Abstract
Germ plasm, a specialized cytoplasm present at the posterior of the early Drosophila embryo, is necessary and sufficient for germ cell formation. Germ plasm is rich in mitochondria and contains electron dense structures called polar granules. To identify novel polar granule components we isolated proteins that associate in early embryos with Vasa (VAS) and Tudor (TUD), two known polar granule associated molecules. We identified Maternal expression at 31B (ME31B), eIF4A, Aubergine (AUB) and Transitional Endoplasmic Reticulum 94 (TER94) as components of both VAS and TUD complexes and confirmed their localization to polar granules by immuno-electron microscopy. ME31B, eIF4A and AUB are also present in processing (P) bodies, suggesting that polar granules, which are necessary for germ line formation, might be related to P bodies. Our recovery of ER associated proteins TER94 and ME31B confirms that polar granules are closely linked to the translational machinery and to mRNP assembly.
Keywords: polar granules, ribonucleoprotein particles, processing bodies, post-transcriptional control, germ cell specification
Introduction
Early in metazoan development germ line stem cells are set aside from the future somatic cells. These cells are necessary for gametogenesis and thus reproduction of the animal. In many insects and some vertebrate species, germ cell formation depends on the germ plasm, a specialized cytoplasm found at the posterior of the embryo (Saffman and Lasko, 1999). An essential link between germ plasm and germ cells was postulated over a century ago (Weismann, 1885), and this link has been well substantiated in subsequent experiments. For example, in the Drosophila embryo, destruction of the germ plasm by UV irradiation impairs germ cell formation, and injection of germ plasm at ectopic sites leads to germ cell formation, showing that the germ plasm is necessary and sufficient for germ cell formation (Geigy, 1931; Illmensee and Mahowald, 1974, 1976).
The germ plasm is morphologically distinct from the somatic cytoplasm (Mahowald, 2001). In Drosophila, germ plasm is enriched in mitochondria and contains large structures, referred to as polar granules, that are enriched in RNA and ribosomes (Mahowald, 1961). After fertilization of the egg, maternally synthesized RNAs are protected in the germ plasm from degradation, suggesting that polar granules store RNA in a translationally repressed state until needed for primordial germ cell development (Tadros and Lipshitz, 2005). After germ cell formation, most RNA is lost from polar granules (Mahowald, 2001). Upon their release, these RNAs are either translationally de-repressed or degraded (Thomson and Lasko, 2005).
Maternal-effect mutations in the posterior group (or grandchildless) class display defects in germ cell formation as well as in posterior somatic patterning (Nusslein-Volhard et al., 1987). Molecular characterization of the corresponding genes revealed that many posterior group gene products localize to polar granules (Thomson and Lasko, 2005). One such gene product, Oskar (OSK), is not only essential for normal germ cell formation, but is also able to induce ectopic formation of germ cells at high concentrations (Ephrussi et al., 1991; Ephrussi and Lehmann, 1992). OSK regulates endocytosis and cytoskeletal elements at the posterior of the oocyte, and nucleates polar granule assembly (Vanzo et al., 2007). Two key genes downstream of osk that are required for germ cell formation are vasa (vas) and tudor (tud). vas encodes a DEAD box RNA helicase that is present in polar granules (Hay et al., 1988, 1990; Lasko and Ashburner, 1988, 1990; Liang et al., 1994; Styhler et al., 2002). There is strong evidence that VAS functions in translation initiation via direct binding to the translation initiation factor eIF5B (Carrera et al., 2000; Johnstone and Lasko, 2004). TUD functions downstream of VAS in the pole plasm assembly pathway (Arkov et al., 2006; Thomson and Lasko, 2004). Unlike strong osk and vas mutations, which are maternal-effect lethal because progeny embryos lack posterior pattern elements, around 15% of embryos produced by tud-null females have normal somatic patterning and hatch to become sterile adults (Thomson and Lasko, 2004).
TUD is a large protein (285 kDa) that contains eleven Tudor domains. The Tudor domain is a protein motif that binds efficiently to other proteins that contain methylated arginine and lysine residues (Thomson and Lasko, 2005). Consistent with this function, a null allele of dart5 (also called capsuleen), which encodes a germline-specific arginine methyl transferase, has a strikingly similar phenotype to tud null mutations (Anne et al., 2007; Gonsalvez et al., 2006). TUD itself is essential for polar granule assembly, but little is known about its specific function in germ cell formation.
Here we describe a biochemical approach to isolate polar granule components. To enrich for components specifically localized to the germ plasm we isolated TUD and VAS containing complexes and analyzed their components. Both complexes contained eIF4A, ME31B, TER94, and AUB, suggesting strongly that these proteins are components of polar granules. We confirmed this by ultrastructural-level analysis of their distribution in early embryos. AUB, ME31B and eIF4A are also found in P bodies, cytoplasmic structures present in yeast to human cells that have been implicated in RNA storage, translational repression, and RNA degradation. Interestingly, our analysis of TER94, an endoplasmic reticulum (ER) component, showed that polar granules not only associate with TER94 but directly with ER. Taken together, these results support a close association between polar granules, mRNPs that regulate RNA stability, and the translational machinery.
Results
Isolation of TUD and VAS containing complexes from early Drosophila embryos
Genetic and molecular analysis identified TUD and VAS as specific regulators of germ plasm assembly. vas mutant females produce embryos that lack germ plasm, including polar granules. In tud alleles of varying severity, there is a strong correlation between the number of germ cells formed, and the size, number and shape of polar granules, suggesting that proper assembly of polar granules is essential for germ cell formation (Amikura et al., 2001; Arkov et al., 2006; Boswell and Mahowald, 1985; Thomson and Lasko, 2004). Therefore, we reasoned that identifying TUD and VAS-associated proteins through immunoprecipitation would shed light on polar granule composition and formation. However, some TUD exists outside polar granules and significant amounts of VAS are found outside the germ plasm in early embryos (Bardsley et al., 1993; Lasko and Ashburner, 1990). We therefore reasoned that proteins that were recovered as associated with both VAS and TUD were most likely to represent bona fide polar granule components.
