Abstract
Stalled ribosomes in bacteria are rescued by the tmRNA system. In this process, the nascent polypeptide is modified by the addition of a short C-terminal sequence called the ssrA tag, which is encoded by tmRNA and allows normal termination and release of ribosomal subunits. In most bacteria, ssrA-tagged proteins are degraded by the AAA+ protease, ClpXP. However, in bacterial species of the genus Mycoplasma, genes for ClpXP and many other proteins were lost through reductive evolution. Interestingly, Mycoplasma ssrA tag sequences are very different from the tags in other bacteria. We report that ssrA-tagged proteins in Mesoplasma florum, a Mycoplasma species, are efficiently recognized and degraded by the AAA+ Lon protease. Thus, retaining degradation of ssrA-tagged translation products was apparently important enough during speciation of Mycoplasma to drive adaptation of the ssrA tag to a different protease. These results emphasize the importance of coupling proteolysis with tmRNA-mediated tagging and ribosome rescue.
Keywords: AAA+ protease, Lon, reductive evolution, ClpX
Stalled ribosomes in bacteria are rescued by tmRNA, the product of the ssrA gene, in a process sometimes called trans-translation (1–4). When ribosomes reach the end of a mRNA lacking a stop codon, protein synthesis ceases, and release factors cannot be recruited to allow disassembly. These ribosomes are eventually rescued by tmRNA, which functions initially as an alanyl-tRNA and next as a surrogate messenger RNA to allow resumption of translation. A stop codon at the end of the tmRNA open reading frame allows normal termination of translation, release of the polypeptide, and recycling of the ribosomal subunits for new rounds of protein synthesis.
As a result of the tagging and ribosome rescue process, polypeptides liberated by the tmRNA system have an ssrA tag at the C terminus. In Escherichia coli, this tag consists of 11 residues with a C-terminal LAA-coo− that targets attached proteins for degradation by ClpXP, an AAA+ protease (1, 5–7). AAA+ proteases are responsible for most intracellular proteolysis and can harness the energy of ATP hydrolysis to degrade native and denatured proteins (for review, see ref. 8). Indeed, ClpXP is able to degrade substrates with high mechanical stability, ensuring elimination of ssrA-tagged proteins, regardless of their folding state. Other AAA+ proteases (ClpAP, FtsH, and Lon) and a non-AAA+ periplasmic protease Tsp also degrade ssrA-tagged proteins under some conditions in E. coli, but ClpXP is responsible for most degradation of ssrA-tagged substrates and is likely to serve the same role in most bacteria (6, 9–14).
Bacteria of the genus Mycoplasma (class Mollicutes) comprise a large group of nonmotile bacteria, characterized by the lack of a cell wall and by small genomes (15). Mycoplasma branched from Gram-positive bacteria by multiple rounds of reductive evolution to reach a genome size of 0.45–1.35 Mbp and are thought to be the smallest self-replicating organisms (16). During genome minimization, Mycoplasma discarded many genes and became largely parasitic organisms that rely on their hosts for many nutrients. Nevertheless, Mycoplasma retained the tmRNA tagging and ribosome rescue system. Interestingly, however, the ssrA tags encoded by the tmRNA molecules in most Mycoplasma are very different from those found in other bacteria (Table 1). Moreover, most Mycoplasma genomes encode only two AAA+ proteases, Lon and FtsH, and have lost the genes for ClpXP, ClpAP, HslUV, and Tsp. In the absence of ClpXP, it is possible that the tmRNA system in Mycoplasma is uncoupled from proteolysis. Alternatively, the unusual Mycoplasma ssrA tag could serve as a degradation signal for the endogenous Lon or FtsH proteases.
Table 1.
