Abstract
Importin-β mediates protein transport across the nuclear envelope through the nuclear pore complex (NPC) by interacting with components of the NPC, called nucleoporins, and a small G protein, Ran. Although there is accumulated knowledge on the specific interaction between importin-β and the Phe-Gly (FG) motif in the nucleoporins as well as the effect of RanGTP on this interaction, the molecular mechanism by which importin-β shuttles across the nuclear envelope through the NPC is unknown. In this study, we focused on four binding pockets of importin-β for the FG motifs and characterized the interaction using a single-molecule force-measurement technique with atomic-force microscopy. The results from a series of importin-β mutants containing amino acid substitutions within the FG-binding pockets demonstrate that the individual FG-binding pockets have different affinities to FG-Nups (Nup62 and Nup153) and different sensitivities to RanGTP; the binding of RanGTP to the amino-terminal domain of importin-β induces the conformational change of the entire molecule and reduces the affinity of some of the pockets but not others. These heterogeneous characteristics of the multiple FG-binding pockets may play an important role in the behavior of importin-β within the NPC. Single-molecule force measurement using the entire molecule of an NPC from a Xenopus oocyte also implies that the reduction of the affinity by RanGTP really occurs at the nucleoplasmic side of the entire NPC.
Keywords: nuclear import, nuclear pore complex, nuclear transport, scanning probe microscopy, single-molecule force measurement
Macromolecular transport between the cytoplasm and the nucleoplasm is mediated by a large protein complex called the nuclear pore complex (NPC), which is embedded in the nuclear envelope (NE) (1). The NPC has an octameric structure and is composed of ≈30 different kinds of subunits termed nucleoporins (Nups) (reviewed in ref. 2). Approximately one-third of the Nups carry phenylalanine-glycine (FG) motifs (3), and the Nups carrying FG motifs (FG-Nups) play important roles in forming the central channel of the NPC. The studies using x-ray crystallography and other biophysical approaches have demonstrated that the domains rich in the FG motifs do not have any particular secondary or tertiary structures (4, 5) and interact with each other through hydrophobic attraction (6). Because a single NPC contains ≈100 FG-Nups, it has been speculated that the FG motif-rich domains fill the central channel of the NPC to form an entropic barrier and prevent the proteins from freely traveling through.
FG-Nups interact with transport mediators such as importin-β (7). Importin-β mediates the translocation of the protein that carries a specific basic amino acid sequence motif called the classical nuclear localization signal (cNLS) from the cytoplasm to the nucleus by making a complex with an adaptor protein, importin-α (reviewed in ref. 8). Importin-β also mediates the import of non-NLS proteins, such as ribosomal proteins and transcription factors, by directly binding to them (9–11). The transport cycle of importin-β is regulated by a small GTPase, Ran. The GTP-bound form of Ran (RanGTP) is abundant in the nucleoplasm, whereas the GDP-bound form (RanGDP) is rich in the cytoplasm, and importin-β preferentially binds to RanGTP (12). In the current model of the nuclear transport, the importin-β-cargo complex reaches the nucleoplasm after passing through the NPC, encounters RanGTP and releases the cargo into the nucleoplasm, and then importin-β returns back to the cytoplasm together with RanGTP (reviewed in refs. 13 and 14).
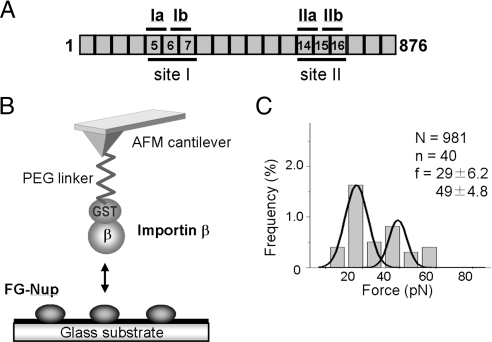
X-ray crystallography has revealed that importin-β consists of 19 tandem HEAT repeats arranged to form a superhelical structure (Fig. 1A) and interacts with other molecules through different regions within the molecule (reviewed in ref. 15). The FG motifs bind to the outer surface of the helix (4, 16, 17), and RanGTP binds to the inner surface of the helix and induces a conformational change of the entire molecule (18–20). Based on the crystal structure (4), the interaction between importin-β and the FG-Nups occurs through two different binding pockets existing in residues 168–297 (site I) (Fig. 1A). One pocket is composed of HEAT repeats 5 and 6 (pocket Ia), and the other is composed of HEAT repeats 6 and 7 (pocket Ib). Another study using site-directed mutagenesis has shown that importin-β carries other FG-binding pockets within HEAT repeats 14–16 between residues 602 and 726 (site II) (Fig. 1A) (21). A 3D structural similarity between these domains (HEAT 5–7 and HEAT 14–16) has suggested that site II also contains two FG-binding pockets; one between HEAT repeats 14 and 15 (pocket IIa) and the other between HEAT repeats 15 and 16 (pocket IIb) (21).
