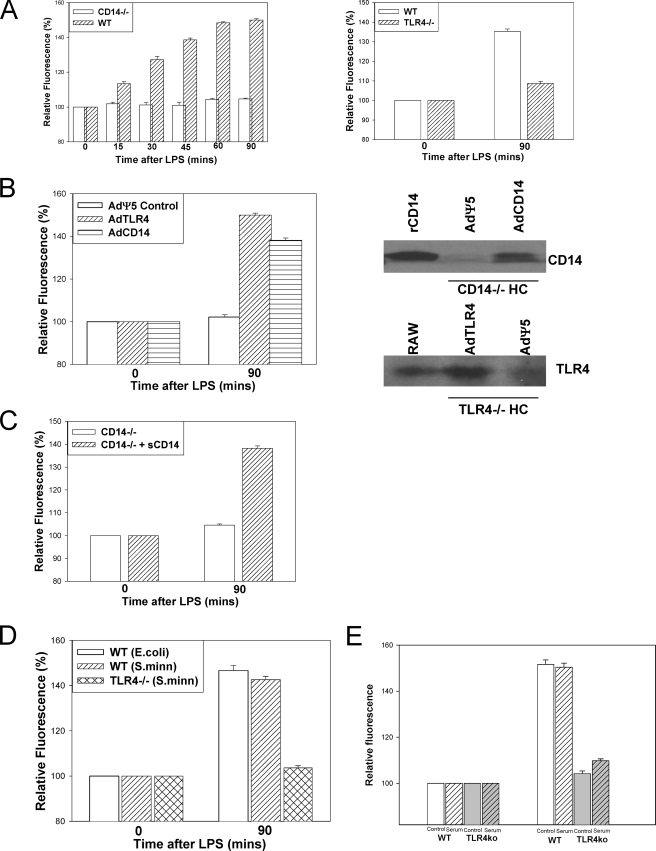
To assess the role of CD14 and TLR4 in LPS uptake, hepatocytes isolated from livers of both WT (C57BL/6) and CD14-/- mice or WT (C57BL/10) and TLR4-/- (C57BL/10ScN) were exposed to 100 ng/ml Alexa-488 Fluor™ E. coli LPS. No LPS uptake was observed in CD14-/- or TLR4-/- hepatocytes after 90 min, compared with WT (Fig. 2A). Even after 4 h there was still no uptake of LPS into either CD14-/- or TLR4-/- hepatocytes (data not shown). These results were confirmed by immunofluorescence using anti-E. coli LPS antibody (data not shown), where we observed no uptake of LPS observed in CD14-/- or TLR4-/- hepatocytes using this method.
FIGURE 2.
Uptake of LPS in isolated hepatocytes is CD14- and TLR4-dependent. A, fluorescent LPS uptake in hepatocytes from WT, CD14-/-, or TLR4-/- mice. Fluorescence is relative to background determined by Olympus Provis fluorescent microscopy (×40 magnification) (n = 4/time point, representative images). B, left panel, LPS uptake after 90 min in CD14-/- hepatocytes pretreated for 24 h with control virus (AdΨ5) or virus expressing full-length CD14 (AdCD14) or TLR4-/- hepatocytes pretreated for 24 h with control virus or virus expressing TLR4 (AdTLR4). Right panels, expression of CD14 and TLR4 confirmed by immunoblot. CD14-/- hepatocytes were pretreated with 1 μg of recombinant soluble CD14 (sCD14) for 1 h before the addition of fluorescent LPS for 90 min. C, uptake of LPS was visualized by fluorescent microscopy, and relative quantitation was performed using Metamorph™. D, hepatocytes were isolated from WT (C57BL/10) and TLR4-/- hepatocytes and treated with 100 ng/ml S. minnesota LPS for 90 min. Hepatocytes were isolated from WT (C57BL/10) and TLR4-/- hepatocytes. Cell culture medium was replaced with media containing 10% fetal calf serum. E, fluorescent LPS (100 ng/ml) was then added for 90 min, and LPS uptake was visualized by fluorescent microscopy. Relative quantitation of LPS uptake was done using Metamorph™.
To establish that the lack of hepatic LPS uptake observed was due to the absence of CD14 or TLR4, we restored expression of CD14 or TLR4 in hepatocytes isolated from CD14-/- or TLR4-/- mice, respectively. We infected CD14-/- hepatocytes with adenovirus containing constructs for full-length CD14 (AdCD14) or control empty vector (AdΨ5), and TLR4-/- hepatocytes were infected with adenovirus containing constructs for full-length TLR4 (AdTLR4) or control adenovirus. In both cases restoration of either CD14 or TLR4 in CD14-/- or TLR4-/- hepatocytes, respectively, was sufficient to enable subsequent LPS uptake by these cells compared with control treated cells (Fig. 2B). In each case the successful expression of CD14 or TLR4 was confirmed by Western blot of cell lysates.
CD14 occurs naturally in two forms: membrane-bound glycosylphosphatidylinositol-linked CD14 and soluble CD14. Soluble CD14 is thought to enable many cells that do not express membrane-bound CD14 to bind LPS and initiate cell signaling via TLR4 or potentially aid in the uptake of LPS into these cells (21). To determine whether soluble CD14 could enhance LPS uptake, CD14-/- hepatocytes were pretreated for 1 h with 1 μg of recombinant soluble CD14 prior to administration of fluorescent LPS. Soluble CD14 alone enabled LPS uptake into CD14-/- hepatocytes, although to a slightly lower level than if membrane-bound CD14 were also present (Fig. 2C).
We also determined uptake of another smooth variant of LPS derived from S. minnesota (Alexa-488 Fluor™ S.minnesota LPS). Experiments similar to those described above were performed using the same dose (100 ng/ml) of Alexa-488 Fluor™ S. minnesota LPS added to WT and TLR4-/- hepatocytes with subsequent fixation and visualization of uptake by fluorescent microscopy. Uptake of S. minnesota LPS was also TLR4-dependent, similar to uptake of E. coli LPS (Fig. 2D). Uptake of E. coli LPS in WT and TLR4-/- hepatocytes cultured in medium containing serum was also not different from uptake in the usual serum-free conditions (Fig. 2E). This indicates that a soluble factor in the serum cannot substitute for cell surface TLR4.