We used a protein crosslinking agent to stabilize complexes before immunoprecipitating. The crosslinkers we used (see Materials and Methods) have different length spacer arms, but all were BMH (bismaleimidohexane) type crosslinkers, homobifunctional crosslinkers that crosslink sulfhydryl groups. This results in crosslinking of proteins that contain two cysteines within the range of the spacer arms. BMH type crosslinkers do not react with nucleic acids and thus only stabilize direct protein-protein associations.
To ensure specificity, we used affinity-purified antibodies to isolate TUD or VAS containing complexes. As controls, pre-immune serum precipitations were performed. As shown in Figure 1, no discernable bands were present after pre-immune serum precipitations at the same size as the TUD containing bands that were observed in TUD immunoprecipitation lanes. We nevertheless excised areas of pre-immune control lanes that corresponded to the location of bands excised in TUD immunoprecipitation lanes, and we recovered several proteins, most of which are of high molecular weight. We excluded these proteins from consideration as authentic TUD complex components. In addition to immunoprecipitation with antibodies directed against TUD and VAS we also immunoprecipitated the product of the mini-tudΔ3 transgene. Mini-TUDΔ3 is a truncated form of TUD that contains six of the eleven Tudor domains (Arkov et al., 2006). Females that lack endogenous TUD and express only Mini-TUDΔ3 produce embryos with germ plasm and functional germ cells, albeit in reduced numbers (Arkov et al., 2006; J.Y. Wang and R.L., unpublished data). As Mini-TUDΔ3 contains an HA tag, we used an anti-HA monoclonal antibody to immunoprecipitate Mini-TUDΔ3 containing complexes from early embryos. As a control, we used the same antibody for immunoprecipitations from GFP-HA expressing embryos. Any proteins found in the GFP containing complexes were not further considered.
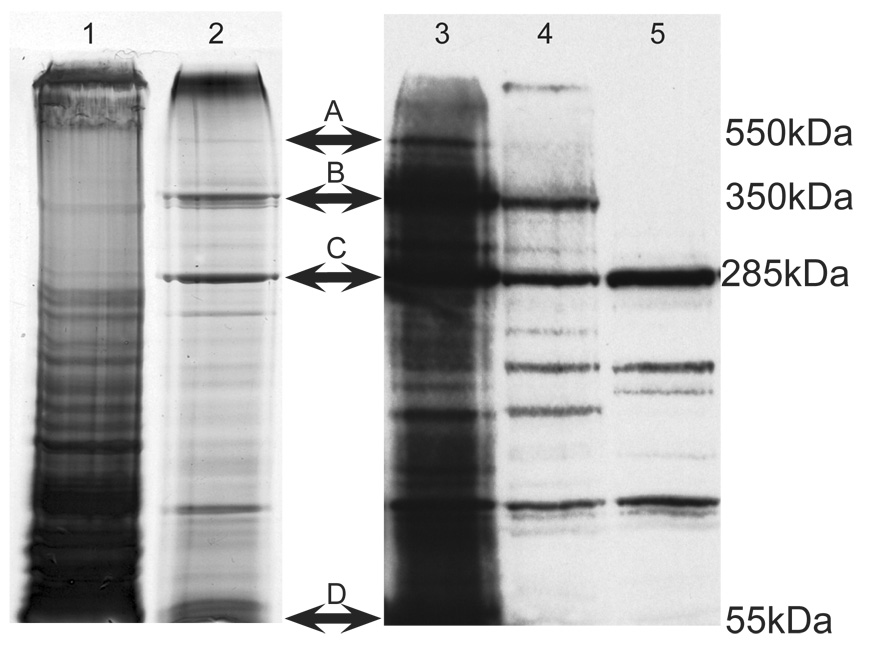
Figure 1. Isolation of TUD complexes.
Figure 1. Isolation of TUD complexes. Lanes 1 and 2 are silver-stained 4.5% SDS-PAGE of immunoprecipitates from BMH-crosslinked embryos using pre-immune serum (lane 1) or anti-TUD (lane 2). Bands A and B were reproducible in TUD immunoprecipitates, and these bands were excised and microsequenced. In some isolations uncrosslinked or "free" TUD was present (C). Lanes 3–5 are immunoblots using the anti-TUD antiserum. Lanes 3 and 4 were loaded with crosslinked extracts subjected to TUD immunoprecipitation, lane 3 was loaded with less total protein than lane 4. Lane 5 was loaded with extract that was immunoprecipitated but not BMH-crosslinked. Immunoglobulin-G (IgG) heavy chain is present at the bottom of the gel (D).