Fig. 1.
Single-molecule force measurement between importin-β and Nup153-C. (A) A schematic illustration of full-length mouse importin-β (residues 1–876). HEAT repeats involved in the pocket formation are indicated. (B) A schematic representation of the single-molecule force measurement system with AFM. The purified Nup153-C was immobilized on a glass substrate. GST-fused importin-β was noncovalently attached to a glutathione-coupled AFM cantilever. The interaction between importin-β and Nup153-C was characterized by monitoring the deflection of the cantilever when the cantilever approaches and then retracts from the glass surface. (C) The histogram of the rupture force obtained in the force measurement. The histogram was fitted with Gaussian distribution. The total number of approaches (N), the number of approaches in which the specific interaction was detected (n) and the mean values of the peaks (f) are also shown.
In this study, we characterized the individual FG-binding pockets of importin-β by using a single-molecule force measurement technique by atomic-force microscopy (AFM). Amino acid substitutions were introduced into importin-β to abolish the individual pockets and the strength of the importin-β–FG-Nup interaction was evaluated at a single-molecule level.
Results
Single-Molecule Force Measurement Between Importin-β and Nup153.
The experimental system for single-molecule force measurement using the AFM is described in Fig. 1B. The amino-terminal fragment of Nup153 (Nup153-C, residues 895-1475), which contains 25 FG motifs, was expressed in bacteria as a GST fusion protein and purified by affinity chromatography. After cleaving off the GST moiety, it was attached to the surface of a cover glass (see Materials and Methods). To attach importin-β to an AFM cantilever, we used a cantilever to which glutathione was covalently coupled via a PEG linker (22), so that purified GST-fused importin-β could be attached to this cantilever. The single-molecule force measurement was performed with the AFM, and the rupture force was obtained from force-extension curves. Statistically, two different populations of rupture force were detected; small (29 ± 6.2 pN) and large force (49 ± 4.8 pN) (mean ± standard deviation) (Fig. 1C). Because the larger force could still be observed when the concentration of importin-β on the cantilever was reduced, it can be speculated that it was not derived from “double pulling” (two importin-β–Nup153 interactions were broken simultaneously) but rather from multiple binding pockets within a single importin-β.
The characteristics of FG-binding pockets were further examined by using an amino-terminal fragment (residues 1–446, termed Imp β-N), which contains pockets Ia and Ib, and a carboxyl-terminal fragment (residues 447–876, termed Imp β-C), which contains pockets IIa and IIb. The rupture force between Imp β-N and Nup153-C showed a single peak (29 ± 9.4 pN) (Fig. 2A), whereas the interaction between Imp β-C and Nup153-C showed double peaks (29 ± 5.1 and 48 ± 4.5 pN) (Fig. 2E), as seen in the full-length importin-β (Fig. 1C). A series of amino acid substitutions were introduced into Imp β-N and Imp β-C fragments to abolish the pockets in these fragments: I178A and F217A within pocket Ia (Imp β-NΔIa), Y255A and I263R within pocket Ib (Imp β-NΔIb), L612D within pocket IIa (Imp β-CΔIIa), and F688A within pocket IIb (Imp β-CΔIIb), which correspond to the amino acid substitutions in human importin-β (I178A, F217A, Y255A, I263R, L612D, and F688A) that reduced the affinity for the FG motifs without affecting the binding to RanGTP (21).
Fig. 2.