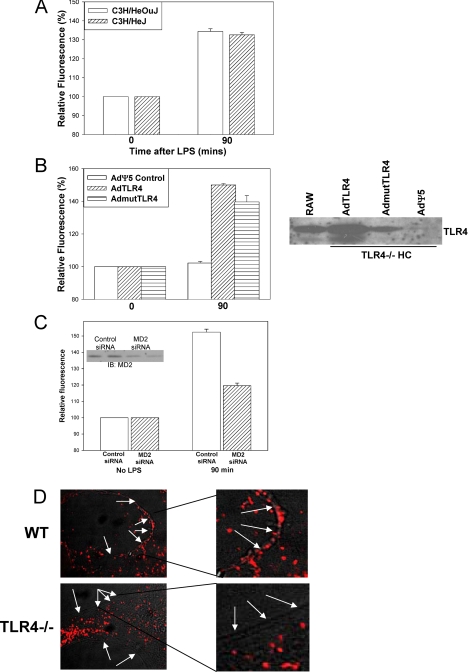
LPS Uptake into Hepatocytes Occurs with Wild Type or Mutated TIR Domains of TLR4—We investigated whether mutations in the TIR domain and alterations of downstream signaling through TLR4 prevented LPS uptake. To do this we used C3H/HeJ mice, which have a point mutation within the TIR domain of TLR4 that prevents association of MyD88 and other downstream signaling mediators after LPS stimulation (11), and their control mice C3H/HeOuJ. To our surprise we found similar levels of LPS uptake into hepatocytes from C3H/HeJ and C3H/HeOuJ mice (Fig. 3A). Once again we used immunofluorescence with anti-LPS antibody to confirm our results (data not shown).
FIGURE 3.
LPS uptake in hepatocytes is not dependent on TLR4 signaling through TIR domain. A, uptake of fluorescent LPS in hepatocytes from C3H/HeOuJ and C3H/HeJ (TLR4 mutant) mice. B, left panel, LPS uptake at 90 min in hepatocytes from TLR4-/- mice pretreated for 48 h with adenovirus expressing TLR4 (AdTLR4), mutant TLR4 (AdmutTLR4), or control (AdΨ5) (×40 magnification, n = 4/experimental group per time point). Right panel, expression of TLR4 confirmed by immunoblot. C, LPS uptake at 90 min in WT hepatocytes pretreated with siRNA targeted against MD2 or a nontargeting control. Inset, knockdown of MD2 confirmed by immunoblot of whole cell lysates. D, isolated hepatocytes from WT and TLR4-/- mice immunostained for MD2 (Cy3, red). The images were taken using confocal microscopy in phase contrast with merged MD2 staining (Cy3, red). The white arrows indicate positions of cell membrane as shown by the phase contrast image. The right panels are magnified portions of the left panels.
To further confirm our results, we used adenoviral vectors to express wild type TLR4 (AdTLR4), TLR4 containing the same point mutation, P712H, in the TIR domain found in C3H/HeJ mice (AdmutTLR4), or AdΨ5 control empty vector virus in TLR4-/- hepatocytes. Cells expressing either TLR4 or the mutated nonsignaling form of TLR4 were both able to take up LPS to a similar degree, whereas the control transduced cells did not take up LPS (Fig. 3B). These data suggest that TLR4 at the cell surface is involved in LPS uptake, although the uptake is not dependent on traditional signaling through TLR4 and MyD88. This implies an alternative and novel signaling pathway initiated by the binding of LPS at the cell surface LPS receptor complex to initiate LPS internalization.
The mechanism involving TLR4 at the cell surface but that did not require signaling through TLR4 for LPS uptake to occur was not immediately apparent. We hypothesized that TLR4 was involved in allowing MD2 to be expressed at the cell surface allowing LPS binding with subsequent initiation of LPS uptake. Hepatocyte MD2 expression was decreased using an siRNA approach. Immunoblot analysis of whole cell lysates (Fig. 3C, inset) confirmed over 50% reduction in the MD2-siRNA-treated cells compared with nontargeting control siRNA-treated cells. The reduction of MD2 expression was associated with a significant decrease in the uptake of Alexa-488 Fluor™ E. coli LPS (Fig. 3C). This suggested that uptake is dependent on MD2 forming part of the LPS receptor complex.
There have been recent studies investigating the role of MD2 in LPS signaling. MD2 rather than TLR4 binds LPS (22-26), but it has also been suggested that MD2 is required for TLR4 to be targeted to the cell membrane (27). We therefore investigated whether TLR4 protein was also important for MD2 to localize to the cell membrane in hepatocytes. We used immunofluorescence and confocal microscopy to determine the location of MD2 in WT, TLR4-/-, C3H/HeJ, and C3H/HeOuJ hepatocytes immunostained for MD2. Hepatocytes from WT (Fig. 3D), C3H/HeOuJ, and C3H/HeJ mice (not shown), all of which take up LPS, had MD2 at the cell membrane as well as within the cell. Hepatocytes from TLR4-/- mice expressed MD2 within the cell but did not express membrane-associated MD2 (Fig. 3D). These data support a role for TLR4 enabling cell surface expression of MD2 in hepatocytes to allow LPS binding and subsequent uptake.
LPS Uptake in Hepatocytes Is Dependent on Activation of p38 MAPK through TIRAP Independently of MyD88—Because we had shown that CD14/TLR4/MD2 were required for LPS uptake, we next investigated whether any of the main pathways known in LPS signaling were also used for LPS uptake. We hypothesized that a signaling adaptor associated with TLR4 might be activated through a novel pathway separate from TLR4-TIR domain signaling. This could allow differential regulation of TLR4 signaling and LPS uptake within the cell. TIRAP and MyD88 are signaling proteins, which associate with TLR4 through the TIR and are involved in one of the main signaling pathways triggered by LPS through TLR4 (14, 28).