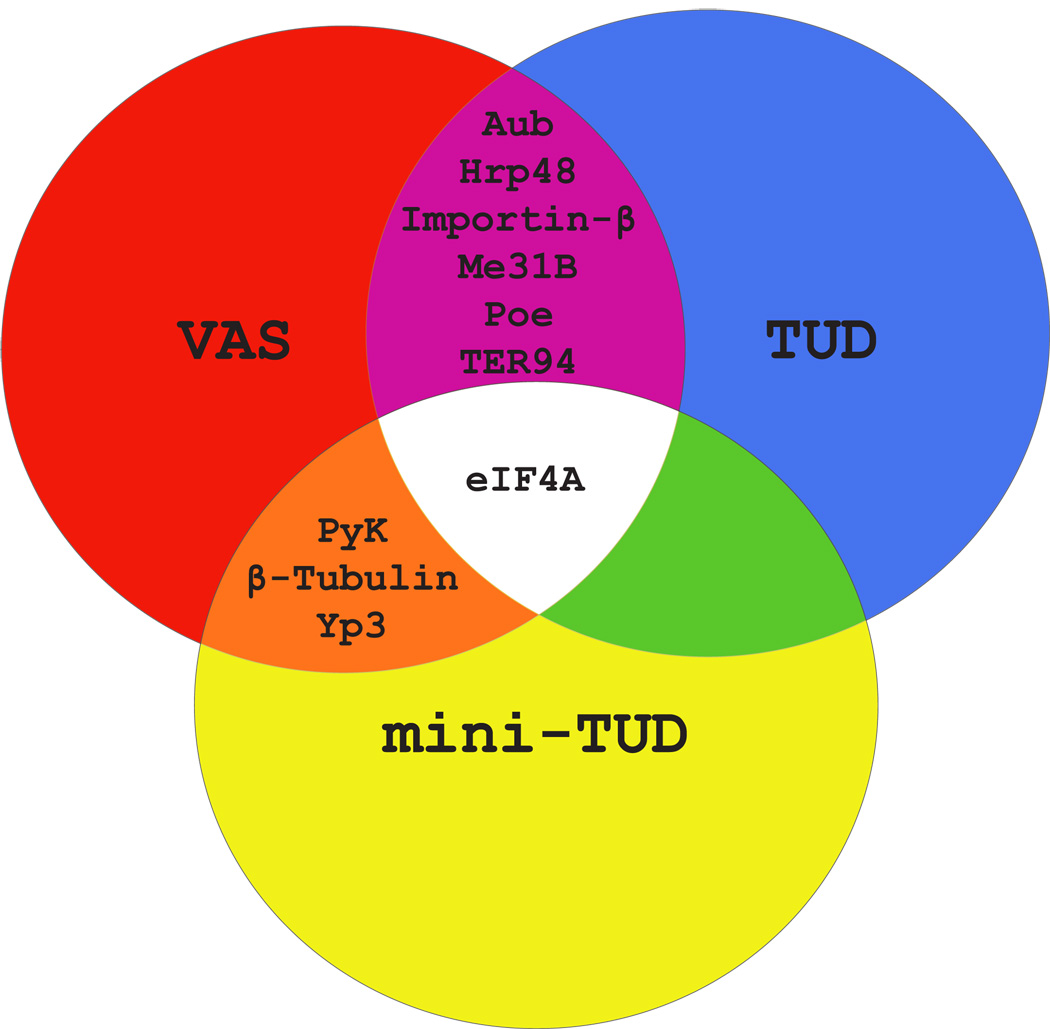
Seven TUD-containing complexes from four independent immunoprecipitation experiments were purified from gels and analyzed by microsequencing. We also analyzed nine independently purified Mini-TUDΔ3-containing complexes. We compared the proteins we identified to those that were co-purified in a similar manner with VAS (Liu et al., 2003). Proteins present in either VAS or TUD containing complexes and proteins common to both are summarized in a Venn diagram in Figure 2, and all proteins recovered more than once in these experiments are listed in Supplementary Table 1.
Figure 2. TUD and VAS protein complexes contain some common components.
A Venn diagram proteins present in more than one of the three types of complex (TUD, mini-TUDΔ3 and VAS).
Seven proteins, AUB, ME31B, HRP48, POE, Importin-β, eIF4A and TER94, were isolated in both the VAS and TUD complexes (Figure 2). AUB is a member of the PIWI subclass of the Argonaute family of proteins, which have been implicated in a specialized RNA pathway controlling repeat-associated small interfering RNAs (rasiRNAs, also called piRNAs) (Gunawardane et al., 2007). Weak aub mutants exhibit a posterior group phenotype similar to tud and vas mutant embryos, and AUB is enriched in germ plasm (Harris and Macdonald, 2001). The DEAD-box RNA helicase ME31B is associated with Drosophila ovarian mRNPs that are involved in translational repression of a subset of mRNAs, including osk mRNA, as they are transported during oogenesis (Nakamura et al., 2001).
We recovered eIF4A, a DEAD-box protein and a general translation initiation factor, in both VAS and TUD complexes, and in three independent immunoprecipitations for mini-TUDΔ3 associated proteins. eIF4A with eIF4E and eIF4G make up the eIF4F complex, which interacts with the 5’ cap structure and is required for the binding and recruitment of mRNAs to a ribosome for translation (Gingras et al., 1999). Independent of its general function in translational initiation, recent evidence also suggests a role for eIF4A in repressing the Drosophila Dpp (TGF-β) signaling pathway (Li et al., 2005; Li and Li 2006).
TER94, a transitional ER membrane associated AAA ATPase, was also present in both the VAS and TUD protein complexes. TER94 homologues in yeast and mammals are required for the export of proteins from the ER into the cytosol (Latterich et al., 1995; Roy et al., 2000), and for the transport of ubiquitinated proteins from the ER to the proteasome (Ye et al., 2001). In the Drosophila germline, TER94 has a role in the posterior accumulation of OSK and in fusome biogenesis (Leon and McKearin, 1999; Ruden et al., 2000). We also identified Purity of Essence (POE), Importin-β, β-Tubulin, yolk protein 3 (YP3), Pyruvate Kinase (PyK) and HRP48 in both the TUD and VAS complexes. HRP48 is enriched at the posterior pole of Drosophila oocytes and regulates osk mRNA localization and translation (Huynh et al., 2004; Yano et al., 2004). Importin-β, while involved in nuclear import, is abundant in the cytoplasm of both nurse cells and oocytes in late states of oogenesis, and in early embryos (Lippai et al., 2000). It is noteworthy that we also recovered Importin-α, but only in the VAS complex (Supplemental Table 1). Existing mutant alleles of poe are male sterile, due to defective spermatid individualization (Fabrizio et al., 1998). poe mRNA is also expressed in ovaries, suggesting a possible wider function for POE in germ line development (Gil et al., 2001; Stapleton et al., 2002). β-Tubulin, YP3, and PyK were isolated several times (5, 5, and 8 respectively) in Mini-TUDΔ3 and VAS complexes, although not in immunoprecipitations for full-length TUD (Supplemental Table 1). YP3 is an extremely abundant protein, which may make it difficult to fully remove from our immunoprecipitations, although we did not recover it in control experiments. We have not further investigated the association of PyK and β-tubulin with polar granules. Thus, by combining purification results from VAS and TUD complexes we were able to select a small number of shared proteins that may play a role in germ cell formation or function.