Single-molecule force measurements between importin-β mutants and Nup153-C. The rupture force between Nup153-C on the glass substrate and various importin-β mutants on an AFM cantilever was measured by AFM as described in Fig. 1B. The results were summarized in the histograms. The schematic illustrations of the importin-β constructs are shown in the histograms. Imp β-N (A), Imp β-NΔIa (B), Imp β-NΔIb (C), Imp β-NΔIaIb (D), Imp β-C (E), Imp β-CΔIIa (F), Imp β-CΔIIb (G), and Imp β-CΔIIaIIb (H). The histograms were fitted with Gaussian distribution. N, n, and f are indicated as in Fig. 1C.
The force measurement between Nup153-C and Imp β-NΔIa showed the rupture force only at ≈30 pN (Fig. 2B), indicating that the interaction between pocket Ib and a single FG motif was ruptured with ≈30 pN in our experimental conditions, whereas Imp β-NΔIb showed only a small peak at ≈30 pN (Fig. 2C). These results suggest that pocket Ia has lower affinity to the FG motifs than pocket Ib. No specific interaction was detected when both pockets Ia and Ib were mutated (Imp β-NΔIaIb) (Fig. 2D). Microwell plate assay also demonstrated that pocket Ia has lower affinity to the FG motifs than pocket Ib [supporting information (SI) Table S1]. Therefore, in the amino-terminal FG-binding site, pocket Ib is the major binding pocket for Nup153 and pocket Ia does not strongly interact with Nup153.
When the interactions between the Imp β-C mutants (Imp β-CΔIIa and Imp β-CΔIIb) and Nup153-C were examined, both of the mutants showed rupture force only at ≈30 pN and not at ≈50 pN (Fig. 2 F and G). No specific interaction was detected when both pockets IIa and IIb were abolished (Imp β-CΔIIaIIb) (Fig. 2H). These results indicate that the ≈30-pN force was derived from the interaction between a single FG-binding pocket and a single FG motif, and the ≈50-pN force, which was observed in Imp β-C, was derived from a simultaneous rupture event of two pockets with the FG motifs.
Some of the FG-Binding Pockets Have Different Binding Properties to Different FG-Nups.
We performed force measurements using Nup62, which localizes in the central channel of the NPC (23). The histogram of the rupture force between full-length importin-β and Nup62-FG fragment (Nup62-N, residues 1–178) showed a similar distribution to that from Nup153 (Fig. S1A). The measurements using a series of importin-β mutants demonstrated that the carboxyl-terminal binding pockets interacted with Nup62 in a similar manner to Nup153; the Imp β-CΔIIa and Imp β-CΔIIb showed the rupture force only at ≈30 pN, and the wild-type Imp β-C showed two peaks at ≈30 pN and ≈50 pN (Fig. S1 F–I). This result demonstrates that the interaction between the carboxyl terminal FG-binding pockets and FG motifs are similar in different FG-Nups. However, the mutants in the amino-terminal binding pockets showed interesting characteristics regarding rupture force against different FG-Nups; Imp β-NΔIb showed the rupture force mainly at ≈30 pN, but Imp β-NΔIa did not show any particular force against Nup62 (Fig. S1 B–E), which is the opposite case of Nup153 (Fig. 2 A–D). These results demonstrate that pockets Ia and Ib have different affinities to different FG-Nups.
RanGTP Affects FG-Binding Pockets Within Importin-β in a Different Manner.
RanGTP localizes mainly in the nucleoplasmic side of the NPC and affects the importin-β-FG-Nup interaction (12). Because Nup153 has been thought to be involved in the termination step of importin-β transport through the NPC (24, 25), the effect of RanGTP on the Nup153-importin-β interaction was investigated.