ssrA-tag sequences and AAA+ proteases in different bacteria
| Type | Bacterium | ClpXP | ClpAP | HslUV | Lon | FtsH | ssrA-tag sequence |
|---|---|---|---|---|---|---|---|
| Gram-negative | |||||||
| α-proteobacteria | C. crescentus | ● | ● | ● | ● | ● | AANDNFAEEFAVAA |
| β-proteobacteria | N. gonorrhoeae | ● | ● | – | ● | ● | AANDETYALAA |
| γ-proteobacteria | E. coli | ● | ● | ● | ● | ● | AANDENYALAA |
| δ-proteobacteria | M. xanthus | ● | ● | ● | ● | ● | AANDNVELALAA |
| ε-proteobacteria | H. pylori | ● | ● | ● | ● | ● | AVNNTDYAPAYAKAA |
| Gram-positive | |||||||
| Actinobacteria | M. tuberculosis | ● | C | – | – | ● | AADSHQRDYALAA |
| Firmcutes/Clostridia | C. botulinum | ● | C | — | ● | ● | AANDNFALAA |
| Firmcutes/Bacilli | B. subtilis | ● | C | ● | ● | ● | AGKTNSFNQNVALAA |
| Firmcutes/Lactobacilli | S. pyogenes | ● | C | – | ● | ● | AAKNTNSYALAA |
| Mollicutes | |||||||
| Mycoplasma | M. florum | – | – | – | ● | ● | AANKNEENTNEVPTFMLNAGQANYAFA |
| Mycoplasma | U. parvum | – | – | – | ● | ● | AAENKKSSEVELNPAFMASATNANYAFAY |
| Mycoplasma | M. genitalium | – | – | – | ● | ● | ADKENNEVLVDPNLIINQQASVNFAFA |
| Mycoplasma | M. pneumoniae | – | – | – | ● | ● | ADKNNDEVLVDPMLIANQQASINYAFA |
| Mycoplasma | P. asteris | – | – | – | ● | ● | AGNNKQTVTNTQDFAGQTPVYQMNFANSFSSQLAFA |
| Mycoplasma | E. dolichum | – | C | – | ● | ● | AGKTKFANIFGANQSVAFAA |
| Other | |||||||
| Cyanobacteria | P. marinus | ● | C | – | – | ● | AANKIVSFSRQTAPVAA |
| Aquificae | A. aeolicus | ● | C | ● | ● | ● | AAPEAELALAA |
| Thermotogae | T. maritima | ● | C | ● | ● | ● | AANEPVAVAA |
| Deinococcus | D. radiodurans | ● | ● | – | ● | ● | AGNQNYALAA |
●, bacterium contains enzyme; –, bacterium does not contain enzyme; C, bacterium contains ClpCP, a relative of ClpAP. ClpCP does not appear to degrade ssrA-tagged proteins (10).
Because Lon is cytoplasmic, whereas FtsH is membrane-bound, it seemed most likely to us that Lon plays a major role in degrading ssrA-tagged proteins in Mycoplasma. To test this idea, we focused on the ssrA tag sequence and the Lon protease from Mesoplasma florum, a nonpathogenic and nonparasitic Mycoplasma with a genome size of 793 kb. M. florum shares many characteristics with its pathogenic cousins, but it can be cultured without special safety precautions. Here, we show that the ssrA tag sequence of M. florum (mf-ssrA) is efficiently recognized by the M. florum Lon protease (mf-Lon). Appending this tag to the C terminus of native or denatured proteins resulted in their rapid proteolysis by mf-Lon. Furthermore, mf-Lon did not degrade proteins bearing the E. coli ssrA tag (ec-ssrA), and E. coli Lon (ec-Lon) did not efficiently degrade proteins bearing the mf-ssrA tag. These results indicate that gene loss in M. florum forced coevolution of both the ssrA tag and Lon protease to permit efficient and specific degradation, supporting an important role for degradation of ssrA-tagged proteins in the evolutionary fitness of bacteria.
Results
Unusual Properties of ssrA Tags in Mycoplasma.
The ssrA tag sequences of diverse bacterial species usually exhibit similarity, especially at their C terminus, which is important for interaction with ClpXP (Table 1; refs. 6 and 7). This observation, together with experiments in Gram-negative E. coli and Gram-positive Bacillus subtilis (6, 10, 11, 14), suggests that ClpXP is the key protease responsible for degradation of ssrA-tagged substrates in most bacteria. Interestingly, the ssrA tags in most Mycoplasma species are much longer than in other groups of bacteria and do not terminate with the normal LAA-coo− ClpX recognition motif (Table 1). For example, in E. coli and B. subtilis, the ssrA tag sequences are 11 and 14 residues long and end with NYALAA-coo− and NVALAA-coo−, respectively. By contrast, the M. florum ssrA tag consists of 27 aa and terminates with ANYAFA-coo−. The presence of several aromatic residues in this region of the M. florum tag is reminiscent of sequences that target certain substrates to ec-Lon (13, 17).
Activity of mf-Lon in E. coli.