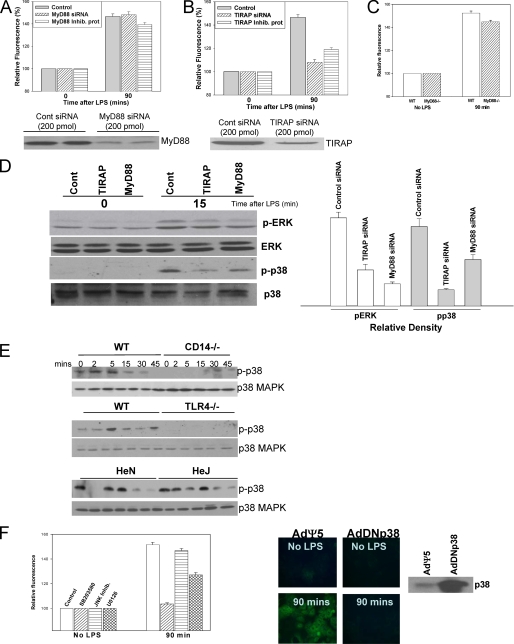
Two separate approaches were used to assess the role of MyD88 and TIRAP in LPS uptake: knockdown of MyD88 or TIRAP using siRNA and suppression of MyD88 or TIRAP function using inhibitory peptides. Nontargeting control siRNA or control peptide was also used as appropriate. Twenty-four hours after siRNA treatment MyD88 protein levels were decreased by more than 75% (Fig. 4A, lower panel), and TIRAP levels decreased by more than 50% (Fig. 4B, lower panel). Additionally LPS-induced activation (phosphorylation) of ERK was significantly decreased using either MyD88 or TIRAP siRNA compared with control (Fig. 4D). Knockdown of MyD88 or inhibition of MyD88 with the inhibitory protein had no significant effect on the uptake of LPS (Fig. 4A). We further confirmed these results using hepatocytes isolated from MyD88-/- mice compared with WT (Fig. 4C). However, either knockdown of TIRAP with siRNA or inhibition of TIRAP using the inhibitory protein significantly decreased the uptake of LPS in hepatocytes (Fig. 4B). It has previously been shown that TIRAP, but not MyD88, interacts with wild type TLR4 as well as the P712H mutated
FIGURE 4.
LPS uptake in hepatocytes is dependent on TIRAP and p38 MAPK activation. A, LPS uptake at 90 min in WT hepatocytes pretreated for 24 h with either siRNA targeting MyD88 or control nontargeting siRNA or pretreated for 24 h with MyD88 inhibitory peptide or control peptide for 24 h. Lower panel, knockdown of MyD88 with siRNA confirmed by immunoblot. B, LPS uptake in WT hepatocytes pretreated with either TIRAP siRNA or control nontargeting siRNA for 24 h or pretreated with TIRAP inhibitory peptide or control peptide for 24 h. Lower panel, knockdown of TIRAP with siRNA confirmed by immunoblot. C, LPS uptake in WT (C57BL/6) and MyD88-/- hepatocytes. D, left panel, Western blot of whole cell lysates from isolated WT hepatocytes pretreated for 24 h with either MyD88, TIRAP, or control siRNA ± LPS for 15 min and immunoblotted for phospho-ERK, total ERK phospho-p38, or total p38 MAPK. Right panel, relative band density for Western blot: pERK and pp38 compared with control. E, Western blot of whole cell lysates from WT, CD14-/-, TLR4-/-, C3H/HeOuJ, and C3H/HeJ hepatocytes to identify activation (phosphorylation) of p38 MAPK and ERK after 100 ng/ml E. coli LPS up to 45 min. F, left panel, inhibition of p38 MAPK by pretreatment with the inhibitor SB203580, but not treatment with JNK inhibitor or U0126 (MEK1/2 inhibitor), prevented uptake of Alexa Fluor™ E. coli LPS (100 ng/ml) into WT hepatocytes (n = 4/experimental group per time point). Right panels, LPS uptake in hepatocytes from WT mice pretreated for 48 h with adenovirus expressing dominant negative p38 MAPK (AdDNp38) or control (AdΨ5) (×40 magnification, images representative of four separate sets of experiments). Expression of p38 was confirmed by immunoblot.
TLR4 found in C3H/HeJ (29). These data therefore confirm a potential role for TIRAP and not MyD88 signaling in the initiation of LPS uptake.
Additionally, we determined phosphorylation of p38 MAPK and ERK in hepatocytes where TIRAP or MyD88 were inhibited. Phosphorylation of p38 MAPK was decreased to a larger extent with inhibition of TIRAP compared with cells where MyD88 was inhibited and with control treated cells (Fig. 4D). These data therefore suggest that TIRAP is required for LPS uptake and signaling through TIRAP-activated p38 MAPK. This activation is not dependent on MyD88, suggesting a signaling role for TIRAP independent of MyD88. This effect of TIRAP inhibition on p38 MAPK activation and LPS uptake suggested a potential role for p38 MAPK in LPS uptake in hepatocytes. The MAPK proteins ERK, JNK, and p38 MAPK are well known to play a major role in LPS signaling and also in pathways involving internalization of receptors and endocytosis (30, 31). Hepatocytes from WT (C57BL/6) and CD14-/- mice, WT (C57BL/10) and TLR4-/- mice, and C3H/HeJ and C3H/HeOuJ mice were exposed to 100 ng/ml E. coli LPS for up to 45 min. ERK and p38 MAPK activation (phosphorylation) was assessed by Western blot. WT hepatocytes phosphorylated p38 MAPK early in response to LPS (Fig. 4E). CD14-/- and TLR4-/- hepatocytes, which were unable to take up LPS in previous experiments, were also unable to phosphorylate p38 MAPK (Fig. 4E), although both cell types were able to activate ERK similarly to WT. However, C3H/HeJ mice that we had previously shown to internalize LPS were also able to phosphorylate p38 MAPK (Fig. 4E). This was unexpected given the mutation in the intracellular portion of TLR4 that prevents TLR4 signaling in response to LPS in C3H/HeJ mice but is consistent with previously published data indicating that C3H/HeJ mice are able to phosphorylate p38 MAPK in response to LPS (32). These data suggested that p38 MAPK activation may be required for internalization of LPS, although activation of p38 MAPK was not dependent on signaling through TLR4.