RanGTP binds to the amino-terminal region of importin-β and induces conformational change (18–20). Our microwell plate assay also demonstrated that the full-length importin-β and Imp β-N showed similar KD values against RanGTP (3.7 ± 0.81 and 7.7 ± 1.9 for full-length importin-β and Imp β-N, respectively, Table S1). In the single-molecule force measurement in the presence of saturating concentration of RanGTP (200 nM), the interaction between Imp β-N and Nup153-C was not significantly affected (Fig. 3A), and, as expected, the interaction between Imp β-C and Nup153-C was not affected by RanGTP (data not shown). However, in the case of the interaction between full-length importin-β and Nup153-C, the addition of RanGTP abolished the ≈50-pN rupture force, and only the ≈30-pN force was observed (compare Figs. 1C and 3B). Because the ≈50-pN-peak was derived from the simultaneous occupation of pockets IIa and IIb in the carboxyl-terminal FG-binding site (Fig. 2 E–H), RanGTP binding to the amino-terminal region of importin-β might affect one of the carboxyl-terminal FG-binding sites.
Fig. 3.
The effect of RanGTP on the FG-binding pockets of importin-β. The rupture force between Nup153-C and importin-β was measured by AFM in the presence (A, B, and E) or absence (D) of RanGTP, and the results were summarized in the histograms. The importin-β constructs used were Imp β-N (A), full-length importin-β (B), Imp βΔIaIb (D and E). The histograms were fitted with Gaussian distribution. N, n, and f are indicated as in Fig. 1C. A schematic illustration of Imp βΔIaIb is shown in C.
To confirm this possibility, an additional mutant of the full-length importin-β that lacks pockets Ia and Ib (Imp βΔIaIb) was constructed (Fig. 3C). The single-molecule force measurement in the absence of RanGTP revealed two peaks in the rupture force histogram (27 ± 7.4 and 50 ± 7.6 pN) (Fig. 3D), consistent with the result from the Imp β-C fragment (Fig. 2E). On the other hand, the measurement in the presence of RanGTP showed only a single rupture force at ≈30 pN, and the ≈50-pN force completely disappeared (Fig. 3E). These results indicate that RanGTP binding to the amino terminus can affect the carboxyl-terminal binding site via an allosteric effect and reduce the affinity to the FG motifs. A similar result was obtained when the nonhydrolyzable GTP analogue GppNHp was used instead of RanGTP (data not shown), demonstrating that the binding of RanGTP to importin-β itself, and not GTP hydrolysis, affects the interaction.
Single-Molecule Force Measurement Between Importin-β and the NPC in the Native NE.
A NE dissected from a Xenopus oocyte was spread onto a glass surface. Through fine micromanipulation, both cytoplasmic and nucleoplasmic faces of the NE could be placed on the glass surface (26). The AFM observation of these specimens revealed a clear difference in the surface structure (Fig. 4 A and B). The nucleoplasmic face of the NE showed nuclear lamina and the NPC, whereas the cytoplasmic face showed the NPC without lamina. A quantitative immunoblot analysis demonstrated that this NE specimen retains a significant amount of Nup153 (Fig. S2). After the imaging, the same specimen was subjected to the force measurement with importin-β attached to an AFM cantilever in the same procedure as described in Fig. 1B. The rupture force between importin-β and the nucleoplasmic side of the NE distributed around 30 pN (31 ± 7.3 pN) and 50 pN (54 ± 8.3 pN) (Fig. 4C), consistent with the result obtained from purified Nup153-C (Fig. 1C). In addition, RanGTP-bound form of importin-β showed only single peak at ≈30 pN (35 ± 7.5 pN) (Fig. 4D), demonstrating that, in the nucleoplasmic face of the NPC, some of the pockets of importin-β are inactivated by RanGTP, although others can still interact with the NPC.
Fig. 4.
Single-molecule force measurements between importin-β and the NPC. The NE was isolated from Xenopus laevis and imaged by AFM in contact mode in liquid. (A and B) The cytoplasmic side (A) and the nucleoplasmic side (B) of the NE were observed. The enlarged images are shown in the Insets. (Scale bars, 500 nm.) (C–E) The same specimen was used to measure the interaction with importin-β. The cantilever used for the imaging was switched to the cantilever carrying full-length importin-β, and then the force measurement was performed with the nucleoplasmic side (C) and the cytoplasmic side of the NE (E). Some of the Nups contain Ran binding site(s) (38–40), although the effect of Ran binding to these Nups on the NPC function is still unknown. To avoid effects, if any, of Ran on the NPC function and/or structure, the importin-β-attached cantilever was first incubated with RanGppNHp, and then the force measurement was performed against the nucleoplasmic side of the NE in the Ran-free solution (D). The rupture force is summarized in the histogram. The histograms were fitted with Gaussian distribution. The peak values (f) are also indicated in the histogram.