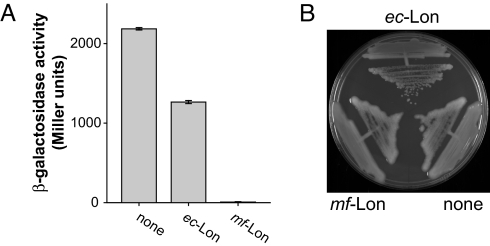
We cloned the lon gene from genomic M. florum DNA, replaced its TGA codons, which encode tryptophans in M. florum but are stop codons in E. coli, with TGG codons, and found that mf-Lon could be expressed in E. coli (data not shown). We then coexpressed plasmid-encoded mf-Lon with a variant of β-galactosidase bearing a C-terminal mf-ssrA tag in an E. coli strain MC4100, which also contained the normal set of AAA+ proteases. Almost no β-galactosidase activity was detected in these cells (Fig. 1A). By contrast, high β-galactosidase activity was observed when ec-Lon was substituted for mf-Lon in the plasmid vector. β-Galactosidase activity was highest when a mock vector was used (Fig. 1A). These results suggested that the mf-Lon protease recognizes and degrades the mf-ssrA-tagged protein much better than do the ec-Lon, ClpXP, ClpAP, HslU, or FtsH enzymes.
Fig. 1.
Phenotypes of mf-Lon expression in E. coli. (A) E. coli strain MC4100 was transformed with a plasmid expressing an IPTG-inducible β-galactosidase-mf-ssrA fusion protein and a second plasmid expressing either ec-Lon, mf-Lon, or neither enzyme. Intracellular β-galactosidase-mf-ssrA was assayed by enzymatic cleavage of X-Gal as described in Materials and Methods. Values are averages (±1 SD; n = 5). (B) Complementation of the mucoid phenotype of a Δlon strain by ec-Lon but not mf-Lon. E. coli strain JT4000 (MC4100 Δlon) was transformed with plasmids expressing ec-Lon, mf-Lon, or a mock vector, as indicated. Cells were plated on minimal medium (45) agar plates with glycerol (0.4%) as a carbon source and chloramphenicol (10 μg/ml). In both panels, ec-Lon and mf-Lon were expressed at basal levels under control of an l-arabinose promoter (PBAD) without added l-arabinose. Arabinose induction of the expression of either Lon enzyme resulted in growth arrest.
To test whether the mf-Lon enzyme could recognize a substrate normally degraded by ec-Lon, we performed a complementation assay by using an E. coli lon-null mutant, which exhibits a mucoid phenotype because ec-Lon normally degrades RcsA, a transcriptional activator of capsule synthesis genes (18). As expected, expression of ec-Lon complemented the mucoid phenotype of the lon-null strain (Fig. 1B). Importantly, however, expression of mf-Lon failed to suppress the mucoid phenotype (Fig. 1B). This result provides additional evidence that the specificities of the ec-Lon and mf-Lon proteases differ.
Degradation in Vitro.
To provide evidence for direct recognition, we purified mf-Lon and assayed degradation in vitro of model proteins bearing the mf-ssrA tag. As potential substrates, we used mf-ssrA-tagged variants of the I27 domain of human titin, which has been used in previous studies of degradation by AAA+ proteases (13, 19, 20). In initial studies, we denatured titin-I27-mf-ssrA by carboxylmethylation of its two buried cysteine residues (19) to ensure that degradation would not depend on the unfolding power of the protease. As assayed by SDS/PAGE, this substrate was degraded efficiently by mf-Lon but poorly by ec-Lon (Fig. 2A). As additional specificity controls, we assayed degradation of carboxylmethylated titin-I27 with no degradation tag or with an E. coli ssrA tag. Neither protein was degraded efficiently by mf-Lon. Thus, degradation is highly specific and requires both the mf-Lon protease and the mf-ssrA tag of the substrate.
Fig. 2.
