Abstract
Homologous recombination (HR) is a major DNA repair pathway and therefore essential for maintaining the integrity of the genome. HR is catalyzed by proteins encoded by genes of the RAD52 epistasis group, including the recombinase Rad51 and its mediator Rad52. HR proteins fused with green fluorescent protein form foci at damaged DNA reflecting the assembly of repair centers that harbor a high concentration of repair proteins. Rad52 mediates the recruitment of Rad51 and other HR proteins to DNA damage. To understand the mechanism for the assembly of Rad52-dependent DNA repair centers, we used a mutational strategy to identify a Rad52 domain essential for its recruitment to DNA repair foci. We present evidence to implicate an acidic domain in Rad52 in DNA repair focus formation. Mutations in this domain confer marked DNA damage sensitivity and recombination deficiency. Importantly, these Rad52 mutants are specifically compromised for interaction with the single-stranded DNA-binding factor RPA. Based on these findings, we propose a model where Rad52 displaces RPA from single-stranded DNA using the acidic domain as a molecular lever.
In eukaryotes, DNA double strand break (DSB)4 repair is essential for maintaining genetic stability. The major pathway of DSB repair in the yeast Saccharomyces cerevisiae is homologous recombination (HR), and the evolutionarily conserved proteins involved in this process are encoded by members of the RAD52 epistasis group, including RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, RDH54/TID1, RFA1, MRE11, and XRS2 (1). In mitotic cells, most DSBs are eliminated by the synthesis-dependent strand annealing pathway of HR (2). The first step of this pathway involves resection of the ends at the DSB to produce a pair of 3′-single-stranded tails. Subsequently, one of these single-stranded tails invades an intact homologous double-stranded DNA sequence to produce a D-loop. DNA polymerase extends the invading end, hence acquiring DNA information that is complementary to the noninvading end. At this stage, the invading strand is dissociated from the invaded duplex and anneals to the noninvading end of the break. Repair is completed when the break is sealed via additional DNA synthesis and ligation.
In vitro it has been shown that Rad51 can catalyze the DNA strand invasion reaction via a filamentous intermediate called the presynaptic filament (3, 4). The efficiency of this reaction is dependent on several accessory factors, including the heterotrimeric single-stranded DNA-binding protein RPA (3, 5–7). RPA minimizes intramolecular secondary structure in the single-stranded DNA (ssDNA) that would otherwise impede presynaptic filament assembly. Paradoxically, if an amount of RPA sufficient to saturate the ssDNA is added to the in vitro recombination reaction prior to or together with Rad51, it strongly inhibits strand invasion by limiting access of Rad51 to the ssDNA (1, 8). Chromatin immunoprecipitation and cytological studies have also shown that RPA excludes Rad51 from the HR substrate (9, 10–13). The inhibitory effect of RPA on the assembly of the Rad51 presynaptic filament is overcome by recombination mediator proteins, including S. cerevisiae Rad52, the Rad55-Rad57 complex, and the tumor suppressor BRCA2 (1, 6, 14–18). Among the recombination mediator proteins, the mediator function is best understood for S. cerevisiae Rad52, which interacts with Rad51, RPA, and also ssDNA (1, 4).
The analysis of HR proteins fused with GFP by fluorescence microscopy has revealed that they form repair centers at damaged DNA (19). Moreover, experiments using chromatin immunoprecipitation or GFP-tagged proteins coupled with cytological analysis have demonstrated the temporal order of recruitment of the HR proteins to DSBs (10–12). Specifically, the first HR factor to arrive at the DSB is the Mre11-Rad50-Xrs2 complex. Next, when the decision has been made to repair the DSB by HR, the ends at the break are resected, and the resulting single-stranded DNA tails become covered by RPA. Subsequently, Rad52 and then Rad51 and others are recruited to the DSB, with the concomitant exit of RPA. Upon successful repair of the DSB, the repair center is dissolved.
Rad52 plays a central role in the assembly of the DNA repair center via its ability to recruit Rad51 and downstream HR factors. However, the signal that triggers the assembly of the Rad52-containing DNA repair centers is unknown. In the present study, we provide evidence that recruitment of Rad52 to a DNA lesion requires direct interaction with RPA via an acidic domain in the middle region of Rad52. Accordingly, alanine substitution mutations in this domain result in strains that fail to repair DNA damage by HR and display reduced HR rates. Based on our observations, we propose a model to explain the mechanism of the mediator function of Rad52.
MATERIALS AND METHODS
Genetic Methods, Strains, and Plasmids—All media were prepared as described previously (20) with minor modifications, since the synthetic medium contains twice the amount of leucine (60 mg/liter). Standard genetic techniques were used to manipulate yeast strains (20), and transformations were performed according to Ref. 21. All strains are derivatives of W303 (22), except that they are RAD5 and are listed in Table S1. RAD52 mutants fused to YFP were constructed using the cloning-free PCR-based allele replacement method previously described (23, 24). Primers for constructing rad52-Δ287-YFP and rad52-Δ307-YFP are listed in Table S2. Strains expressing the remaining Rad52 truncation YFP fusion proteins have been described previously (25). The Escherichia coli Rosetta strain (Novagen) was used to express Rad52-GST fusion proteins.
Vectors, expressing C-terminally YFP-tagged Rad52 species containing alanine substitution mutations, were derivatives of the CEN-based plasmid, pWJ1213 (26), harboring RAD52 fused to YFP and a HIS3 marker for selection. Specifically, the polylinker of pWJ1213 was removed by a blunt end ligation of ends obtained by cutting pWJ1213 with SacI, followed by removal of the 3′-overhang by T4 polymerase and with XmaI, where the 5′-overhang was filled in by T4 polymerase, to produce the plasmid pWJ1213-ΔXmaI-SacI. Next, the plasmid pRad52-9ala-yfp was made by inserting a fusion PCR fragment containing the rad52 mutations D299A/S300A, S304A/D305A/D306A, and Q308A/D309A/D310A/D311A as well as silent point mutations, which create novel (unique) XbaI and PacI sites flanking the mutated RAD52 region, into the BbvCI-SphI vector fragment of pWJ1213-ΔXmaI-SacI. The plasmids pRad52-2ala-yfp, pRad52-3ala-yfp, and pRad52-4ala-yfp, harboring the mutations D299A/S300A, S304A/D305A/D306A, and Q308A/D309A/D310A/D311A, respectively, were made by annealing pairs of oligonucleotides (see Table S2) containing the desired mutations directly into the XbaI-PacI vector fragment of pRad52-9ala-yfp. The 2 μm-based pYESS10Rad51 contains RAD51 under the control of the GAL1/10 promoter and URA3 for selection (27). pET-11d-RAD52-Q308A/D309A/D310A/D311A-His6 was made from pET-11d-RAD52-His6 (37), using the QuikChange site-directed mutagenesis kit (Stratagene) and oligonucleotides listed in Table S2. Plasmids pGEX-GST-Rad52-(169–327) and pGEX-GST-Rad52-(260–327) were made by inserting a BglII/BamHI fragment of RAD52 encoding residues 169–327 and a PCR fragment encoding residues 260–327 into BamHI-digested pGEX-2T, respectively. Stop codons were introduced into the RAD52 portion of the pGEX-GST-Rad52-(169–327) plasmid by site-directed mutagenesis as described above to generate two additional plasmids, pGEX-GST-Rad52-(169–260) and pGEX-GST-Rad52-(169–221), expressing GST fused to the Rad52 fragments 169–260 and 169–221. The two-hybrid plasmids pGBD-RAD51, pGBD-RAD52, and pGAD-Rad52 were kind gifts from R. Rothstein. The two-hybrid plasmids encoding fusion of Rfa1 (pSLH223), Rfa2 (pRB3), and Rfa3 (pRB5) to the Gal4 DNA binding domain were kind gifts from P. Berg (28). The rad52-Q308A/D309A/D310A/D311A mutation was introduced into pGAD-Rad52 by replacing a BglII-NdeI fragment in pGAD-Rad52 with the corresponding fragment from pRad52-4ala-yfp to produce pGAD-Rad52-Q308A/D309A/D310A/D311A.