Next we determined whether p38 MAPK was critical to LPS uptake in WT hepatocytes. We chemically inhibited p38 MAPK as well as JNK or ERK (MEK1/2) in isolated WT hepatocytes using the specific inhibitors SB203580, JNK inhibitor, and U0126, respectively. Inhibition of p38 MAPK, but not JNK, prevented LPS uptake into hepatocytes (Fig. 4F, left panel). There was some decrease in LPS uptake with the ERK/MEK inhibitor but not to the same level as with the p38 MAPK inhibitor. This may represent some MEK inhibitor suppression of p38 MAPK activation.
To further confirm the importance of p38 MAPK activation in LPS uptake, we used adenoviral vectors containing dominant negative p38 (AdDNp38), which cannot be phosphorylated. It contains a mutation of the dual phosphorylation site from TGY to AGF. Expression of AdDNp38 prevented uptake of Alexa-488 Fluor™ LPS into isolated hepatocytes (Fig. 4F, right panels). Chemical inhibition of p38 MAPK or overexpression of dominant negative p38 MAPK also prevented LPS uptake in C3H/HeJ and C3H/HeOuJ hepatocytes (data not shown).
Interestingly, expression of CD14 using AdCD14 in CD14-/- hepatocytes or expression of TLR4 using AdTLR4 or AdmutTLR4 in TLR4-/- hepatocytes, not only restored LPS uptake in these cells but also restored p38 MAPK signaling (data not shown). Similarly, chemical inhibition of p38 MAPK using SB203580 prevented LPS uptake in CD14-/- hepatocytes expressing CD14 or in TLR4-/- hepatocytes expressing TLR4 or mutant TLR4 (data not shown).
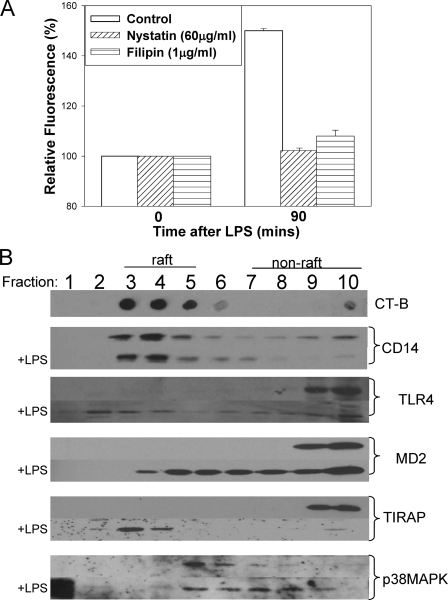
LPS Uptake in Hepatocytes Is Dependent on Formation of the LPS Receptor Complex within Lipid Rafts—Lipid rafts are areas of the cell membrane rich in cholesterol and glycosphingolipids. They have been implicated in multiple signaling and internalization processes within the cell (33, 34). It is thought that lipid rafts form stable areas of the cell membrane that allow interaction between cell surface receptors and signaling molecules inside the cell, as well as allow the formation of relatively large receptor complexes containing multiple proteins (35-37). We hypothesized that lipid rafts would be required to bring together all the elements of the LPS receptor complex needed for LPS uptake. Pretreatment with nystatin or filipin, to disrupt lipid rafts, for 30 min prior to the addition of fluorescent LPS inhibited LPS uptake into isolated hepatocytes (Fig. 5A), supporting an essential role for lipid rafts in LPS uptake into hepatocytes.
FIGURE 5.
LPS uptake in hepatocytes is lipid raft-dependent. A, LPS uptake at 90 min in WT hepatocytes isolated from WT mice pretreated with lipid raft disruptors nystatin, filipin, or control (Me2SO) for 1 h (n = 4/group/time point). B, lipid raft fractions isolated by ultracentrifugation in a discontinuous sucrose gradient from hepatocyte cell lysates, ± 100 ng/ml E. coli LPS for 15 min. Fractions containing lipid rafts were identified by Western dot-blot using horseradish peroxidase-cholera toxin subunit B (CT-B). Western blot of WT hepatocyte fractions separated by SDS-PAGE was immunoblotted for CD14, TLR4, MD2, TIRAP, p38 MAPK ± LPS for 15 min.
To determine whether LPS recognition components associate with lipid rafts in hepatocytes, we isolated lipid raft fractions from WT hepatocytes treated with 100 ng/ml LPS up to 30 min. Cultured hepatocytes were lysed using a mild detergent that keeps lipid raft areas of the membrane intact. Fractions were separated by ultracentrifugation in noncontinuous sucrose gradients, with fractions containing lipid rafts determined on dot-blots by the binding of cholera toxin B to the lipid rafts. Samples from each fraction were then separated by SDS-PAGE and immunoblotted to identify associated proteins in each fraction. CD14 was clearly associated with lipid raft fractions and remained within lipid rafts after the addition of LPS (Fig. 5B). This is consistent with previous findings by others suggesting that glycosylphosphatidylinositol-linked proteins are resident in lipid rafts (37). TLR4 was not associated with lipid raft fractions in resting WT hepatocytes but associated with lipid rafts as quickly as 5 min after the addition of LPS and remained associated for the 30 min studied (Fig. 5B). A similar pattern of association with lipid rafts was observed with MD2 and TIRAP in WT hepatocytes before and after LPS stimulation (Fig. 5B). The association of p38 MAPK followed a different pattern; there was association with lipid rafts in resting hepatocytes, followed by increasing association with nonlipid raft fractions after the addition of LPS (Fig. 5B). These data support our other findings suggesting the formation of a large LPS receptor complex consisting of at least CD14, TLR4, MD2, and TIRAP, which allows internalization of LPS through the activation of p38 MAPK.
Interestingly, in CD14-/- and TLR4-/- hepatocytes we did not see association of members of this LPS receptor complex in lipid rafts after stimulation with LPS, suggesting an inability to form the complex if components are missing (data not shown). However, C3H/HeJ and C3H/HeOuJ cells localized CD14, TLR4 and MD2 to raft fractions after LPS stimulation over similar time courses to WT (data not shown). These data provide further evidence of the need for the formation of the whole LPS receptor complex within the lipid raft to initiate p38 MAPK signaling and LPS uptake.