Mutations in genes encoding polar granule proteins interact
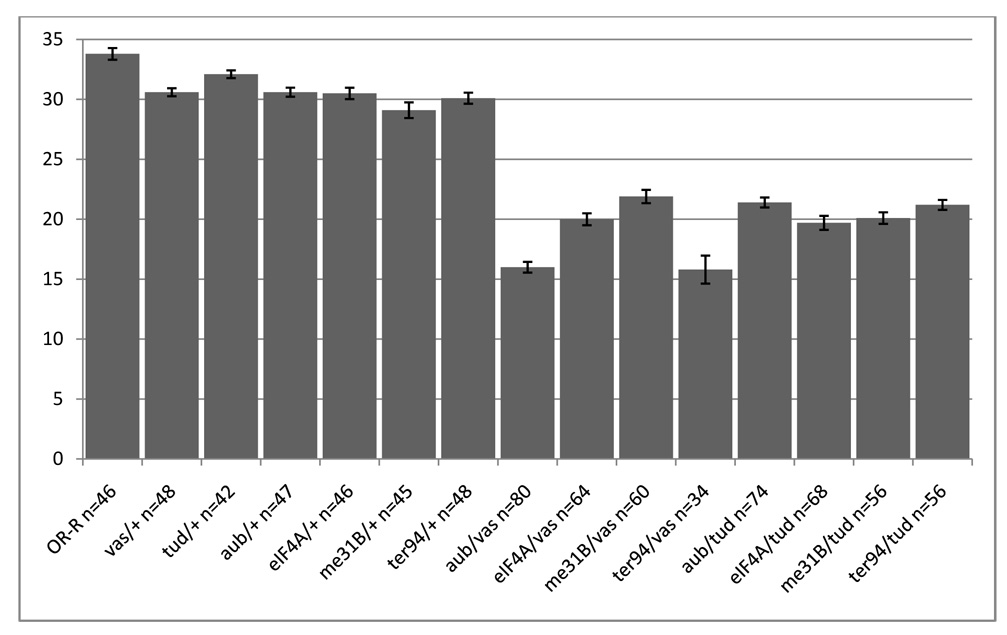
To determine whether the proteins we isolated were functionally related, we analyzed genetic interactions between vas or tud and me31B, ter94, eIF4A and aub. Since germline clones of strong mutants of ter94 (Leon and McKearin, 1999) and null alleles of me31b (Nakamura et al., 2001) do not complete oogenesis, and eIF4A is required throughout development (Galloni and Edgar, 1999; Li and Li, 2006), we were unable to directly test the effect of these genes on pole cell formation. Instead, we assessed germ cell formation in females bearing transheterozygous combinations of mutant alleles of pairs of these genes. We found a significant numerical reduction in germ cells in embryos from females carrying ter94, eIF4A, aub and me31B transheterozygous combinations with tud and vas, while reduction in gene dosage of each single gene had only small effects (Figure 3). Germ cell number is a very sensitive assay to detect interactions between germ plasm components (Liu et al. 2003). We therefore tested genetic interactions between mutations in genes encoding proteins common to the TUD and VAS complexes. Mutations in aub, me31B, eiF4a and Ter94 in combination with mutations in either tud or vas led to a significant reduction in the number of germ cells, while embryos from females heterozygous for only one of these mutations showed only a slight reduction in germ cell number (Figure 3). This synergy is specific for common TUD/VAS complex components. Mutations affecting genes encoding components of only one complex, such as Ef1α48D, which was recovered in the VAS complex, but not in TUD or mini-TUD complexes, did not show this interaction. The genetic interactions between TUD/VAS complex components therefore suggest a common function for these genes in germ cell formation.
Figure 3. aub, me31B, eIF4A and TER94 mutations interact genetically with vas and tud.
Embryos (stage 10) were collected from females of the indicated genotype, stained with anti-VAS, and pole cells were counted. n represents the total number of embryos scored per genotype, and the bar graphs show the average number of pole cells and standard error. Females heterozygous for aub, me31B, eIF4A, or TER94 and either vasPH165 or tudtux46 produce embryos with significantly fewer pole cells than wild type or females heterozygous for only a single mutation. The differences between singly heterozygous females and transheterozygous combinations with vasPH165 or tudtux46 are statistically significant as determined by Student t-test (all p values < 0.001).
Several common components of the VAS and TUD complexes localize to polar granules
In order to determine whether proteins that associate and genetically interact with both VAS and TUD would be enriched on polar granules we analyzed the distribution of these components in the germ plasm. Confocal microscopy does not provide the resolution or magnification needed to unambiguously demonstrate that a protein is a bona fide polar granule component. The best resolution of confocal microscopy on sectioned tissue is approximately 200nm (Robinson et al., 2001), which is approximately the size of a polar granule. Indeed, using confocal microscopy and immuno-electron microscopy (EM) studies, TUD and VAS seem to co-localize in the germ plasm and polar granules. Immuno-EM studies suggest that TUD is not only associated with polar granules but also with mitochondria (Amikura et al., 2001; Bardsley et al., 1993).