Viability after γ-Irradiation and Exposure to Methyl Methanesulfonate (MMS)—γ-Irradiation was performed as previously described (29) using the γ-cell irradiator at the Radiation Research Department at Risø National Laboratory (Roskilde, Denmark). MMS (M4016 from Sigma) sensitivity was assayed by incubating cells for 0, 10, or 20 min in 0.5% MMS according to the method described by Prakash and Prakash (30). Cultures intended for γ-irradiation and cultures treated with MMS were spotted as 10-fold serial dilutions on YPD plates or, in the case of strains transformed with a plasmid, on the proper selective medium. Viability of strains was evaluated after 3 days of incubation at 30 °C.
Epifluorescence Microscopy and Quantification of Focus Formation—Strains were grown in SC medium or, for strains transformed with a plasmid, in the SC medium variant that provided the proper selection to maintain the plasmid. In addition, galactose (2%) was used as the sole carbon source in Rad51 overexpression experiments. Cells were grown at 23 °C in 3-ml cultures to an A600 of 0.3. Cultures were either evaluated for the presence of foci directly (spontaneous foci) or after 15 min of exposure to 0.5% methyl methanesulfonate (MMS-induced foci). In the latter experiments, cells were washed twice with water, resuspended in the appropriate medium, and incubated at 23 °C for 30 min prior to epifluorescence microscopy. For all microscopy, cells were immobilized on a glass slide by mixing cells with a 37 °C solution of 1.2% (w/v) low melting agarose (NuSieve 3:1 from FMC) containing the appropriate medium. Live cell images were captured with a cooled Evolution QEi monochrome digital camera (Media Cybernetics Inc.) mounted on a Nikon Eclipse E1000 camera (Nikon). Images were captured using a Plan-Fluor ×100, 1.30 numerical aperture objective lens. The illumination source was a 103-watt mercury arc lamp (Osram, Germany). The fluorophore YFP was visualized using a band pass YFP filter (EX500/20, DM515, BA520 combination filter; Nikon). Exposure time for Rad52-YFP was 1.5 s with a 12.5% neutral density (ND8) filter in place to reduce photobleaching. For each field of cells, 9–11 fluorescent images were obtained at 0.4-μm intervals along the z axis to allow inspection of all focal planes of each cell. Small and large budded cells were defined as cells where the daughter cell was either less or more than 30% of the mother cell, respectively. Cells where the nucleus was located in the bud neck were considered large budded cells. “No budded cells” were cells that either did not have a bud or cells that were still together but where the two nuclei were fully separated. For each cell type, at least 150 cells were examined to quantitate the percentage of cells containing at least one focus in that particular population of cells.
Determination of Spontaneous and Induced Mitotic Recombination Rates—Spontaneous intergenic mitotic HR between leu2-ΔEcoRI and leu2-ΔBstEII heteroalleles was measured in diploid strains as previously described (31).
Protein Expression and Purification—Plasmids encoding Rad52 derivatives were individually transformed into the E. coli Rosetta strain. Expression of Rad52 species was induced by isopropyl 1-thio-β-d-galactopyranoside, and His6-tagged Rad52 species were purified to near homogeneity as described previously (32).
DNA Binding Assay—The ability of Rad52 species to bind single-stranded DNA was evaluated using the oligonucleotide, 80-mer, and to bind double-stranded DNA by using a substrate obtained by annealing two 40-mer oligonucleotides (see Table S2). The oligonucleotides for the DNA binding assay were purified from a 15% polyacrylamide gel, as described previously (33). 80-mer and 40-mer oligonucleotides were labeled at the 5′-end with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). Varying amounts of Rad52 or Rad52-Q308A/D309A/D310A/D311A (29–450 nm) protein were incubated with 32P-labeled 40-mer (10 nm nucleotides) and 80-mer (10 nm nucleotides) at 37 °C in 10 μl of buffer (40 mm Tris-HCl, pH 7.8, 50 mm KCl, 1 mm dithiothreitol, and 100 μg/ml bovine serum albumin) for 15 min. In the control experiment, the reaction mixture was deproteinized with 0.5% SDS and 500 μg/ml proteinase K at 37 °C for 5 min to release Rad52-bound DNA. The reaction mixtures (6 μl) were resolved by electrophoresis at 4 °C using 10% native polyacrylamide gels in TAE buffer. Dried gels were analyzed by phosphorimaging.
GST Pull-down Assay—RPA (7 μg) was preincubated with φX174 ssDNA (30 μm nucleotides) in 29 μl of buffer P (20 mm Tris-HCl, pH 7.5, 150 mm KCl, 1 mm dithiothreitol, 0.5 mm EDTA, and 0.01% Igepal) at 37 °C for 10 min before the addition of 1 μl of GST-tagged Rad52 fragment (7 μg). Following a 30-min incubation at 4 °C, the reactions were mixed with 10 μl of glutathione-Sepharose-4B beads and incubated for an additional 30 min at 4 °C. After washing the beads twice with 150 μl of buffer P, the bound proteins were eluted with 30 μl of 3% SDS. A 10-μl sample of the supernatants, wash samples, and SDS eluates were subjected to SDS-PAGE analysis. Proteins were visualized by staining the gel with Coomassie Blue.
Yeast Two-hybrid Assay—The assay was performed as described by Bendixen et al. (34), using the method of Rose and Botstein (35) to quantitate β-galactosidase activity. Total protein concentration for each extract was determined by using the Bradford protein assay (Bio-Rad) to allow calculation of the specific β-galactosidase activity. At least three trials were performed to determine the β-galactosidase activity for each protein-protein interaction. For each value, background obtained from the empty plasmid, pGBD, in combination with either pGAD-Rad52 or pGAD-Rad52-Q308A/D309A/D310A/D311A was subtracted. Student's t test was used to evaluate whether two values were significantly different.