In contrast to the nucleoplasmic side of the NE, the cytoplasmic side of the NE showed a single peak at around 40 pN (40 ± 8.4 pN) (Fig. 4E). This result implies that the cytoplasmic components of the NPC behave against importin-β differently from other FG-Nups. We examined the interaction between purified Nup358 (one of the major components of the cytoplasmic face of the NPC) and importin-β. However, the rupture force between these proteins showed similar pattern to Nup62 and Nup153; it showed two peaks at ≈30 and ≈50 pN (Fig. S3). Thus, the current possibilities include (i) a single ≈40 pN-rupture force may come from interaction between importin-β and other FG-Nups on the cytoplasmic side of the NE, and (ii) Nup358-binding protein(s) (27, 28) may affect the interaction.
Discussion
In this report, we performed single-molecule force measurements to characterize the four FG-binding pockets of importin-β. The data obtained here all indicate that some pockets have similar affinity to different FG-Nups but others show different properties to different FG-Nups (Fig. 2 and Fig. S1). Furthermore, RanGTP has a different effect to each binding pocket (Fig. 3). These lines of evidence all imply the existence of multiple heterogeneous binding pockets on the surface of importin-β. The mechanism and the significance of this characteristic are discussed below.
RanGTP-Induced Conformational Change of Importin-β Differentially Affects FG-Binding Pockets.
The single-molecule force measurement using Nup153 in the presence of RanGTP demonstrated that each FG-binding pocket is affected by RanGTP in a different manner; the interaction between FG-binding site I and the FG motifs was hardly affected (Fig. 3A), whereas the interaction of FG-binding site II was significantly reduced upon RanGTP binding (Fig. 3 D and E). The experiment using the nucleoplasmic side of the NE also demonstrated that at least some of the pockets lose their affinity to the NPC in the presence of RanGTP (Fig. 4D), indicating that the inhibition of the pocket by RanGTP could occur in the whole NPC.
The effect of RanGTP on the importin-β-Nup153 interaction can be attributed to a conformational change of the entire importin-β molecule induced by RanGTP binding (4, 20). Based on the crystal structures of RanGTP-unbound and -bound forms of Kap95p, yeast homologue of importin-β (17, 20), the structures of pockets Ib and IIa are not largely affected by RanGTP binding, whereas pockets Ia and IIb become narrower (Fig. 5 A and B), which is likely to hinder the binding of the FG motif. This comparison explains well the result of our force measurement in Fig. 3. The disappearance of the second peak in Imp βΔIaIb is possibly due to the inhibition of pocket IIb by RanGTP-induced conformational change (Fig. 3 D and E). On the other hand, because pocket Ia has a low affinity to the FG motif even in the absence of RanGTP, the effect of RanGTP binding was not prominent, resulting in no obvious difference between the rupture force histograms in the absence and presence of RanGTP (Figs. 2A and 3A). RanGTP-induced conformational change in each pocket is summarized in Fig. 5C.
Fig. 5.
RanGTP-induced conformational change of FG-binding pockets. (A and B) The 3D structure of mouse importin-β was predicted from the crystal structure of yeast importin-β Kap95p (17, 20) complex with (red) and without (blue) RanGTP. The FG-binding site I (HEAT 5–7) (A) and the FG-binding site II (HEAT 14–16) (B) are shown. The amino acid side chains that have been expected to interact with the FG motifs (L174, T175, I178, E214, F217, M219, Q220, C223, Y255, P258, A259, A262, and I263 in site I, M608, A609, L612, Y646, A649, K651, P652, G655, F688, D690, E691, Q694 and L695 in site II) are shown (17, 21). (C) An illustration of the importin-β-Nup153 interaction and the effect of RanGTP. Upon binding of RanGTP to the amino-terminal domain, pocket Ia is inactivated via an allosteric effect, whereas pocket Ib is not affected. The binding of RanGTP also affects pocket IIb, but not IIa, through a far-reaching allosteric effect.