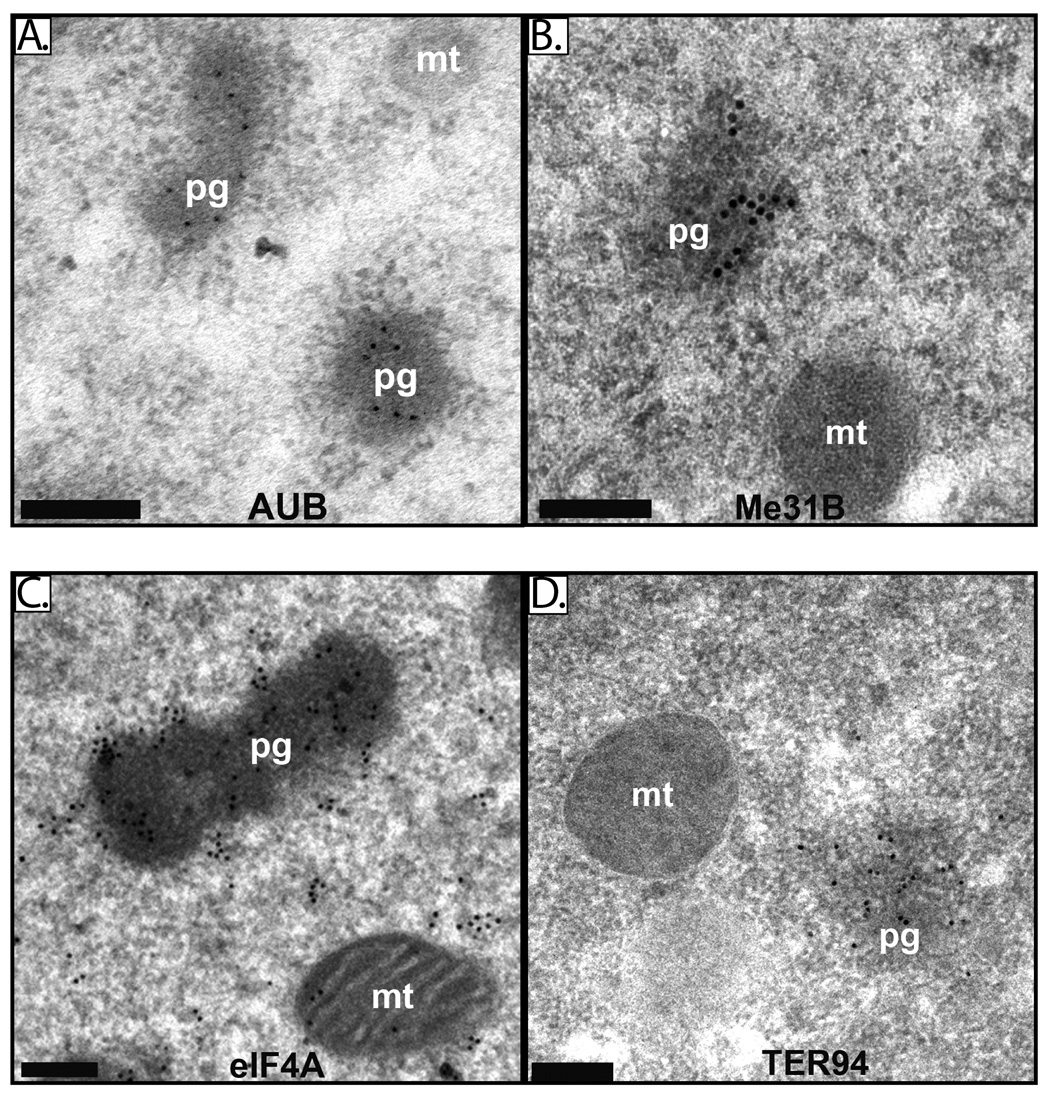
While previous experiments using light microscopy did not reveal enrichment of ME31B or eIF4A at the posterior of the early embryo (Hernandez et al., 2004; Nakamura et al., 2001). using immuno-EM we found that all four proteins are highly enriched in polar granules (Figure 4A–D). By counting gold particles, we estimate that polar granules contain approximately five times the cytoplasmic concentration of eIF4A, as 61% of eIF4A corresponding gold particles localized to polar granules in a section where polar granules comprised approximately 12% of the total area. For VAS, polar granules are approximately 8-fold enriched, as 71% of gold particles localized to polar granules, where polar granules comprised 9% of the total area examined. A more stringent blocking agent (BSA, casein, and ovalbumin rather than BSA alone; see Materials and Methods) resulted in an apparent 10-fold enrichment for VAS on polar granules (90% of VAS-gold particles on polar granules in a section where polar granules comprised 9% of the total area). Under these conditions, TER94 was 7.5-fold enriched on polar granules (75% of gold particles in 10% of the area), ME31B was 7-fold enriched on the polar granules (88% of gold particles in 13% of the area), and AUB was essentially restricted to polar granules (100% of the gold particles on polar granules). We also observed ME31B and eIF4A in electron dense structures outside the pole plasm but these structures were not as large or as dense as germline polar granules (data not shown).
Figure 4. AUB, Me31B, eIF4A and TER94 are components of polar granules.
Electron micrographs of immunostained embryos (stages 1–2) illustrate enrichment of AUB (A), Me31B (B) eIF4A (C) and TER94 (D) on polar granules. Polar granules are denoted as “pg” and mitochondria as “mt”. Black bar = 0.2µM.
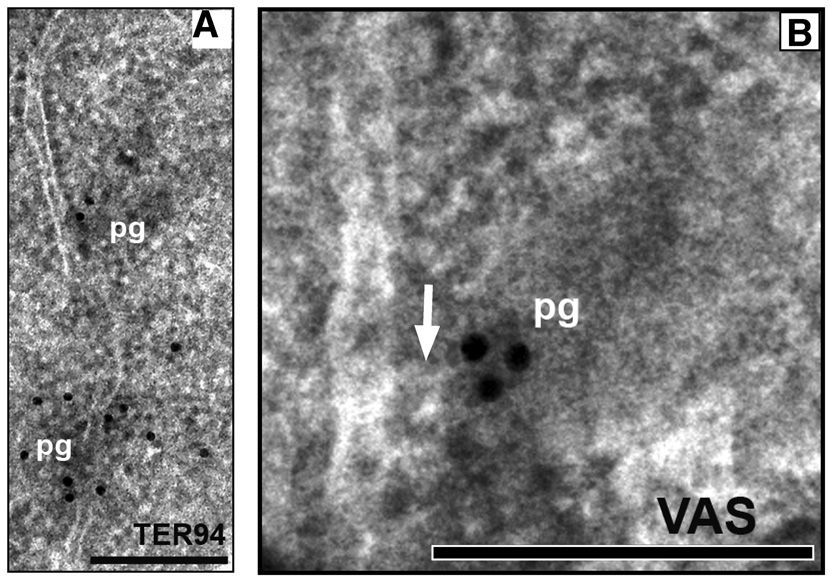
Since TER94 and Me31B associate with ER, we next asked if polar granules are associated with ER (Decker and Parker, 2006; Squirrell et al., 2006; Wilhelm et al., 2005). Using immuno EM analysis, we found several cases where polar granules seemed to be in direct contact with ER structures. Both TER94 and VAS marked polar granules in close association with ER structures (Figure 5).
Figure 5. Polar granules contact ER.