Degradation activity and properties of mf-Lon. (A) SDS/PAGE assays of degradation of carboxylmethylated titin-I27 (5 μM) with different tags by mf-Lon or ec-Lon (150 nM hexamer). (B) Degradation of fluorescein-labeled titin-I27-mf-ssrA (5 μM) by mf-Lon (150 nM hexamer) required ATP and was not observed with ADP, without nucleotide, or in the absence of enzyme. Proteolysis resulted in an increase in fluorescence. (C) Rates at different temperatures for degradation of carboxylmethylated 35S-labeled titin-I27-mf-ssrA (3 μM) by mf-Lon (30 nM hexamer) were determined at pH 7.5 by release of acid-soluble peptides. (D) Rates at different pH values for degradation of carboxylmethylated 35S-labeled titin-I27-mf-ssrA (3 μM) by mf-Lon (30 nM hexamer) were determined at 30°C. The maximal degradation rate is higher than in C because the sodium phosphate buffer (50 mM) used in the temperature experiments was slightly inhibitory. (E) Michaelis–Menten plots. Steady-state rates of degradation of an unfolded substrate (carboxylmethylated 35S-titin-mf-ssrA) or a native substrate (35S-titin-mf-ssrA) by mf-Lon (10 nM hexamer) were assayed by release of acid-soluble peptides and plotted as a function of substrate concentration. The solid curves are fits (R2 ≥ 0.99) to the Hill form of the Michaelis–Menten equation (V = Vmax·[S]n/(Kmn + [S]n). (F) The titin-I27 protein (4 μM; filled circles) and titin-I27-mf-ssrA protein (4 μM; open circles) had the same thermal stability, as assayed by changes in CD ellipticity at 228 nm. Ellipticity data were fitted to a two-state model by nonlinear regression (46). (Inset) CD spectra at 25°C show that both titin variants (40 μM) are natively folded. The differences in the two spectra result from the contribution of the long unstructured mf-ssrA tag to the CD signal.
The degradation assays shown in Fig. 2A contained ATP. To test whether mf-Lon degradation required this nucleotide, we used a fluorescent unfolded variant of titin-I27-mf-ssrA, which allowed continuous assay of the degradation rate. As shown in Fig. 2B, mf-Lon degraded this fluorescent substrate in the presence of ATP but not in the presence of ADP or without nucleotide. E. coli ClpXP did not degrade the unfolded fluorescent variant of titin-I27-mf-ssrA under any conditions (data not shown). To establish the effect of environmental conditions on mf-Lon activity, we measured degradation of carboxylmethylated titin-I27-mf-ssrA at different temperatures and pH values. Maximal activity was observed at 30°C between pH 6.5 and 8.5 (Fig. 2 C and D). M. florum, which was isolated from a lemon tree flower, grows at temperatures that rarely exceed 30°C, and the observed temperature dependence is therefore consistent with its normal physiology.
mf-Lon Is a Fast and Powerful Enzyme.
To measure kinetic parameters for degradation by mf-Lon, steady-state rates were determined at different concentrations of protein substrates. For the unfolded substrate, carboxylmethylated titin-I27-mf-ssrA, Michaelis–Menten analysis gave a Km of 3.5 ± 0.3 μM (Fig. 2E). This value is similar to the Km of ClpXP for substrates bearing the E. coli ssrA tag (19, 21–23). Vmax for the mf-Lon degradation reaction was surprisingly fast (11.5 ± 0.3 min−1 Lon6−1), approximately three times faster than either ec-Lon or ec-ClpXP degrades unfolded titin (13, 19).
To determine whether mf-Lon could denature and degrade a stably folded substrate, we assayed proteolysis of native titin-I27-mf-ssrA. The titin-I27 protein is remarkably resistant to mechanical unfolding and has been used as a stringent test of the unfolding power of AAA+ proteases (13, 19, 20, 24, 25). The mf-Lon enzyme degraded native titin-I27-mf-ssrA efficiently, with a Km of 3.7 ± 0.4 μM and a Vmax of 4.3 ± 0.1 min−1 enz−1 (Fig. 2E). Vmax for degradation of the native substrate was lower than for the unfolded variant, indicating that unfolding is the rate-limiting step in degradation.
Interestingly, mf-Lon degraded native titin-I27-mf-ssrA ≈8-fold faster than ec-Lon degraded a similar titin-I27 substrate with a different degradation tag (13). This result was not caused by a destabilizing effect of the mf-ssrA tag on the thermodynamic stability of native titin. As measured by changes in CD ellipticity, untagged titin-I27 and titin-I27-mf-ssrA had almost identical thermal stabilities (Fig. 2F). These results suggest that mf-Lon is a more powerful unfoldase than ec-Lon. Moreover, mf-Lon degraded native titin-I27-mf-ssrA ≈16-fold faster than ec-ClpXP degraded titin-I27-ec-ssrA (19).
Peptide Degradation and Recognition Determinants.