DNA Strand Exchange Reaction—In the standard reaction, Rad51 (10 μm) was incubated with ssDNA (30 μm nucleotides) in 10 μl of buffer R (35 mm K-MOPS, pH 7.2, 1 mm dithiothreitol, 50 mm KCl, 2.5 mm ATP, and 3 mm MgCl2) for 5 min at 37 °C. After the addition of RPA (1.2 μm) in 0.5 μl, the reaction mixtures were incubated at 37 °C for another 5 min before the incorporation of double-stranded DNA (30 μm nucleotides) in 1 μl and 1 μl of 50 mm spermidine hydrochloride. After 100 min of incubation at 37 °C, the reaction was terminated by adding an equal volume of 1% SDS containing 1 mg/ml proteinase K, followed by a 10-min incubation at 37 °C. The deproteinized samples (12 μl) were run in 0.9% agarose gels in TAE buffer, stained with ethidium bromide for 30 min, and then destained for at least 4 h in a large volume of water. Images were recorded in a NucleoTech Gel documentation system and analyzed with the software provided. To examine recombination mediator activity, reaction mixtures containing Rad51 and RPA with or without the indicated amount of Rad52 or Rad52-Q308A/D309A/D310A/D311A were incubated on ice for 10 min before φX ssDNA was added. Following a 10-min incubation at 37 °C, linear double-stranded DNA and spermidine were incorporated. The reaction mixtures were incubated and analyzed as above. In the time course experiments, the reaction mixtures were scaled up accordingly, and 6-μl portions of these mixtures were withdrawn for analysis, as above.
RESULTS
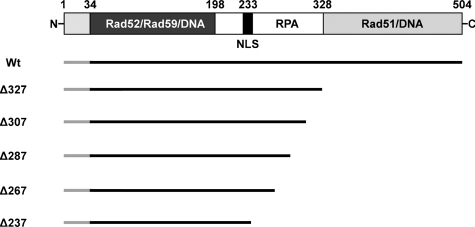
Identification of a Rad52 Region Required for DNA Repair Center Formation—An important aspect of Rad52 in recombinational repair is its recruitment to a repair focus at damaged DNA during the S-phase of the cell cycle (19, 24). To understand this process, we have used a mutational strategy to identify the Rad52 domain required for its recruitment to DNA repair centers (Fig. 1). We have previously observed that a C-terminally truncated Rad52-YFP fusion protein, Rad52-Δ237-YFP, enters the nucleus (25). Visual inspection of a strain expressing this Rad52 species by fluorescence microscopy revealed that it rarely forms spontaneous or MMS-induced Rad52 repair foci (Fig. 2, A and B). Using Rad52-Δ237-YFP as a starting point for locating the region in Rad52 required for its recruitment to a repair focus, we initially constructed four additional mutant strains expressing YFP-tagged C-terminally truncated Rad52 species, of which the largest species terminates at residue 327 (Δ327) (see Fig. 1).
FIGURE 1.
Rad52 truncation mutations analyzed in this study. A schematic representation of S. cerevisiae Rad52, including the position of known functional domains, is shown at the top. The region covering residues 1–33 is not expressed (55). The dark gray region spanning amino acid residues 34–198 corresponds to the region of Rad52 that is highly evolutionarily conserved. The regions in Rad52 that are involved in protein-protein interactions and in binding to DNA are indicated. NLS, position of the nuclear localization signal in Rad52 (25). A diagram showing all individual Rad52 deletion species relative to wild-type Rad52 is presented below. All Rad52 species are C-terminally extended by YFP (not shown in the figure): Rad52-YFP (Wt), Rad52-Δ327-YFP (Δ327), Rad52-Δ307-YFP (Δ307), Rad52-Δ287-YFP (Δ287), Rad52-Δ267-YFP (Δ267), and Rad52-Δ237-YFP (Δ237).
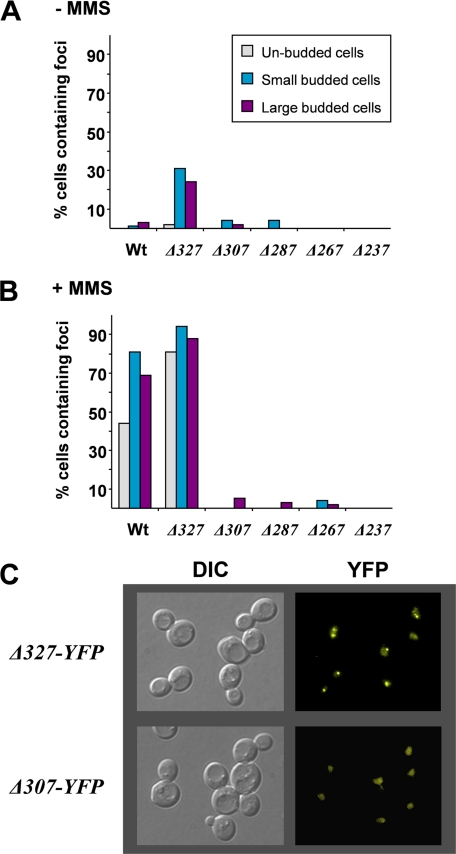
FIGURE 2.
Identification of truncated Rad52 mutants unable to form MMS-induced repair foci. A, quantification of the ability of rad52 mutant strains to form spontaneous foci. The percentage of cells that contains at least one focus was determined, and the results are presented in a histogram for three different cell types. Gray bars, unbudded cells; blue bars, small budded cells; purple bars, large budded cells. B, quantification of the ability of rad52 mutant strains to form MMS-induced foci. Results are presented as described in A. C, differential interference contrast (DIC) and fluorescence microscopy of rad52-Δ327-YFP and rad52-Δ307-YFP. Pictures obtained by fluorescence microscopy are pseudocolored images.
By analyzing cells displaying a small bud, a large bud, or no bud (representing S, G2, and G1 cells, respectively), we observed that most wild-type RAD52-YFP and rad52-Δ327-YFP cells generate Rad52 foci after the induction of DNA damage by MMS irrespective of the cell cycle stage (Fig. 2, B and C). Interestingly, the percentage of cells containing spontaneous and MMS-induced foci is generally higher in rad52-Δ327-YFP strains compared with wild-type strains (Fig. 2, A and B), and the Rad51 interaction domain (36) and a newly discovered DNA binding domain residing in the C terminus of Rad52 (37) are apparently not important for DNA repair focus formation. In contrast, when strains expressing Rad52 species terminating at amino acid 237, 267, 287, or 307 were analyzed, cells containing mutant Rad52 protein foci were rarely observed after MMS treatment (Fig. 2, B and C). In fact, for each of the other rad52 truncation mutant strains, the percentage of cells with foci is reduced more than 10-fold compared with wild type, irrespective of the cell cycle stage investigated. In agreement with this, it has recently been reported that a C-terminal Rad52 truncation terminating at residue 284 does not form foci (38).
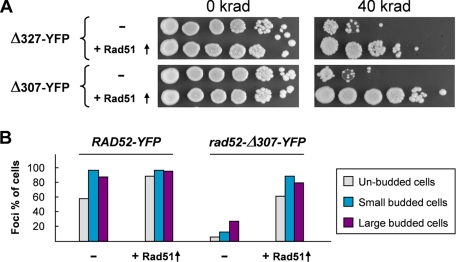
Strains that express C-terminally truncated Rad52 species terminating at residues in the range of 169–327 have previously been shown to be MMS-sensitive. However, the MMS sensitivity of these strains, except the one expressing a Rad52 species terminating at amino acid 169, can be fully or partially suppressed by Rad51 overexpression (36, 39). For example, it was shown that the rad52-Δ292 and rad52-Δ327 truncation mutations are both efficiently suppressed. In this light, it is unexpected that we find that rad52-Δ327-YFP, but not rad52-Δ307-YFP strains, form foci, and it raises the question of whether the YFP moiety of Rad52-Δ307-YFP impairs one or more of the remaining Rad52 functions in this truncated Rad52 species. However, this possibility is eliminated by our finding that the γ-ray and MMS sensitivity of rad52-Δ307-YFP can also be suppressed by overexpression of Rad51 (Fig. 3A and Fig. S1). At first glance, this result implies that Rad52 repair focus formation may not be a necessary prerequisite for MMS damage repair. However, when MMS-treated rad52-Δ307-YFP cells overexpressing Rad51 were visually inspected, a high number of Rad52 foci were observed (Fig. 3B). Together, these results indicate that the region between or close to residues 307 and 327 is required for efficient Rad52 focus formation, but that this function can be compensated by the overexpression of Rad51.