A. Electron micrograph of germ plasm of an early embryo (stage 1–2) showing contact between ER and two electron dense polar granules immunostained for TER94. B. Electron micrograph of germ plasm of an early embryo (stage 1–2) immunostained for VAS showing contact between ER and polar granule. The ER appears to protrude toward the polar granule (arrow). Polar granules are denoted as “pg”. Black bar = 0.2µM.
Discussion
Isolation of novel polar granule components
Little is understood of the molecular events that link the assembly of germ plasm to the formation of germ cells. There is a strong correlation between polar granule formation and germ cell formation, yet their functional relationship is still unclear. In an attempt to understand polar granule formation and function we set out to isolate polar granule components with a biochemical approach; we analyzed proteins common to both TUD and VAS complexes. These complexes were isolated by crosslinking proteins from early embryonic extracts followed by anti-TUD or anti-VAS immunoprecipitation; proteins found in both complexes were then immunolocalized using EM. Using this method we confirmed that AUB is a polar granule component (Wilson et al., 1996) and identified three new polar granule components: ME31B, TER94 and eIF4A. Through genetic interaction analysis in transheterozygous embryos we showed that decreasing the levels of VAS or TUD along with either AUB, ME31B, TER94 or eIF4A reduces germ cell number. This approach both identified novel polar granule components and implicated novel processes in germ cell formation.
Polar granules contain P body components
The presence of AUB, ME31B and eIF4A in polar granules supports the hypothesis that polar granules and P bodies are structurally, and perhaps functionally, related. Our recovery of CUP, an ME31B-interacting protein, in the TUD complex (Supplemental Table 1) further supports this. Similar parallels have recently been found for the mouse chromatoid body, an electron dense structure in the male germline with similarities to Drosophila polar granules and nurse cell nuage (Grivna et al., 2006). RNAs in P bodies are stored in a translationally quiescent state and can later be either degraded or translationally activated in response to physiological cues (Decker and Parker, 2006). Translational repression in P bodies occurs at the level of mRNA recruitment to the ribosome, and through miRNA silencing pathways. The polar granule components we identified suggest their involvement in both types of post-transcriptional regulation. AUB has been implicated in processing of germ line specific piRNAs (Brennecke et al., 2007; Gunawardane et al., 2007; Lim and Kai, 2007). Our findings that AUB associates with polar granules implicates piRNAs in germ cell formation, as has a previous study (Megosh et al., 2006). On the other hand, VAS and eIF4A have been closely linked with translational regulation and are not known to participate in miRNA silencing pathways.
The RNA-rich nature of early polar granules supports the idea that specific germ line-specific mRNAs are stored in polar granules in a translationally repressed state. Subsequently, these RNAs are translated and their function may be required for germ cell formation and further development. How could general translational repression mediated by polar granules be overcome? Conceivably VAS could be a key factor. VAS, a highly conserved polar granule component with homologues in other species involved in germline formation, binds directly to eIF5B. Disrupting the eIF5B-VAS interaction abrogates germ cell formation, presumably due to the loss of the ability of VAS to initiate translation of yet unidentified mRNAs (Johnstone and Lasko, 2004). Thus, VAS may act as a germline specific mRNA translation derepression factor. Other tissue specific factors could adapt a P body to a specific function or cell line. Identification of mRNAs that localize in polar granules and are dependent on VAS for their translation will no doubt provide more insight into this mechanism.
Evidence for an association between polar granules and ER
Ultrastructural analysis of proteins found in the VAS and TUD containing complexes revealed that polar granules were often in close proximity or in contact with ER. Supporting such a link, TER94 and ME31B were present in both TUD and VAS complexes, and are enriched in polar granules. Further work is required to elucidate what proportion of polar granules associate with ER, and whether this association is stage dependent. The presence of TER94, an ER exit site marker, with VAS, ME31B, AUB and eIF4A in the same structure suggests that ER exit sites directly associate with the translational machinery with both activating and repressing factors. Polar granules may form at ER exit sites, which could provide a mechanism for the localization and assembly of mRNPs required for the translational regulation of their constituent mRNAs. There is evidence that P bodies associate with ER exit sites. In the Drosophila ovary, Trailer Hitch (TRAL) associates with ER exit sites and associates with P body components such as ME31B and CUP (Wilhelm et al., 2005). The C. elegans homologue of TRAL, Car-1, associates with DCAP-1, a P-body marker, and car-1 mutations affect ER assembly (Squirrell et al., 2006). We recovered a single TRAL peptide in one immunoprecipitation with TUD (data not shown), perhaps lending additional support to an association between polar granules and ER exit sites.
Biochemical purification of polar granules
Repeated attempts to biochemically isolate polar granules were made over 30 years ago, before the advent of modern analytical techniques that allow the identification of very small amounts of protein. From this work a major polar granule component of approximately 95 kDa was identified (Waring et al., 1978). The nature of this protein was not determined although TER94 has approximately the same molecular mass, as does PIWI, a 97-kDa likely polar granule component that eluded our screens. PIWI associates with VAS, a polar granule component, as well as with components of the miRNA machinery (Megosh et al., 2006). PIWI RNA and protein are enriched in germ plasm and piwi mutants have defects in germ cell formation. Our screen also did not identify OSK, which was shown by a yeast two-hybrid screen to bind directly to VAS. This may be because these proteins were not present in high enough abundance for detection in our assay. Alternatively, as the reactive ends of the crosslinkers we used specifically crosslink cysteine residues, they would not stabilize a particular protein-protein interaction unless a pair of cysteine residues is within the range of the crosslinker. Our work demonstrates that a molecular approach can be a powerful complement to genetics, and that purification schemes based on two independent reagents can reduce signal-to-noise problems that are inherent in co-immunoprecipitation experiments. Molecular approaches such as this one also have the capacity to identify proteins involved in a developmental process that are encoded by genes with multiple functions, or required for cellular viability, that will therefore elude phenotype-based genetic screens.