LPS Uptake in Hepatocytes Requires β2-Integrins—The data above suggest that activation of p38 MAPK via TIRAP is critical to LPS internalization through a pathway that is TLR4-TIR domain signaling-independent. However, we have also shown that CD14, TLR4, and MD2 are important in LPS uptake initiation. It has been shown in studies of macrophages and monocytes that LPS signaling occurs following the formation of an LPS receptor complex that includes multiple proteins in addition to CD14/TLR4/MD2 (36, 37). CD11b/CD18, a member of the β2-integrin family, has been associated with LPS signaling and with the LPS receptor complex in macrophages (38). CD11b/CD18 is a transmembrane signaling protein known to bind LPS and was originally postulated to be the main LPS receptor before the discovery of TLR4 (39-41). More recently CD11b/CD18 has been shown to be required for TIRAP to localize to the plasma membrane (42). CD11b/CD18 therefore has the potential to initiate signaling separately from TLR4 as part of an LPS receptor complex and also to potentially activate TIRAP.
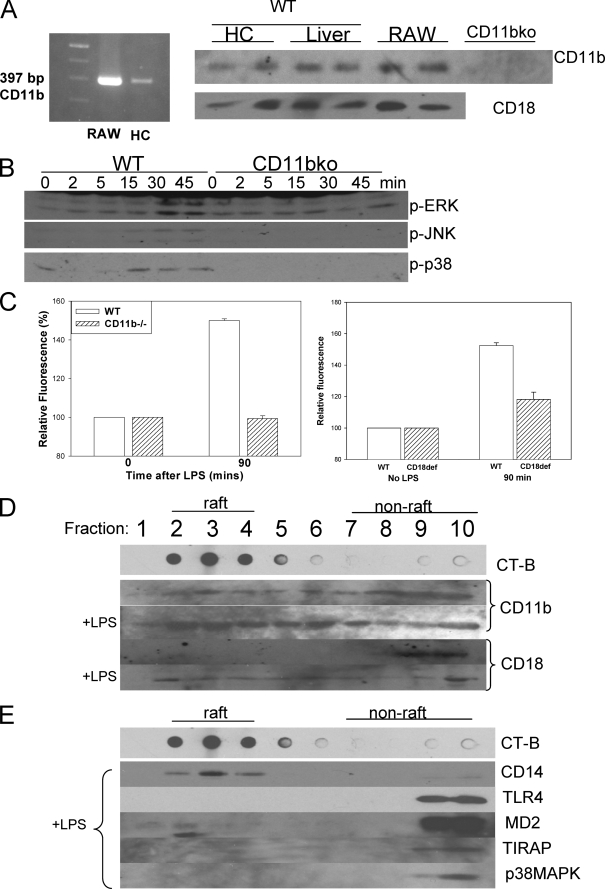
CD11b/CD18 has not previously been shown to be expressed in hepatocytes. Using reverse transcription-PCR we identified mRNA expression of CD11b in mouse hepatocytes (Fig. 6A), using RAW cells as a positive control. We also used identified CD11b and CD18 protein expression by Western blot in whole liver and whole cell lysate of isolated hepatocytes (Fig. 6A). To determine whether β2-integrins are important in LPS uptake, we used CD11b-/- mice (B6.B129S4Itgam) that do not express the CD11b/CD18 complex. Previous studies by others using these mice have shown decreased sensitivity of macrophages to LPS and reduced cellular responses to LPS (38, 43). Similarly, we found that there was a decreased activation of ERK, JNK, and p38 MAPK compared with WT in isolated CD11b-/- hepatocytes treated with 100 ng/ml LPS (Fig. 6B). There was also little NFκB activation in CD11b-/- hepatocytes compared with WT in response to LPS (data not shown). As above, we investigated uptake of fluorescent LPS into isolated hepatocytes from WT and CD11b-/- mice. There was no significant uptake of LPS into CD11b-/- hepatocytes compared with uptake seen in WT hepatocytes (Fig. 6C). CD18-deficient (B6.129Itgb2) mice have significantly reduced levels of CD18, although some mRNA and protein can be detected (44). Uptake of fluorescent LPS in CD18-deficient hepatocytes was also decreased (Fig. 6C). Altogether these data suggest that CD11b/CD18 is involved as part of the LPS receptor complex to enable LPS uptake and activation of p38 MAPK independently of signaling through TLR4.
FIGURE 6.
Activation of p38 MAPK requires β2-integrins and β2-integrin localization to lipid rafts. A, left panel, CD11b mRNA expression in RAW cells (positive control) and mouse WT hepatocytes (HC) using reverse transcription-PCR. Right panel, CD18 and CD11b protein expression by Western blot in RAW cells (positive control), WT liver tissue lysates, and cell lysates from isolated WT and CD11b-/- hepatocytes. B, Western blot of whole cell lysates from WT and CD11b-/- (CD11bko) hepatocytes to show activation (phosphorylation) of ERK, JNK, and p38 MAPK after addition of 100 ng/ml E. coli LPS. LPS uptake at 90 min in hepatocytes isolated from WT, CD11b-/-, and CD18-deficient mice. C, the cells were fixed and visualized by fluorescent microscopy, and fluorescence relative to background was determined (n = 4/group/time point). D, lipid raft fractions isolated from WT hepatocytes ± LPS for 15 min separated by SDS-PAGE and immunoblotted for CD11b or CD18. E, lipid raft fractions from CD11b-/- hepatocytes ± LPS for 15 min separated by SDS-PAGE and immunoblotted for CD14, TLR4, MD2, TIRAP, and p38 MAPK.
We then looked to see whether CD11b and CD18 were associated with lipid rafts after LPS stimulation of WT hepatocytes. We found that CD11b associated with both lipid raft and nonlipid raft fractions, and this pattern was not altered by the addition of LPS (Fig. 6D). However, CD18 was associated with nonlipid raft fractions prior to the addition of LPS but moved into the raft as early as 5 min after the addition of LPS and remained there for the 30 min studied (Fig. 6D). We also investigated protein association with lipid rafts in isolated hepatocytes from CD11b-/- cells. Again CD14 associated with the lipid raft both before and after LPS stimulation (Fig. 6E). Interestingly, however, proteins that were found to associate with lipid raft fractions in WT hepatocytes after LPS did not do so after LPS stimulation in CD11b-/- hepatocytes, including TLR4, MD2, and TIRAP (Fig. 6E). This suggests that the LPS receptor complex was unable to form within the lipid raft in the absence of CD11b. This is therefore further evidence that CD11b/CD18 is an important signaling partner in the LPS receptor complex initiating signaling for LPS internalization separate from TLR4.