Importin-β returns to the cytoplasm together with RanGTP after it releases the cargo in the nucleoplasm (29, 30). Our result indicates that the pocket Ib and pocket IIa can still interact with the FG motifs after importin-β binds to RanGTP. These interactions might be involved in the outward movement of importin-β from the nucleoplasm to the cytoplasm.
Importin-β Is Composed of Multiple Heterogeneous Binding Pockets for FG Motifs.
Our single-molecule force measurement on the N-terminal binding pockets (Ia and Ib) indicated that these pockets have different affinities to Nups153 and Nup62 (Figs. 2 B–D and S1 C–E), implying that at least some of the pockets have different affinities to different FG motifs. The FG motifs are often found in short hydrophobic amino acid clusters such as GLFG and FxFG. Although the phenyl side chain in the FG motif plays a central role in the interaction with importin-β (4, 16, 17), the neighboring amino acids, which vary considerably among different Nups, also directly interact with the pockets (17, 31). In particular, a lysine residue has been shown to play an important role in the determination of the affinity (31). There are several lysine residues around seven FG motifs within Nup153-C, whereas there is only one in Nup62-N. FG-binding site I, but not site II, might be more sensitive to these lysine residues. Interestingly, the distribution of lysine residues neighboring the FG motifs in all of the FG-Nups indicates that the FG-Nups located in the central channel of the NPC (Nup62, Nup54, Nup58) contain a low percentage of neighboring lysine residues, whereas those located in the nucleoplasmic (Nup50 and Nup153) and cytoplasmic (Nup358) ends have a high percentage of neighboring lysine residues (Table S2). These facts lead us to a hypothesis that the N-terminal binding pockets may play an important role in the directional movement of importin-β from the cytoplasmic side to the central channel and from the central channel to the nucleoplasmic side of the NPC.
Because FG-rich domains of FG-Nups interact with each other and form a FG-meshwork within the NPC (6), it is likely that importin-β goes through this mesh by interacting with the FG motifs through multiple binding pockets. Considering the rapid transport of cargo proteins through the NPC in semiintact cells [>1,000 molecules per NPC per second (32, 33)], individual binding pockets of importin-β must rapidly repeat the association and dissociation with and from FG motifs in the process of transport. Our results suggest that one of the pockets of importin-β can jump from one FG motif to another, while other pockets remain associated with FG motifs. Alternatively, the FG-binding pockets in importin-β communicate with each other, such that the interaction of one binding pocket with an FG motif induces a change in importin-β conformation and facilitates the dissociation of other binding pockets from other FG motifs.
Recently, the crystal structure of Kap95p and Nup1p (yeast homolog of Nup153) fragments demonstrated that not only the pockets within HEAT 5–6 and 6–7 but also the pocket within HEAT 7–8 interact with a phenylalanine residue (17). Molecular dynamics as well as our unpublished result from molecular simulation also suggested that the FG motifs could bind to some of the outer clefts of importin-β composed of two adjacent HEAT repeats (“pocket”) (34). Thus, it is possible that importin-β contains other FG-binding pockets. These multiple binding pockets with heterogeneous binding properties to different Nups, together with the effect of RanGTP, may be involved in the directional transport.
Materials and Methods
DNA Constructs.
The plasmid encoding His-tagged human Ran was a kind gift from T. Horigome at Niigata University. The plasmid encoding GST-tagged mouse importin-β was described in a previous study (35). The cDNAs of importin-β amino- and carboxyl-terminal fragments (residues 1–446 and 447–876, respectively), the amino terminal region of rat Nup62 (residues 1–178) and human Nup153-carboxyl-terminal region (residues 895-1475) were amplified by PCR from the full-length cDNA clones (the cDNA clones of rat Nup62 and human Nup153 were kind gifts from K. Ullman at Utah University). The amplified PCR fragments were subcloned into the expression vector pGEX-6P (GE Healthcare) with appropriate restriction enzymes. Site-directed mutagenesis of importin-β (I178A, F217A, Y255A, I263R, L612D, and F688A) was performed by using the GeneTailor Site-Directed Mutagenesis system (Invitrogen) in accordance with the manufacturer's instructions.
Expression and Purification of Recombinant Proteins.