To test whether mf-Lon recognized the mf-ssrA tag in the absence of an attached protein substrate, we synthesized a peptide corresponding approximately to the C-terminal half of the tag sequence (the full-length tag could not be synthesized easily). In this peptide, a para-aminobenzoic acid (PABA) fluorophore was placed at the N terminus, and a nitrotyrosine (nY), which serves as a quencher, was inserted at position 5. The peptide sequence was PABA-TFML-nY-NAGQANYAFA-coo− and is hereafter called FQ14-mf-ssrA. Proteolytic cleavage between the fluorophore and quencher results in enhanced fluorescence. By this assay, the FQ14-mf-ssrA peptide was degraded rapidly by mf-Lon in the presence of ATP (Fig. 3A). Degradation did not occur in the presence of ADP and was very slow without nucleotide (Fig. 3A). We conclude that the C-terminal half of the mf-ssrA tag is sufficient for recognition by mf-Lon.
Fig. 3.
Degradation of an mf-ssrA peptide by mf-Lon. (A) Degradation of the FQ14-mf-ssrA peptide (5 μM) by mf-Lon (0.15 μM hexamer) was assayed by increased fluorescence in the presence of 2 mM ATP, 2 mM ADP, without nucleotide, or without enzyme in the presence of 2 mM ATP. (B) WebLogo representation (http://weblogo.berkeley.edu/logo.cgi; ref. 47) of ssrA tag sequences from Mycoplasma and from other Gram-positive bacteria. Sequences of ssrA tags were obtained from the tmRNA website (http://www.indiana.edu/∼tmrna/; ref. 48) or from genomic sequences by using the program ARAGORN (49). (C) The Y→D/F→D substitutions of the aromatic residues in the C-terminal pentapeptide of FQ14-mf-ssrA (5 μM) slowed degradation by mf-Lon (0.15 μM hexamer) dramatically. The N→D substitution in the C-terminal FQ14-mf-ssrA pentapeptide had little effect on degradation under the same conditions.
In Mycoplasma ssrA tags, the region of highest information content is near the C terminus, where most sequences share the consensus NΦAΦA (Φ = F/Y/L) (Fig. 3B). The C terminus of the M. florum ssrA tag is NYAFA-coo−. When we replaced this C-terminal sequence with NDADA-coo− in an FQ14-mf-ssrA variant, the resulting peptide was degraded poorly by mf-Lon (Fig. 3C). This result suggested that the substituted aromatic residues play important roles in recognition of the mf-ssrA tag by mf-Lon. Aromatic residues have also been implicated in peptide recognition by ec-Lon (13). Changing the NYAFA sequence of FQ14-mf-ssrA to DYAFA-coo− did not slow degradation by mf-Lon significantly (Fig. 3B).
Discussion
Evolutionary Implications of the Proteolytic Specificity Switch.
The tmRNA system is encoded in all fully sequenced bacterial genomes, which currently number >700. The apparently universal conservation of this system points to an important biological role. However, the entire tmRNA system is dispensable for viability in many bacteria (for review, see refs. 2–4). Moreover, although tmRNA is genetically essential in Neisseria gonorrhoeae, degradation of the protein products of tmRNA tagging is not required for viability (26). Nevertheless, our results indicate that degradation of ssrA-tagged proteins is important for the fitness of bacteria. First, ssrA tagging was retained in M. florum, despite extreme evolutionary events involving massive gene loss. Second, to maintain degradation in the absence of ClpXP, the ssrA tag of M. florum had to adapt to allow recognition by a different protease. This switch in proteolytic specificity presumably required changes both in the tmRNA-encoded degradation tag and in the Lon protease.
The switch from ClpXP to Lon degradation of ssrA-tagged proteins represents a case of nonorthologous gene displacement (27). It is plausible that the initial event in the proteolytic specificity switch in M. florum was loss of ClpXP, which is principally responsible for degrading ssrA-tagged proteins in most other bacteria. However, the selective force driving coevolution of Lon and the ssrA tag to allow efficient degradation must have been a fitness advantage for mutants with improved ability to degrade the protein products of tmRNA tagging. Because ssrA-tagged proteins are generally incomplete proteins, they are typically unfolded and nonfunctional (28, 29). Thus, degrading these tagged proteins should allow productive recycling of their constituent amino acids to prevent deleterious effects caused by aggregation or other factors. Presumably, the same fitness advantages led to the initial evolution of ClpXP degradation of ssrA-tagged proteins. Indeed, many bacteria contain adaptor proteins that enhance ClpXP degradation of ssrA-tagged proteins and/or that redirect degradation of ssrA-tagged proteins from ClpAP to ClpXP (7, 30–33). Some of these adaptors recognize a portion of the ssrA tag, and it is possible that the long ssrA tags of Mycoplasma also serve as docking sites for adaptor proteins.