FIGURE 3.
Effects of Rad51 overexpression in rad52 truncation mutants. A, overexpression of Rad51 suppresses the γ-ray sensitivity of rad52-Δ327-YFP and rad52-Δ307-YFP strains. Strains transformed with a Rad51 overexpression plasmid (+Rad51 ↑) or empty vector (-) were spotted as serial 10-fold dilutions on SC-Ura plates containing galactose and irradiated with the indicated dose. Pictures were captured after 2 days of incubation. B, overexpression of Rad51 leads to formation of MMS-induced Rad52 foci in rad52-Δ307-YFP strains. Strains RAD52-YFP and rad52-Δ307-YFP transformed with a Rad51 overexpression plasmid (+Rad51 ↑) or the corresponding empty plasmid (-) were quantified for focus formation. The percentage of cells containing at least one focus was determined, and the results are presented in a histogram for three different cell types. Gray bars, unbudded cells; blue bars, small budded cells; purple bars, large budded cells.
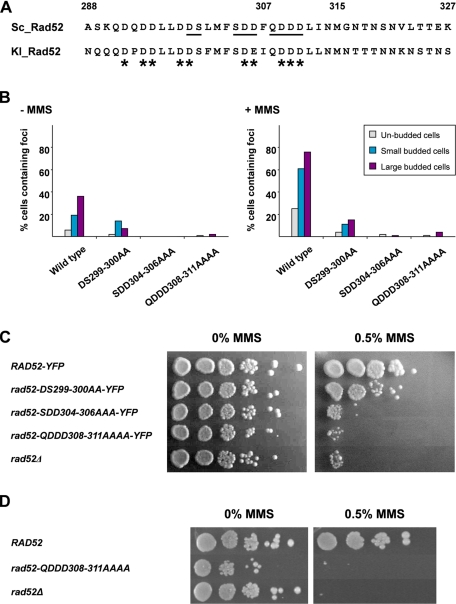
Acidic Amino Acid Residues in the Middle Part of Rad52 Are Important for Rad52 Repair Center Formation—We have previously observed that Rad52 from Kluyveri lactis is efficiently sorted to the nucleus of S. cerevisiae rad52Δ strains (25). Inspection of this strain demonstrated that the K. lactis Rad52-YFP fusion protein forms repair foci after MMS treatment (Fig. S2). This is in agreement with the fact that K. lactis Rad52 partially suppresses the MMS sensitivity of rad52Δ null mutant strains (36). We therefore compared the sequence regions of K. lactis and S. cerevisiae Rad52 corresponding to residues 288–327 in the S. cerevisiae Rad52 protein sequence with the aim of identifying conserved features and residues that could potentially be involved in mediating Rad52 focus formation (Fig. 4A). Interestingly, we observed that this region harbors several conserved acidic and hydrophilic residues. To investigate the possibility that these conserved residues are critical for focus formation, we first constructed a plasmid that expresses a Rad52-YFP species wherein nine of the acidic/hydrophilic residues within a 14-residue region (see Fig. 4A) were replaced with alanine residues. This plasmid was transformed into a rad52Δ strain, and the transformants were inspected by fluorescence microscopy. Importantly, almost no spontaneous or MMS-induced Rad52 foci formed in these cells (data not shown). These observations strongly support the premise that this acidic/hydrophilic region of Rad52 is indispensable for Rad52 focus formation. In addition, these results also allowed us to conclude that the failure of Rad52-Δ307-YFP to form foci is due to loss of a specific Rad52 function rather than to nonspecific/steric interference of the YFP moiety.
FIGURE 4.
Mutations in the acidic region of Rad52 impair its ability to form MMS-induced DNA repair foci and its ability to repair MMS-induced damage. A, sequence alignment of amino acid residues 288–327 from S. cerevisiae to the corresponding sequence of K. lactis Rad52. Conserved acidic residues are indicated by asterisks, and mutated residues are underlined. B, the ability of Rad52 mutants to form repair foci after MMS treatment. A rad52Δ strain was transformed with single copy plasmids encoding wild-type or mutant Rad52 species fused to YFP and propagated in SC-His prior to MMS treatment and microscopy. The percentage of cells that contains at least one focus was determined, and the results are presented in a histogram for three different cell types. Gray bars, unbudded cells; blue bars, small budded cells; purple bars, large budded cells. C, complementation assay to measure the ability of Rad52 mutants to rescue the MMS sensitivity of rad52Δ strains. A rad52Δ strain was transformed with plasmids encoding RAD52-YFP, rad52-D299A/S300A-YFP (rad52-DS299-300AA-YFP), Rad52-S304A/D305A/D306A-YFP (Rad52-SDD304-306AAA-YFP), Rad52-Q308A/D309A/D310A/D311A-YFP (Rad52-QDDD308-311AAAA-YFP, or an empty plasmid, as indicated. Transformants were treated with 0 or 0.5% MMS for 10 min as indicated and spotted as serial 10-fold dilutions on SC-His plates. D, survival of RAD52 and Rad52-Q308A/D309A/D310A/D311A (Rad52-QDDD308-311AAAA) strains after MMS treatment. Strains were grown in YPD prior to exposure to either 0 or 0.5% MMS for 15 min and spotted as serial 10-fold dilutions on YPD plates.
We constructed additional alleles of RAD52 to further investigate the importance of the acidic region in Rad52 focus formation. Specifically, plasmids expressing the Rad52 mutants Rad52-D299A/S300A-YFP, Rad52-S304A/D305A/D306A-YFP, and Rad52-Q308A/D309A/D310A/D311A, which cover different portions of the nine critical residues identified above, were constructed (see Fig. 4A). Following the introduction of these plasmids into rad52Δ cells, the cellular localization of the three Rad52 mutant proteins was determined. Cells that expressed Rad52-D299A/S300A-YFP were able to assemble spontaneous Rad52 foci (Fig. 4B), albeit at a reduced level. Specifically, in the populations of small budded and large budded cells, the numbers of cells with Rad52 foci were 1.4- and 5-fold reduced compared with cells expressing wild-type Rad52-YFP from a plasmid. Moreover, after MMS treatment, the number of cells containing Rad52-D299A/S300A-YFP foci did not increase dramatically, indicating that this mutant species forms Rad52 foci at a reduced rate or, alternatively, that it fails to be recruited to MMS-induced damage (Fig. 4B). In contrast, hardly any of the cells expressing Rad52-S304A/D305A/D306A-YFP or Rad52-Q308A/D309A/D310A/D311A-YFP formed any spontaneous or MMS-induced Rad52 foci (Fig. 4B). Importantly, these effects are not due to decreased Rad52-YFP concentrations as judged by a Western blot analysis (Fig. S3A). We conclude from these results that the acidic region between residues 299 and 311 is important for the recruitment of Rad52 to a repair center.