Methods
Isolation of the TUD and VAS containing complexes from early embryos
The TUD complex was isolated as described in detail (Liu et al. 2003, supplemental information) with the following modifications: 0–1 hr embryos were used, and the cross-linking agent BMH was used at 100 µg/ml. We used a combination of 130 µg antibodies generated against amino acids 1–554 of TUD and 130 µg of antibodies raised against amino acids 1824–2515 of TUD (Thomson and Lasko, 2004). Thus 260 µg rabbit polyclonal anti-TUD antibodies were used for every 10 ml of 10 mg/ml total embryonic extract. The TUD complexes were immunoprecipitated once, run on a 4.5% SDS-PAGE gel, stained with Coomassie blue, and bands were excised. Multiple elution and immunoprecipitation steps were not necessary. In parallel, pre-immune serum controls were performed on cross-linked embryo extracts. To ensure that we were excising bands only enriched in TUD immunoprecipitate lanes we used 2.6 mg of pre-immune serum for control immunoprecipitations. No clear bands were seen in pre-immune controls corresponding to those seen in TUD isolations, but areas of gels containing pre-immune controls that corresponded to a band found in TUD immunoprecipitated lanes were excised and sequenced. The TUD polyclonal antibodies we used did not reproducibly recognize any specific antigens of extracts from tud null embryos or ovaries as determined by western immunoblots. As well, the TUD antibodies used did not show any localized signal in tud null embryos or ovaries (data not shown).
Isolation of Mini-TUDΔ3 containing complexes from early embryos
For generation of mini-TUDΔ3 protein complexes, 0–3 hr embryos laid by mini-tudΔ3; tud1/tud1 females were homogenized in lysis buffer (PBS, 10% glycerol) in the presence of protease inhibitors (Roche) as described (Liu et al., 2003), and the final protein concentration was adjusted to 10 mg/ml. The protein crosslinker BM(PEO)3, [1,11-bismaleimidotriethylene glycol] (Pierce) was added to 1 ml of embryo extracts to a final concentration of 300 µg/ml and extracts were incubated at room temperature for 30 min with subsequent incubation at 4°C for 5 hrs. Cross-linked mini-TUDΔ3 complexes were immunoprecipitated with 50 µl of anti-HA affinity matrix (Roche) in the presence of 2% Triton X-100 at 4°C overnight. The matrix was washed as described (Liu et al., 2003) and the mini-TUDΔ3 complexed were eluted with 50 µl of HA peptide solution (1 mg/ml in PBS, 0.1 mM EDTA) at 37°C for 15 min. Eluted mini-TUDΔ3 complexes were gel-purified and proteins present in the complex were identified by mass spectrometry. As a control, extracts from embryos expressing HA-tagged GFP were used (P. Rangan, A.L.A., R.L., unpublished data). HA-GFP protein was not incorporated into higher molecular weight complexes under cross-linking conditions used to generate mini-TUDΔ3 complexes. In addition, no proteins migrating as mini-TUDΔ3 complexes were detected in the HA-GFP control after immunoprecipitation. Nevertheless, blank control gel bands were excised and subjected to mass spectrometry in parallel with identifying mini-TUDΔ3 complex components.
Drosophila strains and analysis of trans-heterozygotes
The following alleles were used in this work: tudtux46 (tud null allele) (Thomson and Lasko, 2004), vasPH165 (Styhler et al., 1998), me31BΔ1, ter9426-8 (Ruden et al., 2000), aubHN and eIF4A1013 (Galloni and Edgar, 1999) flies. As control, all mutant alleles were crossed to Oregon-R wild-type flies to count germ cell numbers in a heterozygous background. Flies were grown at 25°C, females of the desired genotype were put in cages and crossed to males of desired genotype, 4–6 hour-old eggs were collected and immunostained with rabbit α-VAS, and numbers of germ cells were counted in stage 10 embryos.
Immuno-EM
Embryos were dechorionated in 50% bleach for 1 minute, fixed for 1 minute in 25% glutaraldehyde, and then vitelline membranes were manually removed using tungsten needles. Embryos were then fixed for 30 minutes in fixative containing concentrations between 0.1% glutaraldehyde and 3% formaldehyde to 2% glutaraldehyde and 8% formaldehyde. Lower concentrations of both glutaraldehyde and formaldehyde were used to maintain antigenicity of desired proteins while higher concentrations were used to maintain morphology. Embryos were then dehydrated, embedded in LR white (EMS), then sectioned. Resulting grids were incubated with antibodies using standard protocols, summarized as follows. Grids were preincubated on 2% glycine, then blocked for 5 minutes on PBS + 2% BSA or BSA casein ovalbumin (BCO) and then incubated with primary antibody in blocking solution. The grids were washed with PBS, blocked with BCO or PBS + BSA again, and incubated with secondary gold-labeled antibodies diluted in the blocking solution. The grids were washed with PBS, and then stained with uranyl acetate followed by a Reynold’s lead treatment. Antibodies against AUB, VAS, ME31B and TER94 worked well with the stringent blocking agent BCO, while with anti-eIF4A, we used PBS and BSA.
Supplementary Material
A list of all proteins that were isolated more than one in VAS, mini-TUD, and TUD complexes. Numbers in parentheses represent the number of peptides sequenced, if more than one number is presented, then each number represents the number of peptides sequenced from each immunoprecipitation experiment in which the particular protein was identified.