How complicated was the switch from ClpXP to Lon in proteolysis of ssrA-tagged substrates in the ancestor of Mycoplasma? E. coli Lon recognizes substrates with ec-ssrA tags weakly (12, 13). If the same were true for Lon in the bacteria from which M. florum evolved, then this would provide a low level of starting activity after loss of ClpXP. Moreover, the C-terminal sequences of Mycoplasma tags are sufficiently similar to those in other Gram-positive bacteria (Table 1; Fig. 3B) that only a few mutations would be required to convert a tag from the major Gram-positive group into a tag with the Mycoplasma consensus and thus presumably to improve Lon affinity. Our results show that some residues in the C-terminal consensus region of the M. florum ssrA tag are important determinants of mf-Lon recognition.
Lon Is a Powerful Protein Unfoldase.
Lon plays a major role in the degradation of misfolded and damaged proteins in bacteria and in the organelles of eukaryotes (34). As a consequence, its ability to act as a robust protein unfoldase has generally been underappreciated. Using a tagged native substrate (titin-I27) that is very stable to mechanical denaturation, we found that mf-Lon appears to be a more powerful enzyme than either ec-Lon or ec-ClpXP. Under conditions of substrate saturation, each of these AAA+ proteases degraded tagged variants of native titin-I27 more slowly than the tagged unfolded protein. Thus, in each instance, enzymatic unfolding of the native substrate appears to be the rate-limiting step in the degradation reaction. However, mf-Lon degraded a tagged variant of titin-I27 ≈8-fold more rapidly than ec-Lon and nearly 16-fold faster than ec-ClpXP. Moreover, mf-Lon also degraded unfolded titin-I27 substantially faster than either ec-Lon or ec-ClpXP. The mf-Lon enzyme may have evolved into a faster and more powerful enzyme to compensate for the absence of ClpAP, ClpXP, and HslUV in M. florum. The only remaining AAA+ protease in this organism is FtsH, and its E. coli ortholog has been shown to have a weak unfoldase activity (35).
We anticipate that mf-Lon and other Mycoplasma Lon enzymes will play numerous biological roles both in regulation and in maintaining protein quality control. In E. coli and Salmonella, for example, lon mutants exhibit multiple defects, including difficulties in cell division, excessive capsule synthesis, poor survival after DNA damage, failure to degrade damaged proteins, and reduced virulence (34, 36–38).
An Experimental Windfall.
Although Lon was one of the first ATP-dependent proteases to be purified and studied, mechanistic studies of other AAA+ proteases subsequently progressed far more rapidly. A scarcity of soluble well behaved Lon substrates for biochemical experiments and structural studies has contributed to sluggish progress in understanding how this key enzyme recognizes, unfolds, translocates, and degrades substrates. By contrast, mechanistic studies of other AAA+ proteases, like ClpXP and ClpAP, were greatly accelerated because fusion of the ec-ssrA tag to any well behaved protein resulted in a good substrate, allowing steady-state kinetic constants to be determined, the effects of substrate stability and dynamics to be probed, and the design of novel substrates to address specific experimental questions (6, 19, 21–23, 35, 39–42). Our finding of a specific and tight interaction between mf-Lon and the mf-ssrA tag provides an exceptional opportunity to deepen understanding of Lon structure, function, and mechanism.
Materials and Methods
Plasmids.
The LacZ-mf-ssrA protein contained the entire mf-ssrA tag fused to the C terminus of the E. coli LacZ protein and was cloned under control of the lacZ promoter into pSH21, which constitutively expresses the LacI repressor. After changing all of the TGA codons to TGG codons, the gene encoding mf-Lon was cloned into pBAD33. The ec-Lon enzyme was expressed from pBAD33-lon (43). Variants of the human titin-I27 domain were expressed from pSH21, under transcriptional control of a T7 promoter.
Proteins and Peptides.