Failure to Form Rad52 Foci Impairs DNA Repair and Recombination—Next, we determined whether the impaired ability of the three alanine substitution mutants to form Rad52 foci is associated with a DNA repair defect. Accordingly, the transformants analyzed above were treated with MMS, and their survival was assessed. Strains expressing Rad52-S304A/D305A/D306A-YFP and Rad52-Q308A/D309A/D310A/D311A-YFP, which fail to form Rad52 foci, are very MMS-sensitive (Fig. 4C). In comparison, Rad52-D299A/S300A-YFP, which retains some ability to form foci, is only mildly sensitive to MMS. One of the mutations, rad52-Q308A/D309A/D310A/D311A, was integrated into the endogenous RAD52 locus. A Western blot analysis showed that the level of Rad52 in this mutant strain is similar to that of a wild-type strain (Fig. S3B). Next, rad52-Q308A/D309A/D310A/D311A was analyzed for its ability to survive DNA damage induced by MMS. In agreement with the failure of Rad52-Q308A/D309A/D310A/D311A-YFP to complement the rad52Δ phenotype when it is expressed from a plasmid, we observed that the rad52-Q308A/D309A/D310A/D311A strain is very MMS-sensitive (Fig. 4D). We and others have previously identified mutations in RAD52 that separate its DSB repair and homologous recombination functions (40, 41). We therefore investigated the ability of rad52-Q308A/D309A/D310A/D311A to mediate interchromosomal leu2-ΔBstEII/leu2-ΔEcoRI heteroallelic recombination (29, 40, 42). In the homozygous rad52-Q308A/D309A/D310A/D311A diploid strain, LEU2 prototrophs were formed at a low rate, 3.4 × 10-8. Compared with the recombination rates we have previously determined for isogenic rad52Δ/rad52Δ and RAD52/RAD52 strains using the same assay (43), we find that it is more similar to the rate obtained in the absence of Rad52, 0.6 × 10-8, than that obtained in the presence of Rad52, 1.1 × 10-6. Together, these observations indicate that failure to form spontaneous and MMS-induced foci results in severe defects in DNA repair and recombination.
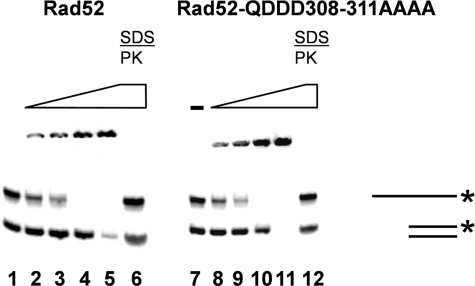
The Ability of Rad52 to Bind DNA and Perform Single Strand Annealing Is Not Influenced by the Acidic Rad52 Domain—To understand the failure of Rad52-Q308A/D309A/D310A/D311A-YFP to form repair foci at the molecular level, we purified wild-type Rad52 and Rad52-Q308A/D309A/D310A/D311A to near homogeneity (see “Materials and Methods”). First, we investigated whether the mutation influences the ability of Rad52 to interact with DNA. Accordingly, a DNA mobility shift assay was used to determine whether the Q308A/D309A/D310A/D311A mutation affects the ability of Rad52 to bind single- and double-stranded DNA substrates. We found that the mutation has little or no effect on the DNA binding specificity of Rad52, since the amount of protein required to shift the DNA species is the same for mutant and wild-type Rad52 (Fig. 5). As expected (41), for both mutant and wild-type Rad52, more protein is required to shift the double-stranded DNA compared with single-stranded DNA. Next, we investigated whether the mutation affects the ability of Rad52 to anneal complementary oligonucleotides, which is a well known attribute of Rad52 (1, 4, 7, 41, 44). We did not notice any difference between wild-type and mutant Rad52 proteins in terms of the amount of annealed product (Fig. S4). Taken together, the results indicate that the mutation in the acidic domain of Rad52 has no impact on the DNA binding or DNA annealing activities of Rad52.
FIGURE 5.
The Rad52-Q308A/D309A/D310A/D311A (QDDD308-311AAAA) mutant protein efficiently binds DNA. DNA binding was evaluated in a gel shift assay. A radiolabeled double-stranded 40-mer and a radiolabeled single-stranded 80-mer were preincubated with increasing concentrations (29–450 nm) of wild-type Rad52 or Rad52-Q308A/D309A/D310A/D311A protein. Samples were loaded in lanes 2–5 (Rad52) and lanes 8–11 (Rad52-Q308A/D309A/D310A/D311A), respectively, and analyzed by SDS-PAGE. No Rad52 (-) was included in reactions loaded in lanes 1 and 7. Samples loaded in lanes 6 and 12 correspond to those loaded in lanes 5 and 11, except that they were deproteinized by SDS and proteinase K treatment (SDS/PK) prior to loading.
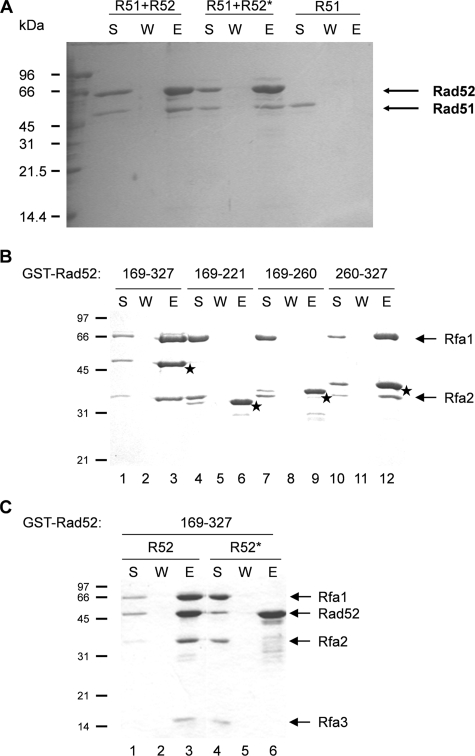
The Ability of Rad52 to Interact with Rad51 Is Not Impaired by Mutation of the Acidic Rad52 Domain—In our analysis, the truncation mutation Rad52-Δ327-YFP forms foci at a high frequency (Fig. 2, A–C). This Rad52 fragment does not interact with Rad51 (36, 41), indicating that the Rad51 binding domain is not required for Rad52 to form repair foci. On the other hand, the fact that overexpression of Rad51 stimulates Rad52-Δ307-YFP focus formation raised the possibility that the acidic domain in Rad52 somehow could interact with Rad51. After all, the region identified in the present study is right at the border of the fragment proposed to contain the Rad51 binding domain (see Fig. 1). To investigate this possibility, we expressed Rad52-Q308A/D309A/D310A/D311A as a C-terminally His6-tagged protein in E. coli and purified it to homogeneity. Next, pull-down experiments were performed by incubating Rad51 either with His6-tagged wild-type or His6-tagged mutant Rad52 immobilized on nickel beads. Following extensive washing of the beads, the bound proteins were eluted with SDS and analyzed. Rad51 efficiently associated with wild type as well as with mutant Rad52, and no Rad51 was visible in the wash fractions (Fig. 6A). As expected, Rad51 did not associate with the nickel beads in the absence of Rad52 (Fig. 6A). This conclusion is supported by our observation that Rad51 and Rad52-Q308A/D309A/D310A/D311A efficiently interact in a yeast two-hybrid assay (Table 1). Together, these results demonstrate that the Q308A/D309A/D310A/D311A mutation does not compromise the ability of Rad52 to interact with Rad51.