GST-fused proteins were expressed and purified as described (22). The GST portion of the fusion proteins were cleaved off with an appropriate protease (thrombin from Sigma, PreScision protease from GE Healthcare), if necessary. Nup62-N was further purified by gel filtration chromatography (Superdex 75 HR; GE Healthcare) equilibrated with phosphate buffer solution. Expression and purification of RanGTP are described in SI Text. The purified protein was finally dialyzed against a transport buffer (20 mM Hepes, 110 mM CH3COOK, 5 mM CH3COONa, 2 mM (CH3COO)2Mg, 0.5 mM EGTA, 2 mM DTT, 1 mM PMSF) and concentrated by Centricon (Millipore) if necessary.
Single-Molecule Force Measurement by AFM.
A clean glass slide (76 × 26 mm) was treated with 3-aminopropyltriethoxysilane (APTES) (Sigma) as described (22), and purified Nup153-C (10 nM) was mixed with 1-Ethyl-3[3-dimethylamino-propyl]carbodiimide hydrochloride (EDC) (500 ng/ml) and immediately dropped onto the amino-functionalized glass slide, and then incubated for 2 h at room temperature. For Nup62, purified Nup62-N (30 nM) was dropped onto the nonfunctionalized glass slide and then incubated at 4°C overnight. The cover glass was washed with nuclear isolation medium (NIM; 10 mM NaCl, 90 mM KCl, 2 mM MgCl2, 1.1 mM EGTA, 10 mM Hepes-KOH) and immediately used in the experiment.
The glutathione-coupled cantilever was prepared as described (22), incubated with purified GST-fused importin-β at 4°C for 30 min, and then washed with the NIM three times. Force measurement was performed with a Molecular Force Probe 3D (MFP-3D; Asylum Research). A modified cantilever with a spring constant of 0.02 N/m (OMCL TR-400 PSA; Olympus) was routinely used. The force measurement was performed in NIM. All of the measurements were performed at a constant loading rate (2000 pN/s). The trigger channel was set for 20 nm. The approach-retract movement was repeated from 150 to 200 times with a single cantilever, and all of the measurements were completed within 1 h at room temperature. In the experiment using RanGTP, the GST-fusion protein-attached cantilever was preincubated with NIM containing 0.2 μM RanGTP for 20 min at room temperature, and then the force measurement was performed in the same solution. In the experiment using RanGppNHp, the cantilever was incubated for 20 min at room temperature in NIM containing 0.2 μM RanGppNHp and 0.5 mg/ml BSA. After the cantilever was washed several times with NIM, the force measurement was performed in the NIM without RanGppNHp. The obtained data were analyzed with the software accompanying the AFM imaging module (Asylum Research). The force curves were fitted by a worm-like-chain model of the PEG molecule (36), and the force curves that fit to the model were used for the analyses. When multiple rupture events were observed in a single force curve, only the last event was used for the analyses. More detailed explanation about the force measurement with the NPC is described in SI Text.
Preparation of the NE from Xenopus Oocyte.
The NE of a Xenopus oocyte was prepared as described (37), except that the NE was spread on a glass surface coated with poly-l-lysine. In brief, full-grown oocytes were obtained from an adult female Xenopus laevis and a nucleus (germinal vesicle) was isolated from each oocyte with microneedles in NIM. The NE was separated from chromatin, spread on a glass surface, and dehydrated in air after repeated washing with pure water. The NE was then rehydrated in NIM and immediately used for AFM imaging and force measurement. The images were taken in contact mode by a 100-μm-long cantilever with a spring constant of 6 pN/nm (Bio-Lever; Olympus) at a scan rate of 0.5–1 Hz.
Structural Comparison Analysis of Importin-β.
The 3D structures of Kap95p with or without RanGTP were taken from the National Center for Biotechnology Information Entrez Structure database (PDB ID codes 2BKU and 2BPT, respectively). Homology modeling of the 3D structure of mouse importin-β was performed with Prime (Schrödinger) by providing the amino acid sequence of mouse importin-β and the 3D structures of Kap95p. The structural alignments of GTP-bound and unbound forms of mouse importin-β were carried out by DaliLite Pairwise comparison (EMBL-EBI).
Supplementary Material
Acknowledgments.