The ec-Lon protease was purified as described in ref. 13. E. coli ClpXP was a gift from Mary Lee (Massachusetts Institute of Technology). For purification of mf-Lon, a 2-liter culture of E. coli strain ER2566 (New England Biolabs) carrying plasmid pBAD33-mf-lon was grown at 37°C in 2XYT medium supplemented with chloramphenicol (20 μg/ml). At an A600 of 1.0, l-arabinose was added (0.2%, wt/vol), and the culture was grown for an additional 3 h before harvesting cells and freezing them at −80°C. After thawing cells in 25 ml of cold MF buffer [25 mM Hepes (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM DTT], the cell suspension was lysed by using a French press. The lysate was centrifuged (18,000 × g, 30 min), and streptomycin sulfate was added to the supernatant to precipitate nucleic acids. After gentile agitation of the solution at 4 °C for 1 h, the solution was recentrifuged (18,000 × g, 30 min), and the supernatant was decanted and loaded onto a hydroxylapatite column equilibrated with MF buffer at room temperature. The column was washed with MF buffer and developed with a linear potassium phosphate (pH 7.5) gradient from 0 to 0.5 M. Fractions containing mf-Lon were pooled, concentrated with an Amicon concentrator with a 100-kDa cutoff, and loaded onto a 26/60 Sephacryl S300 gel filtration column (GE Healthcare) preequilibrated with MF buffer. Fractions containing mf-Lon at purity >90% were concentrated as described above, aliquoted, and kept frozen at −80°C.
The titin-I27-mf-ssrA protein was purified by using the Impact-CN system (New England Biolabs). The gene encoding titin-I27-mf-ssrA with an N-terminal His6 tag was cloned into pTYB1 and transformed into an E. coli strain ER2566. A culture was grown at 37°C in 2XYT medium supplemented with ampicillin (100 μg/ml). At an A600 of 1.0, IPTG was added (0.5 mM), and the culture was grown for an additional 2.5 h before harvesting cells and freezing them at −80°C. After thawing cells in 25 ml of cold Ni buffer [25 mM Hepes (pH 7.5), 500 mM NaCl, 20 mM imidazole] containing 0.02 mg/ml lysozyme, the cell suspension was lysed by using a French press. The lysate was centrifuged, and the supernatant was mixed in a 50-ml tube with 2 ml of Ni-nitrilotriacetic acid resin (Qiagen; prewashed with Ni buffer) and incubated at 4°C for 15 min. The resin was washed three times with 20 ml of Ni buffer, resuspended in 5 ml of buffer, transferred to a gravity column, and washed once with 10 ml of buffer. The protein was eluted with 3 ml of Ni buffer containing 250 mM imidazole, loaded onto a 10-ml chitin–agarose column, and cleavage, and purification was performed according to the Impact-CN system protocol. Carboxylmethylation and purification of all other titin-I27 variants were performed as described in ref. 19.
FQ14-mf-ssrA peptides were synthesized by the Massachusetts Institute of Technology Biopolymers Laboratory. Peptide concentrations were determined from absorbance at 381 nm (ε = 2,200 M−1 cm−1).
Biochemical and Biophysical Assays.
Unless noted, degradation assays using mf-Lon were performed at 30°C in MF buffer. Degradation assays using ec-Lon were carried out as described in ref. 13. Unless noted, degradation reactions contained ATP (2 mM) and an ATP-regeneration system composed of 20 mM phosphoenolpyruvate and 10 units/ml pyruvate kinase. For degradation of radioactive substrates, trichloroacetic acid precipitation was carried out as described in ref. 6.
β-Galactosidase assays were carried out based on described procedures (44). Cells were grown at 30°C to midlog phase in LB broth supplemented with ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), and IPTG (1 mM), and absorbance at 600 nm was measured. Cells (20 μl) were lysed by mixing with 80 μl of B-PER protein extraction reagents (Pierce) containing PMSF (1 mM) and incubating the solution for 10 min at room temperature. To start the β-galactosidase reaction, 670 μl of 100 mM sodium phosphate (pH 7.4), 10 mM KCl, 1 mM MgCl2, 1 mM DTT, and 1 mg/ml o-nitrophenyl β-d-galactoside were added. After incubation for 10 min at room temperature, 330 μl of 1 M Na2CO3 was added to stop the reaction, and the absorbance at 420 nm was measured. Miller units were calculated as: (1,000 · A420)/(A600 · culture volume · incubation time · 1.61).
CD spectra were taken at 1-nm intervals in a 1-mm path length cuvette on an AVIV 400 instrument [40 μM protein in 10 mM potassium phosphate (pH 7.6), 20 mM KCl]. For thermal denaturation monitored by CD, the protein concentration was 4 μM [10 mM potassium phosphate (pH 7.6), 100 mM KCl]. A 10-mm path length cuvette was used, the heating rate was 1°C/min, and ellipticity at 228 nM was averaged for 10 s after temperature equilibration.