FIGURE 6.
Rad52-Q308A/D309A/D310A/D311A interacts poorly with RPA. A, determination of the ability of Rad51 to interact with Rad52 species using a Rad51 pull-down assay. Purified wild-type Rad52-His6 (R52) and Rad52-Q308A/D309A/D310A/D311A-His6 (R52*) were individually mixed and preincubated with Rad51 (R51) before nickel-agarose beads were added. The beads were washed with buffer and treated with SDS to elute bound wild-type Rad52 and Rad52-Q308A/D309A/D310A/D311A. The supernatant containing unbound Rad52 or Rad52-Q308A/D309A/D310A/D311A (S), the wash (W), and the SDS eluate (E) were analyzed by 10% SDS-PAGE followed by staining with Coomassie Blue. B, the RPA binding domain is located in a central section of Rad52. Different GST-Rad52 fragments were evaluated for their ability to bind RPA using a pull-down assay. The Rad52 residues contained in each fragment are indicated in the top of the panel. The positions of individual GST-Rad52 fragments in the gel are indicated by stars. C, mutation of amino acids 308–311 abolishes the RPA binding ability of GST-Rad52-(169–327). The ability of a wild-type (R52) and a mutant (R52*) fragment to interact with RPA was evaluated in a pull-down assay.
TABLE 1.
Rad52-Q308A/D309A/D310A/D311A interacts poorly with Rfa1 in vivo Interactions between wild-type Rad52 and Rad52-Q308A/D309A/D310A/D311A (Rad52*) and Rad51, Rad52, Rfa1, Rfa2, and Rfa3, respectively, were individually evaluated by measuring the specific β-galactosidase activity in a two-hybrid assay. At least three trials were performed for each protein-protein interaction analyzed.
|
β-Galactosidase activity
|
||||
|---|---|---|---|---|
|
GBD fusion
|
GAD fusion
|
p value | Rad52*/Rad52 | |
| Rad52 | Rad52* | |||
| Rad51 | 1060 ± 270 | 710 ± 48 | 0.081 | 0.67 |
| Rad52 | 390 ± 45 | 310 ± 57 | 0.12 | 0.79 |
| Rfa1 | 97 ± 32 | 21 ± 8.1 | 0.015 | 0.22 |
| Rfa2 | 145 ± 38 | 65 ± 19 | 0.018 | 0.45 |
| Rfa3 | 19 ± 5.1 | 29 ± 11 | 0.25 | 1.5 |
The Acidic Domain in Rad52 Binds RPA—Rad52 also binds RPA (28), and we therefore tested whether this activity is influenced by the Q308A/D309A/D310A/D311A mutation using the yeast two-hybrid assay. Interestingly, when the three subunits of RPA were analyzed individually, the largest subunit, Rfa1, interacted less strongly with the Rad52 mutant than with wild-type Rad52 (Table 1). Interaction with the middle subunit, Rfa2, was slightly impaired, whereas the interaction with the smallest subunit, Rfa3, was unaffected. Hence, the yeast two-hybrid analysis indicates that the mutation of the acidic domain in Rad52 has a major effect on the ability of this protein to interact with the largest subunit of RPA, Rfa1. To strengthen this conclusion, we fused GST to Rad52 fragments and analyzed their ability to interact with RPA in vitro using an affinity pull-down assay with glutathione-Sepharose (which binds GST). Using this approach, we found that the middle portion of Rad52 protein encompassing residues 169–327 is able to interact with RPA in a manner that is enhanced by ssDNA (37) (Fig. 6B). To further delimit the RPA binding domain in Rad52, we expressed different portions (residues 169–221, 169–260, and 260–327) of Rad52 as fusions to GST, purified these GST-tagged Rad52 fragments, and examined them for RPA binding. As shown in Fig. 6B, only the GST fusion that harbors residues 260–327, which encompasses the acidic domain of Rad52, has RPA binding capability. Finally, we examined whether the Q308A/D309A/D310A/D311A mutation ablates RPA interaction in this pull-down assay. Accordingly, a GST-tagged Rad52 fragment spanning residues 169–327 and harboring this mutation was purified to homogeneity and tested for RPA binding. In contrast to the corresponding wild-type Rad52 fragment, the Rad52-Q308A/D309A/D310A/D311A mutant fragment is completely devoid of the ability to associate with RPA (Fig. 6C).
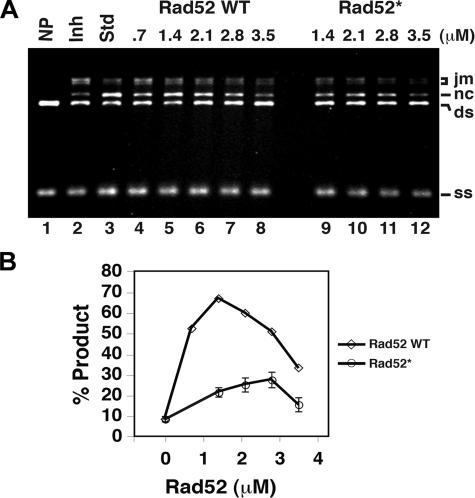
The Rad52 Mediator Activity Depends on the Acidic Rad52 Domain—An important aspect of the function of Rad52 in HR is to overcome the inhibitory effect of RPA on the assembly of the Rad51 presynaptic filament. Since the acidic Rad52 domain is important for RPA interaction, we investigated the possibility that this domain is also important for the mediator activity of Rad52. Hence, we determined the ability of wild-type Rad52 and Rad52-Q308A/D309A/D310A/D311A to mediate Rad51-catalyzed DNA strand exchange under conditions wherein the reaction efficiency is suppressed by co-incubating the ssDNA substrate with Rad51 and the single strand DNA-binding protein RPA (see “Materials and Methods” and Ref. 45). By analyzing the amounts of strand exchange products obtained with different concentrations of wild-type and mutant Rad52 protein, we found that two key aspects of the mediator activity of Rad52 are impaired by the Q308A/D309A/D310A/D311A mutation. Specifically, the maximum level of product formation is much lower with the mutant protein compared with that obtained with wild-type Rad52 (Fig. 7A). Moreover, the addition of twice as much mutant protein is required to reach the maximum level of product (Figs. 7B and S5).
FIGURE 7.