Acknowledgements
We would like to thank Hong Han, Dennis McKearin and Miltiadis Paliouras for α-AUB, α-TER94 and α-ME31B antibodies respectively. Thanks also to Chiara Gamberi, Jan-Michael Kügler, and Caroline Laplante for critical evaluation of our manuscript. Technical assistance from J. Miu and K. Sears from the McGill EM Centre was greatly appreciated. This work was supported by grants from NICHD (R01-HD36631) and NSERC to P. L and by the Howard Hughes Medical Institute investigator and the Kimmel Stem Cell Center to R.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amikura R, Hanyu K, Kashikawa M, Kobayashi S. Tudor protein is essential for the localization of mitochondrial RNAs in polar granules of Drosophila embryos. Mech Dev. 2001;107:97–104. doi: 10.1016/s0925-4773(01)00455-5. [DOI] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- Arkov AL, Wang JY, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–4062. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- Bardsley A, McDonald K, Boswell RE. Distribution of tudor protein in the Drosophila embryo suggests separation of functions based on site of localization. Development. 1993;119:207–219. doi: 10.1242/dev.119.1.207. [DOI] [PubMed] [Google Scholar]
- Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carrera P, Johnstone O, Nakamura A, Casanova J, Jackle H, Lasko P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J Cell Biol. 2006;173:159–163. doi: 10.1083/jcb.200601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Fabrizio JJ, Hime G, Lemmon SK, Bazinet C. Genetic dissection of sperm individualization in Drosophila melanogaster. Development. 1998;125:1833–1843. doi: 10.1242/dev.125.10.1833. [DOI] [PubMed] [Google Scholar]
- Galloni M, Edgar BA. Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development. 1999;126:2365–2375. doi: 10.1242/dev.126.11.2365. [DOI] [PubMed] [Google Scholar]
- Geigy R. Action de l'ultra-violet sûr le pôle germinale dans l'ouef de Drosophila melanogaster. Rev Suisse Zool. 1931;38:187–288. [Google Scholar]
- Gil P, Dewey E, Friml J, Zhao Y, Snowden KC, Putterill J, Palme K, Estelle M, Chory J. BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 2001;15:1985–1997. doi: 10.1101/gad.905201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol. 2006;16:1077–1089. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55:577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development. 1990;109:425–433. doi: 10.1242/dev.109.2.425. [DOI] [PubMed] [Google Scholar]
- Hernandez G, Lalioti V, Vandekerckhove J, Sierra JM, Santaren JF. Identification and characterization of the expression of the translation initiation factor 4A (eIF4A) from Drosophila melanogaster. Proteomics. 2004;4:316–326. doi: 10.1002/pmic.200300555. [DOI] [PubMed] [Google Scholar]
- Huynh JR, Munro TP, Smith-Litiere K, Lepesant JA, St Johnston D. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev Cell. 2004;6:625–635. doi: 10.1016/s1534-5807(04)00130-3. [DOI] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci U S A. 1974;71:1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP. The autonomous function of germ plasm in a somatic region of the Drosophila egg. Exp Cell Res. 1976;97:127–140. doi: 10.1016/0014-4827(76)90662-5. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 1990;4:905–921. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- Latterich M, Frohlich KU, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- Leon A, McKearin D. Identification of TER94, an AAA ATPase protein, as a Bam-dependent component of the Drosophila fusome. Mol Biol Cell. 1999;10:3825–3834. doi: 10.1091/mbc.10.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li WX. A novel function of Drosophila eIF4A as a negative regulator of Dpp/BMP signalling that mediates SMAD degradation. Nat Cell Biol. 2006;8:1407–1414. doi: 10.1038/ncb1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippai M, Tirian L, Boros I, Mihaly J, Erdelyi M, Belecz I, Mathe E, Posfai J, Nagy A, Udvardy A, et al. The Ketel gene encodes a Drosophila homologue of importin-beta. Genetics. 2000;156:1889–1900. doi: 10.1093/genetics/156.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dansereau DA, Lasko P. Fat facets interacts with vasa in the Drosophila pole plasm and protects it from degradation. Curr Biol. 2003;13:1905–1909. doi: 10.1016/j.cub.2003.10.026. [DOI] [PubMed] [Google Scholar]
- Mahowald A. Fine Structure of Pole Cells and Polar Granules in Drosophila Melanogaster. Journal of Experimental Zoology. 1961;151:201–215. [Google Scholar]
- Mahowald AP. Assembly of the Drosophila germ plasm. Int Rev Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Frohnhofer HG, Lehmann R. Determination of anteroposterior polarity in Drosophila. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- Robinson JM, Takizawa T, Pombo A, Cook PR. Correlative fluorescence and electron microscopy on ultrathin cryosections: bridging the resolution gap. J Histochem Cytochem. 2001;49:803–808. doi: 10.1177/002215540104900701. [DOI] [PubMed] [Google Scholar]
- Roy L, Bergeron JJ, Lavoie C, Hendriks R, Gushue J, Fazel A, Pelletier A, Morre DJ, Subramaniam VN, Hong W, et al. Role of p97 and syntaxin 5 in the assembly of transitional endoplasmic reticulum. Mol Biol Cell. 2000;11:2529–2542. doi: 10.1091/mbc.11.8.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden DM, Sollars V, Wang X, Mori D, Alterman M, Lu X. Membrane fusion proteins are required for oskar mRNA localization in the Drosophila egg chamber. Dev Biol. 2000;218:314–325. doi: 10.1006/dbio.1999.9583. [DOI] [PubMed] [Google Scholar]
- Saffman EE, Lasko P. Germline development in vertebrates and invertebrates. Cell Mol Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, Lyons GE, Anderson P, White JG. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, et al. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Lasko P. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev Cell. 2002;3:865–876. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev Dyn. 2005;232:593–608. doi: 10.1002/dvdy.20297. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Tudor and its domains: germ cell formation from a Tudor perspective. Cell research. 2005;15:281–291. doi: 10.1038/sj.cr.7290297. [DOI] [PubMed] [Google Scholar]
- Vanzo N, Oprins A, Xanthakis D, Ephrussi A, Rabouille C. Stimulation of endocytosis and actin dynamics by oskar polarizes the Drosophila oocyte. Dev Cell. 2007;12:543–555. doi: 10.1016/j.devcel.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Waring GL, Allis CD, Mahowald AP. Isolation of polar granules and the identification of polar granule-specific protein. Dev Biol. 1978;66:197–206. doi: 10.1016/0012-1606(78)90284-1. [DOI] [PubMed] [Google Scholar]
- Weismann A. Continuity of the Germ Plasm. In: Poulton E, et al., editors. Essays upon heredity and kindred biological problems. Oxford: Clarendon Press; 1885. [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Yano T, De Quinto SL, Matsui Y, Shevchenko A, Ephrussi A. Hrp48, a Drosophila hnRNPA/B Homolog, Binds and Regulates Translation of oskar mRNA. Dev Cell. 2004;6:637–648. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A list of all proteins that were isolated more than one in VAS, mini-TUD, and TUD complexes. Numbers in parentheses represent the number of peptides sequenced, if more than one number is presented, then each number represents the number of peptides sequenced from each immunoprecipitation experiment in which the particular protein was identified.