Acknowledgments.
We thank E. Alm, B. Cezairliyan, J. Davis, T. Knight, M. Lee, and A. Martin (Massachusetts Institute of Technology), M. Cordes (University of Arizona, Tucson, AZ), and U. Gophna (Tel Aviv University, Tel Aviv, Israel), for materials, advice, and help. This work was supported by National Institutes of Health Grants AI-15706 and AI-16892.
Footnotes
The authors declare no conflict of interest.
References
- 1.Keiler KC, Waller PR, Sauer RT. Role of a peptide-tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 2.Withey JH, Friedman DI. A salvage pathway for protein structures: tmRNA and trans-translation. Annu Rev Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 3.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 4.Keiler KC. Biology of trans-translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- 5.Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J Biol Chem. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxyl-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn JM, et al. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci USA. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauer RT, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 10.Wiegert T, Schumann W. SsrA-mediated tagging in Bacillus subtilis. J Bacteriol. 2001;183:3885–3889. doi: 10.1128/JB.183.13.3885-3889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell CM, Grossman AD, Sauer RT. Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol. 2005;57:1750–1761. doi: 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- 12.Choy JS, Aung LL, Karzai AW. Lon protease degrades transfer-messenger RNA-tagged proteins. J Bacteriol. 2007;189:6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gur E, Sauer RT. Recognition of misfolded proteins by Lon, an AAA+ protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lies MA, Maurizi MR. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J Biol Chem. 2008;283:22918–22929. doi: 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fadiel A, Eichenbaum KD, El Semary N, Epperson B. Front Biosci. 2007;12:2020–2028. doi: 10.2741/2207. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez M, Frank EG, Levine AS, Woodgate R. Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: In vitro degradation and identification of residues required for proteolysis. Genes Dev. 1998;12:3889–3899. doi: 10.1101/gad.12.24.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres-Cabassa AS, Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 20.Burton RE, Baker TA, Sauer RT. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat Struct Mol Biol. 2005;12:245–251. doi: 10.1038/nsmb898. [DOI] [PubMed] [Google Scholar]
- 21.Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- 22.Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J Struct Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Politou AS, Thomas DJ, Pastore A. The folding and stability of titin immunoglobulin-like modules, with implications for the mechanism of elasticity. Biophys J. 1995;69:2601–2610. doi: 10.1016/S0006-3495(95)80131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: A comparison. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin EV. How many genes can make a cell: The minimal-gene-set concept. Annu Rev Genomics Hum Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roche ED, Sauer RT. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J Biol Chem. 2001;276:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- 29.Hong SJ, Lessner FH, Mahen EM, Keiler KC. Proteomic identification of tmRNA substrates. Proc Natl Acad Sci USA. 2007;104:17128–17133. doi: 10.1073/pnas.0707671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 31.Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- 32.Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci USA. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lessner FH, Venters BJ, Keiler KC. Proteolytic adaptor for transfer-messenger RNA-tagged proteins from α-proteobacteria. J Bacteriol. 2007;189:272–275. doi: 10.1128/JB.01387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsilibaris V, Maenhaut-Michel G, Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res Microbiol. 2006;157:701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Herman C, Thévenet D, Bouloc P, Walker GC, D'Ari R. Degradation of carboxyl-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizusawa S, Gottesman S. Protein degradation in Escherichia coli: The lon gene controls the stability of SulA protein. Proc Natl Acad Sci USA. 1983;80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goff SA, Goldberg AL. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- 38.Boddicker JD, Jones BD. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect Immun. 2004;72:2002–2013. doi: 10.1128/IAI.72.4.2002-2013.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 40.Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 42.Reid BG, Fenton WA, Horwich AL, Weber-Ban EU. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc Natl Acad Sci USA. 2001;98:3768–3772. doi: 10.1073/pnas.071043698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen SK, et al. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: Involvement of the yefM–yoeB toxin-antitoxin system. Mol Microbiol. 2004;51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 44.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 45.Davis BD, Mingioli ES. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayr LM, Landt O, Hahn U, Schmid FX. Stability and folding kinetics of ribonuclease T1 are strongly altered by the replacement of cis-proline 39 with alanine. J Mol Biol. 1993;231:897–912. doi: 10.1006/jmbi.1993.1336. [DOI] [PubMed] [Google Scholar]
- 47.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gueneau de Novoa P, Williams KP. The tmRNA website: Reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004;32:104–108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]