This work was supported by a Japanese Ministry of Education, Culture, Sports, Science and Technology Grant-in-Aid for Scientific Research on Priority Areas (to Y.Y., S.H.Y., and K.T.), a Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Basic Research (A) (to K.T.), for Young Scientists (A) (to S.H.Y.), and for JSPS Fellows (to S.O.) and the Japan Science and Technology Agency (Core Research for Evolutional Science and Technology) (to S.H.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802647105/DCSupplemental.
References
- 1.Vasu SK, Forbes DJ. Nuclear pores and nuclear assembly. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 5.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 7.Allen NP, Huang L, Burlingame A, Rexach M. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem. 2001;276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- 8.Mattaj IW, Englmeier L. Nucleocytoplasmic transport: The soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 9.Jakel S, Gorlich D. Importin-beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 11.Lee SJ, et al. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- 12.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 13.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weis K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 15.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 16.Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J Biol Chem. 2002;277:50597–50606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- 17.Liu SM, Stewart M. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J Mol Biol. 2005;349:515–525. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Structural view of the Ran-Importin-beta interaction at 23 A resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 19.Fukuhara N, Fernandez E, Ebert J, Conti E, Svergun D. Conformational variability of nucleo-cytoplasmic transport factors. J Biol Chem. 2004;279:2176–2181. doi: 10.1074/jbc.M309112200. [DOI] [PubMed] [Google Scholar]
- 20.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 21.Bednenko J, Cingolani G, Gerace L. Importin-beta contains a COOH-terminal nucleoporin binding region important for nuclear transport. J Cell Biol. 2003;162:391–401. doi: 10.1083/jcb.200303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimura SH, Takahashi H, Otsuka S, Takeyasu K. Development of glutathione-coupled cantilever for the single-molecule force measurement by scanning force microscopy. FEBS Lett. 2006;580:3961–3965. doi: 10.1016/j.febslet.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 23.Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: From molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- 24.Fahrenkrog B, et al. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol. 2002;140:254–267. doi: 10.1016/s1047-8477(02)00524-5. [DOI] [PubMed] [Google Scholar]
- 25.Gilchrist D, Mykytka B, Rexach M. Accelerating the rate of disassembly of karyopherin. cargo complexes. J Biol Chem. 2002;277:18161–18172. doi: 10.1074/jbc.M112306200. [DOI] [PubMed] [Google Scholar]
- 26.Stoffler D, Goldie KN, Feja B, Aebi U. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J Mol Biol. 1999;287:741–752. doi: 10.1006/jmbi.1999.2637. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama N, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 28.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kose S, Imamoto N, Tachibana T, Yoshida M, Yoneda Y. β-subunit of nuclear pore-targeting complex (importin-β) can be exported from the nucleus in a Ran-independent manner. J Biol Chem. 1999;274:3946–3952. doi: 10.1074/jbc.274.7.3946. [DOI] [PubMed] [Google Scholar]
- 31.Cushman I, Palzkill T, Moore MS. Using peptide arrays to define nuclear carrier binding sites on nucleoporins. Methods. 2006;39:329–341. doi: 10.1016/j.ymeth.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Gelles J, Musser SM. Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA. 2004;101:12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isgro TA, Schulten K. Binding dynamics of isolated nucleoporin repeat regions to importin-β. Structure (London) 2005;13:1869–1879. doi: 10.1016/j.str.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Imamoto N, et al. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- 36.Kienberger F, et al. Static and dynamic properties of single poly(ethylene glycol) molecules investigated by force spectroscopy. Single Mol. 2000;1:123–128. [Google Scholar]
- 37.Kramer A, Ludwig Y, Shahin V, Oberleithner H. A pathway separate from the central channel through the nuclear pore complex for inorganic ions and small macromolecules. J Biol Chem. 2007;282:31437–31443. doi: 10.1074/jbc.M703720200. [DOI] [PubMed] [Google Scholar]
- 38.Loeb JD, Davis LI, Fink GR. NUP2, a novel yeast nucleoporin, has functional overlap with other proteins of the nuclear pore complex. Mol Biol Cell. 1993;4:209–222. doi: 10.1091/mbc.4.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yaseen NR, Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci USA. 1999;96:5516–5521. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.