Recombination mediator activity of Rad52 and Rad52-Q308A/D309A/D310A/D311A. A, the standard reaction (Std; lane 3) involved preincubating the ssDNA with Rad51 to allow for the formation of the presynaptic filament before RPA was added. Co-incubating the ssDNA with Rad51 and RPA resulted in severe inhibition of the DNA strand exchange reaction (Inh; lane 2). The inclusion of the indicated amounts of Rad52 or Rad52-Q308A/D309A/D310A/D311A (Rad52*) during the incubation of ssDNA with Rad51 and RPA led to a varying degree of DNA strand exchange restoration. B, a plot showing percentage of product (nc/(nc + ds) × 100) obtained in the reactions shown in A. ds, double strand; jm, joint molecule; nc, nicked circle; ss, single strand.
DISCUSSION
The recombinational repair of damaged DNA occurs within a center that harbors high concentrations of different repair proteins (9, 19, 24, 46–49). These repair centers can be visualized cytologically as nuclear foci by tagging repair proteins with a fluorescent marker and in situ by immunostaining for repair proteins (13). Importantly, proteins involved in the repair reaction are recruited in a defined temporal order to DNA damage (10–12, 50). For example, Rad52 is recruited to damage prior to Rad51, probably reflective of the role of Rad52 in the replacement of RPA for Rad51. Rad52 associates with DNA, RPA, and Rad51 (see Fig. 1), but it has remained unclear whether any of these attributes are important for repair center assembly. To delineate the mechanistic role of Rad52 in DNA recombination and repair, we have employed a mutational approach to identify a small acidic domain in Rad52 that is required for its recruitment to a repair center. The functional importance of this region of Rad52 is evidenced by the fact that strains expressing Rad52 internal deletion species lacking this domain are impaired for DNA damage repair (39). In the present study, we have shown that substitution of conserved acidic residues within this domain by alanine prevents the recruitment of Rad52 to repair foci. Importantly, we find that such mutant strains exhibit a phenotype similar to that of rad52Δ strains. We have performed biochemical experiments to show that this Rad52 domain promotes RPA binding via an interaction with the largest RPA subunit Rfa1. Accordingly, a direct interaction with Rfa1 is essential for Rad52 focus formation. This is in agreement with the fact that no Rad52 foci are formed after DNA damage in cells lacking Rfa1 (12).
Rad52-YFP forms foci in the absence of Rad51 (12), so it is hardly surprising that Rad52-Δ327-YFP readily forms foci despite the fact that it lacks the capability to bind Rad51. In fact, the number of cells containing a Rad52 focus is higher in rad52-Δ327-YFP compared with RAD52-YFP strains. This is probably due to the accumulation of uncompleted repair events in rad52-Δ327-YFP strains resulting from its failure to recruit Rad51. In agreement with this, overexpression of Rad51 can suppress the MMS sensitivity of rad52-Δ327 (36, 39) and rad52-Δ327-YFP strains (this study). However, it is puzzling that the MMS sensitivity of rad52-Δ292 (39) and rad52-Δ307 (this study) can be fully suppressed by Rad51 overexpression, since these Rad52 mutants lack not only the Rad51 binding domain but also the RPA binding domain that we have identified in this study. A simple model to explain the suppression of these rad52 mutants is that the RPA binding activity is severely, but not completely, impaired by the truncation mutations. This may likely be the case, since Rad52 interacts with all three RPA subunits (28), and the Q308A/D309A/D310A/D311A mutation mainly affects the interaction with one of the subunits, Rfa1. If so, the residual RPA binding activity may still be sufficient for the formation of complexes that harbor ssDNA, RPA, and the Rad52 mutant proteins. However, due to the weakened interaction between RPA and mutant Rad52, these complexes would be short lived, thus leaving sufficient time for efficient recruitment of Rad51 only when Rad51 is overexpressed. In this scenario, we envision that the recruitment of Rad51, at the expense of RPA, exposes binding epitopes at the DNA lesion that enhance the recruitment or retention of the Rad52-Δ307-YFP mutant protein, thus allowing it to accumulate at the lesion and form a repair focus. In addition, we note that recruitment of Rad52-Δ307-YFP to the lesion is probably essential for repair at the later stages of repair, where the Rad52 annealing activity is required for the second ssDNA capture (50–52).
One major function of Rad52 in DNA recombination and repair is to help Rad51 in the displacement of RPA from ssDNA in such a way as to promote the assembly of the presynaptic Rad51 filament (4, 45). In this regard, the functionality of Rad52 depends on its ability to bind Rad51 (36, 39, 42, 53). In this study, we have provided evidence that this important functional attribute of Rad52 is also dependent on its ability to directly interact with Rfa1. In the simplest model to explain this mediator function of Rad52, these protein-protein interactions ensure a sufficiently high local concentration of Rad52 and associated proteins (such as Rad51) at the DNA lesions to promote Rad51 filament formation, hence explaining the need to form a repair focus. However, our finding that the RPA binding domain in Rad52 is highly acidic suggests an active role of Rad52 in RPA displacement. It was recently shown that an acidic amphipathic helix derived from the transactivating domain of p53 associates with the DNA binding cleft of RPA70 by acting as an ssDNA mimic (54). In analogous fashion, ssDNA mimicry may also underlie the recombination mediator function of Rad52. Specifically, the resemblance of the acidic domain of Rad52 to the highly charged backbone of ssDNA may enable Rad52 to act as a competitive inhibitor to weaken the interaction of RPA with ssDNA, thus facilitating the RPA-Rad51 exchange process by acting as a molecular lever.
Supplementary Material
Acknowledgments
We thank Lene Christiansen for excellent technical help, Arne Miller and co-workers at the Radiation Research Department at Risø National Laboratory for access to the γ-cell, and members of the Mortensen laboratory for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants ES07061, GM57814, and P01CA92584 (to P. S.). This work was also supported by the Danish Research Council for Technology and Production Sciences (to U. H. M.), the Hartmann Foundation (to U. H. M.), the Lundbeck Foundation (to U. H. M.), and the Technical University of Denmark (to S. C. L. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
Footnotes
The abbreviations used are: DSB, double strand break; HR, homologous recombination; ssDNA, single-stranded DNA; YFP, yellow fluorescent protein; MMS, methyl methanesulfonate; GST, glutathione S-transferase.