DISCUSSION
Within the liver, hepatocytes form a large mass of cells possessing an LPS recognition system, the purpose of which is largely unknown. Because the liver is the primary site for the clearance of LPS from the circulation, we postulated that hepatocytes express an LPS recognition complex for the purpose of LPS uptake. We show here that CD14, MD2, TLR4, and CD11b/CD18 are all involved in hepatocyte LPS uptake. Furthermore, we demonstrate that uptake is p38 MAPK-dependent and that p38 MAPK activation is initiated by CD11b/CD18 and TIRAP. Although TLR4 is required, uptake does not involve typical signaling via the TIR domain.
LPS clearance by the liver was initially studied over 20 years ago, and uptake of LPS has previously been shown to occur in both hepatocytes and Kupffer cells (45). We now show that pure isolated hepatocytes are able to take up LPS. This suggests that Kupffer cells are not required for uptake to occur in hepatocytes, and this confirms previous experiments by others using gadolinium chloride pretreatment (4). There is well supported evidence in the literature showing that Kupffer cells rapidly take up LPS (1, 2, 46-49). They are also, obviously, extremely important in the cytokine response to LPS and overall innate immune response. Deacylation of LPS also takes place in Kupffer cells (6), as well as in hepatocytes (50). It is possible that Kupffer cells are the primary site for clearance of low, intermittent boluses of LPS that may come from the gut. The slower LPS uptake by hepatocytes would provide a clearance mechanism in the event of higher levels of circulating LPS above a threshold level. However, until now the mechanisms and signaling pathways involved in the process of LPS uptake into hepatocytes have not been extensively studied. Our finding that the LPS receptor complex on hepatocytes is important in LPS uptake provides one physiologic explanation for the expression of TLR4, MD2, CD14, and β2-integrin on this cell type. Our previous work has shown that TLR4 is also involved in hypoxic signaling in hepatocytes (51).
An initial event in any uptake process is binding to elements at the cell surface. This initiates a signaling cascade leading to uptake/endocytosis, which can occur in multiple ways. Both CD14 and MD2 have been shown to bind LPS directly, and it is thought that different LPS subtypes can bind with different affinities to CD14 and MD2, thereby producing some specificity of LPS response (19, 20). However, TLR4 has not been shown to bind directly to LPS (28). How the binding of LPS to CD14 and/or MD2 initiates the activation of signaling through TLR4 remains a mystery, although recent studies have suggested that a conformational change in TLR4 may take place when it associates with CD14/MD2 in the presence of LPS (51). The conformational change presumably would allow the intracellular TIR domain of TLR4 to then interact with intracellular signaling proteins such as MyD88 and TIRAP to initiate the signaling cascade that produces innate responses to LPS.
A novel aspect of our findings is that this signaling cascade through MyD88 is not required to initiate uptake of LPS, but TLR4 itself is required. TLR4 is expressed on the cell surface as a complex with MD2. Additionally, the N-terminal region of TLR4 is thought to be essential to allow MD2 binding in the endoplasmic reticulum and allows receptor translocation to the cell surface (27, 52). Our results suggest that TLR4/MD2/CD14 is important for the formation of a larger LPS receptor complex at the cell surface, which at least includes CD11b/CD18 and which is involved in initiating LPS uptake. CD11b/CD18 have previously been shown to bind LPS (40), and recently the binding sites for LPS have been identified on CD18 (53). This could allow direct LPS signaling through CD11b/CD18 when associated with in the lipid raft as part of the larger LPS receptor complex to activate TIRAP and p38 MAPK and initiate LPS uptake. It is unclear at present how binding of LPS to the initial complex initiates the association of β2-integrins within the lipid raft, although this could also involve conformational changes within the receptor complex itself. Additionally TLR4 activation at the cell surface could activate TIRAP directly, separately from MyD88, to initiate LPS uptake.
For many years studies in macrophages and monocytes have associated β2-integrins with LPS signaling (39-41) and also with the LPS receptor complex within lipid rafts following LPS binding (36, 37). However, until very recently there was no information about the exact role β2-integrins were playing in TLR4 signaling or uptake. Kagan and Medzhitov (42) recently reported that CD11b/CD18 is vital to TLR4 signaling through localization of TIRAP to the plasma membrane. This was the first indication of the direct involvement of β2-integrins in TLR4 signaling. Our findings have now defined a role for CD11b/CD18 within the receptor cluster for the initiation of LPS uptake, which complements its recently described role in TLR4 signaling (42). It is clear from our study that both CD14 and TLR4/MD2 are important for LPS uptake, although signaling through the TLR4-MyD88 pathway is not required. This implies that LPS uptake mechanisms and signaling are separate, with likely separate functions as well as separate regulation of these functions. This understanding will be crucial in identifying the role of LPS uptake during and in further identifying the pathways involved.
We have also shown that signaling does involve TIRAP and subsequent activation of p38 MAPK. TIRAP is well known for its interaction with TLR4 and MyD88 to initiate the MyD88-dependent pathway of signaling for LPS. However, TIRAP has also previously been shown to directly activate signaling pathways through the activation of TRAF6, which can itself activate p38 MAPK (54, 55). This is one possible mechanism of activation of p38 MAPK that is TIRAP-dependent but independent of TLR4/MyD88 signaling. p38 MAPK regulates multiple pathways of endocytosis and phagocytosis often through the regulation of activation of small G-proteins such as Rho, Rac, and Rab (31, 56) or through regulation of MAPK kinases (57). There is therefore a potential link between signaling through CD11b/CD18, activation of p38 MAPK, and subsequent changes in cytoskeleton resulting in endocytosis or phagocytosis.
Our study has clarified which receptors are involved in the process of hepatocyte LPS recognition at the cell surface and subsequent uptake into hepatocytes and correlated that with LPS clearance. We have identified a novel role for CD11b/CD18 within the LPS receptor complex in the lipid raft to activate TIRAP and p38 MAPK to initiate LPS uptake. Our current findings provide a major advance in our understanding of how LPS uptake occurs in hepatocytes, define LPS uptake as a process distinct from LPS signaling, and widen our view of the LPS receptor complex and the roles of proteins and their signaling partners within this complex.
Acknowledgments
We thank Hong Liao, Rick Shapiro, Carol Meiers, Danielle Reiser, and Deb Williams for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM-50441. This work was also supported by an Astellas Young Investigator Award 2007 from the National Foundation for Infectious Diseases and the Infectious Disease Society of America (to M. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: LPS, lipopolysaccharide; TLR, Toll-like receptor; Ad, adenoviral; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; WT, wild type; PBS, phosphate-buffered saline; siRNA, small interfering RNA; MEK, MAPK/ERK kinase.
References
- 1.Mathison, J. C., and Ulevitch, R. J. (1979) J. Immunol. 123 2133-2143 [PubMed] [Google Scholar]
- 2.Freudenberg, M. A., Freudenberg, N., and Galanos, C. (1982) Br. J. Exp. Pathol. 63 56-65 [PMC free article] [PubMed] [Google Scholar]
- 3.Hopf, U., Ramadori, G., Moller, B., and Galanos, C. (1984) Am. J. Emerg. Med. 2 13-19 [DOI] [PubMed] [Google Scholar]
- 4.Mimura, Y., Sakisaka, S., Harada, M., Sata, M., and Tanikawa, K. (1995) Gastroenterology 109 1969-1976 [DOI] [PubMed] [Google Scholar]
- 5.Kmiec, Z. (2001) Adv. Anat. Embryol. Cell Biol. 161 1-151 [DOI] [PubMed] [Google Scholar]
- 6.Shao, B., Lu, M., Katz, S. C., Varley, A. W., Hardwick, J., Rogers, T. E., Ojogun, N., Rockey, D. C., Dematteo, R. P., and Munford, R. S. (2007) J. Biol. Chem. 282 13726-13735 [DOI] [PubMed] [Google Scholar]
- 7.Treon, S. P., Thomas, P., and Broitman, S. A. (1993) Proc. Soc. Exp. Biol. Med. 202 153-158 [DOI] [PubMed] [Google Scholar]
- 8.Liu, S., Khemlani, L. S., Shapiro, R. A., Johnson, M. L., Liu, K., Geller, D. A., Watkins, S. C., Goyert, S. M., and Billiar, T. R. (1998) Infect. Immun. 66 5089-5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vodovotz, Y., Liu, S., McCloskey, C., Shapiro, R., Green, A., and Billiar, T. R. (2001) J. Endotoxin. Res. 7 365-373 [PubMed] [Google Scholar]
- 10.Liu, S., Gallo, D. J., Green, A. M., Williams, D. L., Gong, X., Shapiro, R. A., Gambotto, A. A., Humphris, E. L., Vodovotz, Y., and Billiar, T. R. (2002) Infect. Immun. 70 3433-3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poltorak, A., Smirnova, I., He, X., Liu, M. Y., Van Huffel, C., McNally, O., Birdwell, D., Alejos, E., Silva, M., Du, X., Thompson, P., Chan, E. K., Ledesma, J., Roe, B., Clifton, S., Vogel, S. N., and Beutler, B. (1998) Blood Cells Mol. Dis. 24 340-355 [DOI] [PubMed] [Google Scholar]
- 12.Seglen, P. O. (1976) Methods Cell Biol. 13 29-83 [DOI] [PubMed] [Google Scholar]
- 13.Twining, S. S., Goryshin, I. Y., Bhasin, A., and Reznikoff, W. S. (2001) J. Biol. Chem. 276 23135-23143 [DOI] [PubMed] [Google Scholar]
- 14.Palsson-McDermott, E. M., and O'Neill, L. A. (2004) Immunology 113 153-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tapping, R. I., Gegner, J. A., Kravchenko, V. V., and Tobias, P. S. (1998) Prog. Clin. Biol. Res. 397 73-78 [PubMed] [Google Scholar]
- 16.Kitchens, R. L., Ulevitch, R. J., and Munford, R. S. (1992) J. Exp. Med. 176 485-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fearns, C., Kravchenko, V. V., Ulevitch, R. J., and Loskutoff, D. J. (1995) J. Exp. Med. 181 857-866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyakushima, N., Mitsuzawa, H., Nishitani, C., Sano, H., Kuronuma, K., Konishi, M., Himi, T., Miyake, K., and Kuroki, Y. (2004) J. Immunol. 173 6949-6954 [DOI] [PubMed] [Google Scholar]
- 19.Gangloff, S. C., Zahringer, U., Blondin, C., Guenounou, M., Silver, J., and Goyert, S. M. (2005) J. Immunol. 175 3940-3945 [DOI] [PubMed] [Google Scholar]
- 20.Coats, S. R., Pham, T. T., Bainbridge, B. W., Reife, R. A., and Darveau, R. P. (2005) J. Immunol. 175 4490-4498 [DOI] [PubMed] [Google Scholar]
- 21.Moreno, C., Merino, J., Ramirez, N., Echeverria, A., Pastor, F., and Sanchez-Ibarrola, A. (2004) Microbes. Infect. 6 990-995 [DOI] [PubMed] [Google Scholar]
- 22.Gangloff, M., and Gay, N. J. (2004) Trends Biochem. Sci. 29 294-300 [DOI] [PubMed] [Google Scholar]
- 23.Visintin, A., Latz, E., Monks, B. G., Espevik, T., and Golenbock, D. T. (2003) J. Biol. Chem. 278 48313-48320 [DOI] [PubMed] [Google Scholar]
- 24.da Silva, C. J., and Ulevitch, R. J. (2002) J. Biol. Chem. 277 1845-1854 [DOI] [PubMed] [Google Scholar]
- 25.Viriyakosol, S., Tobias, P. S., Kitchens, R. L., and Kirkland, T. N. (2001) J. Biol. Chem. 276 38044-38051 [DOI] [PubMed] [Google Scholar]