References
- 1.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66 630-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paques, F., and Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63 349-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung, P. (1994) Science 265 1241-1243 [DOI] [PubMed] [Google Scholar]
- 4.Sung, P., Krejci, L., Van, K. S., and Sehorn, M. G. (2003) J. Biol. Chem. 278 42729-42732 [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama, T., Zaitseva, E. M., and Kowalczykowski, S. C. (1997) J. Biol. Chem. 272 7940-7945 [DOI] [PubMed] [Google Scholar]
- 6.Sung, P. (1997) J. Biol. Chem. 272 28194-28197 [DOI] [PubMed] [Google Scholar]
- 7.Sugiyama, T., New, J. H., and Kowalczykowski, S. C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6049-6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung, P. (1997) Genes Dev. 11 1111-1121 [DOI] [PubMed] [Google Scholar]
- 9.Gasior, S. L., Wong, A. K., Kora, Y., Shinohara, A., and Bishop, D. K. (1998) Genes Dev. 12 2208-2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugawara, N., Wang, X., and Haber, J. E. (2003) Mol. Cell 12 209-219 [DOI] [PubMed] [Google Scholar]
- 11.Wolner, B., Van Komen, S., Sung, P., and Peterson, C. L. (2003) Mol. Cell 12 221-232 [DOI] [PubMed] [Google Scholar]
- 12.Lisby, M., Barlow, J. H., Burgess, R. C., and Rothstein, R. (2004) Cell 118 699-713 [DOI] [PubMed] [Google Scholar]
- 13.Lisby, M., and Rothstein, R. (2005) Biochimie (Paris) 87 579-589 [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini, L., and Venkitaraman, A. (2004) Trends Biochem. Sci. 29 310-316 [DOI] [PubMed] [Google Scholar]
- 15.Sung, P., and Klein, H. (2006) Nat. Rev. Mol. Cell. Biol. 7 739-750 [DOI] [PubMed] [Google Scholar]
- 16.Shinohara, A., and Ogawa, T. (1998) Nature 391 404-407 [DOI] [PubMed] [Google Scholar]
- 17.New, J. H., Sugiyama, T., Zaitseva, E., and Kowalczykowski, S. C. (1998) Nature 391 407-410 [DOI] [PubMed] [Google Scholar]
- 18.Benson, F. E., Baumann, P., and West, S. C. (1998) Nature 391 401-404 [DOI] [PubMed] [Google Scholar]
- 19.Lisby, M., Mortensen, U. H., and Rothstein, R. (2003) Nat. Cell Biol. 5 572-577 [DOI] [PubMed] [Google Scholar]
- 20.Sherman, F., Fink, G. R., and Hicks, J. B. (1986) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 21.Gietz, D., St. Jean, A., Woods, R. A., and Schiestl, R. H. (1992) Nucleic Acids Res. 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas, B. J., and Rothstein, R. (1989) Cell 56 619-630 [DOI] [PubMed] [Google Scholar]
- 23.Erdeniz, N., Mortensen, U. H., and Rothstein, R. (1997) Genome Res. 7 1174-1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisby, M., Rothstein, R., and Mortensen, U. H. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8276-8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plate, I., Albertsen, L., Lisby, M., Hallwyl, S. C., Feng, Q., Seong, C., Rothstein, R., Sung, P., and Mortensen, U. H. (2008) DNA Repair 7 57-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng, Q., During, L., de Mayolo, A. A., Lettier, G., Lisby, M., Erdeniz, N., Mortensen, U. H., and Rothstein, R. (2007) DNA Repair 6 27-37 [DOI] [PubMed] [Google Scholar]
- 27.Jiang, H., Xie, Y., Houston, P., Stemke-Hale, K., Mortensen, U. H., Rothstein, R., and Kodadek, T. (1996) J. Biol. Chem. 271 33181-33186 [DOI] [PubMed] [Google Scholar]
- 28.Hays, S. L., Firmenich, A. A., Massey, P., Banerjee, R., and Berg, P. (1998) Mol. Cell. Biol. 18 4400-4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortensen, U. H., Erdeniz, N., Feng, Q., and Rothstein, R. (2002) Genetics 161 549-562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prakash, L., and Prakash, S. (1977) Genetics 86 33-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, J., and Rothstein, R. (1995) Mol. Cell. Biol. 15 1632-1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, B., and Sung, P. (2000) J. Biol. Chem. 275 15895-15904 [DOI] [PubMed] [Google Scholar]
- 33.Trujillo, K. M., and Sung, P. (2001) J. Biol. Chem. 276 35458-35464 [DOI] [PubMed] [Google Scholar]
- 34.Bendixen, C., Gangloff, S., and Rothstein, R. (1994) Nucleic Acids Res. 22 1778-1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose, M., and Botstein, D. (1983) Methods Enzymol. 101 167-180 [DOI] [PubMed] [Google Scholar]
- 36.Milne, G. T., and Weaver, D. T. (1993) Genes Dev. 7 1755-1765 [DOI] [PubMed] [Google Scholar]
- 37.Seong, C., Sehorn, M. G., Plate, I., Shi, I., Song, B., Chi, P., Mortensen, U., Sung, P., and Krejci, L. (2008) J. Biol. Chem. 283 12166-12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorpe, P. H., Marrero, V. A., Savitzky, M. H., Sunjevaric, I., Freeman, T. C., and Rothstein, R. (2006) Mol. Cell. Biol. 26 3752-3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asleson, E. N., Okagaki, R. J., and Livingston, D. M. (1999) Genetics 153 681-692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malone, R. E., Montelone, B. A., Edwards, C., Carney, K., and Hoekstra, M. F. (1988) Curr. Genet. 14 211-223 [DOI] [PubMed] [Google Scholar]
- 41.Mortensen, U. H., Bendixen, C., Sunjevaric, I., and Rothstein, R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 10729-10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boundy-Mills, K. L., and Livingston, D. M. (1993) Genetics 133 39-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lettier, G., Feng, Q., de Mayolo, A. A., Erdeniz, N., Reid, R. J., Lisby, M., Mortensen, U. H., and Rothstein, R. (2006) PLoS Genet. 2 1773-1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinohara, A., Shinohara, M., Ohta, T., Matsuda, S., and Ogawa, T. (1998) Genes Cells 3 145-156 [DOI] [PubMed] [Google Scholar]
- 45.San, F. J., Sung, P., and Klein, H. (2008) Annu. Rev. Biochem. 77 229-257 [DOI] [PubMed] [Google Scholar]
- 46.Liu, Y., Li, M., Lee, E. Y., and Maizels, N. (1999) Curr. Biol. 9 975-978 [DOI] [PubMed] [Google Scholar]
- 47.Tan, T. L., Essers, J., Citterio, E., Swagemakers, S. M., de, W. J., Benson, F. E., Hoeijmakers, J. H., and Kanaar, R. (1999) Curr. Biol. 9 325-328 [DOI] [PubMed] [Google Scholar]
- 48.Scully, R., Chen, J., Plug, A., Xiao, Y., Weaver, D., Feunteun, J., Ashley, T., and Livingston, D. M. (1997) Cell 88 265-275 [DOI] [PubMed] [Google Scholar]
- 49.Essers, J., Houtsmuller, A. B., van Veelen, L., Paulusma, C., Nigg, A. L., Pastink, A., Vermeulen, W., Hoeijmakers, J. H., and Kanaar, R. (2002) EMBO J. 21 2030-2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyazaki, T., Bressan, D. A., Shinohara, M., Haber, J. E., and Shinohara, A. (2004) EMBO J. 23 939-949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugiyama, T., Kantake, N., Wu, Y., and Kowalczykowski, S. C. (2006) EMBO J. 25 5539-5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lao, J. P., Oh, S. D., Shinohara, M., Shinohara, A., and Hunter, N. (2008) Mol. Cell 29 517-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krejci, L., Song, B., Bussen, W., Rothstein, R., Mortensen, U. H., and Sung, P. (2002) J. Biol. Chem. 277 40132-40141 [DOI] [PubMed] [Google Scholar]
- 54.Bochkareva, E., Kaustov, L., Ayed, A., Yi, G. S., Lu, Y., Pineda-Lucena, A., Liao, J. C., Okorokov, A. L., Milner, J., Arrowsmith, C. H., and Bochkarev, A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15412-15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antunez de, M. A., Lisby, M., Erdeniz, N., Thybo, T., Mortensen, U. H., and Rothstein, R. (2006) Nucleic Acids Res. 34 2587-2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.