- 26.Akashi, S., Saitoh, S., Wakabayashi, Y., Kikuchi, T., Takamura, N., Nagai, Y., Kusumoto, Y., Fukase, K., Kusumoto, S., Adachi, Y., Kosugi, A., and Miyake, K. (2003) J. Exp. Med. 198 1035-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy, M. N., Mullen, G. E., Leifer, C. A., Lee, C., Mazzoni, A., Dileepan, K. N., and Segal, D. M. (2004) J. Biol. Chem. 279 34698-34704 [DOI] [PubMed] [Google Scholar]
- 28.Fitzgerald, K. A., Rowe, D. C., and Golenbock, D. T. (2004) Microbes. Infect. 6 1361-1367 [DOI] [PubMed] [Google Scholar]
- 29.Horng, T., Barton, G. M., and Medzhitov, R. (2001) Nat. Immunol. 2 835-841 [DOI] [PubMed] [Google Scholar]
- 30.Cavalli, V., Vilbois, F., Corti, M., Marcote, M. J., Tamura, K., Karin, M., Arkinstall, S., and Gruenberg, J. (2001) Mol. Cell 7 421-432 [DOI] [PubMed] [Google Scholar]
- 31.Dong, C., Davis, R. J., and Flavell, R. A. (2002) Annu. Rev. Immunol. 20 55-72 [DOI] [PubMed] [Google Scholar]
- 32.Kraatz, J., Clair, L., Rodriguez, J. L., and West, M. A. (1999) Shock 11 58-63 [DOI] [PubMed] [Google Scholar]
- 33.Triantafilou, M., Miyake, K., Golenbock, D. T., and Triantafilou, K. (2002) J. Cell Sci. 115 2603-2611 [DOI] [PubMed] [Google Scholar]
- 34.Parton, R. G., and Richards, A. A. (2003) Traffic 4 724-738 [DOI] [PubMed] [Google Scholar]
- 35.Olsson, S., and Sundler, R. (2006) Mol. Immunol. 43 607-612 [DOI] [PubMed] [Google Scholar]
- 36.Schmitz, G., and Orso, E. (2002) Curr. Opin. Lipidol. 13 513-521 [DOI] [PubMed] [Google Scholar]
- 37.Triantafilou, M., and Triantafilou, K. (2002) Trends Immunol. 23 301-304 [DOI] [PubMed] [Google Scholar]
- 38.Perera, P. Y., Mayadas, T. N., Takeuchi, O., Akira, S., Zaks-Zilberman, M., Goyert, S. M., and Vogel, S. N. (2001) J. Immunol. 166 574-581 [DOI] [PubMed] [Google Scholar]
- 39.Ingalls, R. R., and Golenbock, D. T. (1995) J. Exp. Med. 181 1473-1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingalls, R. R., Arnaout, M. A., and Golenbock, D. T. (1997) J. Immunol. 159 433-438 [PubMed] [Google Scholar]
- 41.Flaherty, S. F., Golenbock, D. T., Milham, F. H., and Ingalls, R. R. (1997) J. Surg. Res. 73 85-89 [DOI] [PubMed] [Google Scholar]
- 42.Kagan, J. C., and Medzhitov, R. (2006) Cell 125 943-955 [DOI] [PubMed] [Google Scholar]
- 43.Flo, T. H., Ryan, L., Kilaas, L., Skjak-Braek, G., Ingalls, R. R., Sundan, A., Golenbock, D. T., and Espevik, T. (2000) Infect. Immun. 68 6770-6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, R. W., Ballantyne, C. M., Smith, C. W., Montgomery, C., Bradley, A., O'Brien, W. E., and Beaudet, A. L. (1993) J. Immunol. 151 1571-1578 [PubMed] [Google Scholar]
- 45.Bikhazi, A. B., Jurjus, A. R., Kamal, M. T., Al Housseini, A. M., Saab, R. N., Jaroudi, W. A., and Bitar, K. M. (2001) Comp. Biochem. Physiol C. Toxicol. Pharmacol. 129 339-348 [DOI] [PubMed] [Google Scholar]
- 46.Ge, Y., Ezzell, R. M., Clark, B. D., Loiselle, P. M., Amato, S. F., and Warren, H. S. (1997) J. Infect. Dis. 176 1313-1321 [DOI] [PubMed] [Google Scholar]
- 47.Ge, Y., Ezzell, R. M., Tompkins, R. G., and Warren, H. S. (1994) J. Infect. Dis. 169 95-104 [DOI] [PubMed] [Google Scholar]
- 48.Freudenberg, N., Piotraschke, J., Galanos, C., Sorg, C., Askaryar, F. A., Klosa, B., Usener, H. U., and Freudenberg, M. A. (1992) Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 61 343-349 [DOI] [PubMed] [Google Scholar]
- 49.Praaning-van Dalen, D. P., Brouwer, A., and Knook, D. L. (1981) Gastroenterology 81 1036-1044 [PubMed] [Google Scholar]
- 50.Fukuda, I., Tanamoto, K., Kanegasaki, S., Yajima, Y., and Goto, Y. (1989) Br. J. Exp. Pathol. 70 267-274 [PMC free article] [PubMed] [Google Scholar]
- 51.Fitzgerald, K. A., and Chen, Z. J. (2006) Cell 125 834-836 [DOI] [PubMed] [Google Scholar]
- 52.Fujimoto, T., Yamazaki, S., Eto-Kimura, A., Takeshige, K., and Muta, T. (2004) J. Biol. Chem. 279 47431-47437 [DOI] [PubMed] [Google Scholar]
- 53.Wong, K. F., Luk, J. M., Cheng, R. H., Klickstein, L. B., and Fan, S. T. (2007) FASEB J. 21 3231-3239 [DOI] [PubMed] [Google Scholar]
- 54.Mansell, A., Brint, E., Gould, J. A., O'Neill, L. A., and Hertzog, P. J. (2004) J. Biol. Chem. 279 37227-37230 [DOI] [PubMed] [Google Scholar]
- 55.Matsuzawa, A., Saegusa, K., Noguchi, T., Sadamitsu, C., Nishitoh, H., Nagai, S., Koyasu, S., Matsumoto, K., Takeda, K., and Ichijo, H. (2005) Nat. Immunol. 6 587-592 [DOI] [PubMed] [Google Scholar]
- 56.Blander, J. M., and Medzhitov, R. (2004) Science 304 1014-1018 [DOI] [PubMed] [Google Scholar]
- 57.Zaru, R., Ronkina, N., Gaestel, M., Arthur, J. S., and Watts, C. (2007) Nat. Immunol. 8 1227-1235 [DOI] [PubMed] [Google Scholar]