Abstract
The temporal phosphorylation of cell cycle-related proteins by cyclin-dependent kinases (Cdks) is critical for the correct order of cell cycle events. In budding yeast, CDC28 encodes the only Cdk and its association with various cyclins governs the temporal phosphorylation of Cdk substrates. S-phase Cdk substrates are phosphorylated earlier than mitotic Cdk substrates, which ensures the sequential order of DNA synthesis and mitosis. However, it remains unclear whether Cdk substrates are dephosphorylated in temporally distinct windows. Cdc14 is a conserved protein phosphatase responsible for the dephosphorylation of Cdk substrates. In budding yeast, FEAR (Cdc14 early anaphase release) and MEN (mitotic exit network) activate phosphatase Cdc14 by promoting its release from the nucleolus in early and late anaphase, respectively. Here, we show that the sequential Cdc14 release and the distinct degradation timing of different cyclins provides the molecular basis for the differential dephosphorylation windows of S-phase and mitotic cyclin substrates. Our data also indicate that FEAR-induced dephosphorylation of S-phase Cdk substrates facilitates anaphase progression, revealing an extra layer of mitotic regulation.
Keywords: Cdc14 phosphatase, spindle, Clb5, Clb2, anaphase
Among the 6 B-type cyclins in budding yeast, S-phase-expressed Clb5 is important for DNA synthesis, whereas mitotic cyclin, Clb2, appears during G2/M phases and plays a key role in mitosis. The association with different cyclins endows Cdk1 with substrate specificity, which contributes to the temporal phosphorylation of Cdk substrates. The phosphorylation efficiency of 150 Cdk substrates by Clb5-Cdk1 and Clb2-Cdk1 was compared using in vitro kinase assay (1). The study indicates that some Cdk substrates are phosphorylated more efficiently by Clb5-Cdk1 than by Clb2-Cdk1. Clb2-Cdk1 has been found to possess higher intrinsic kinase activity, allowing efficient phosphorylation of mitotic Cdk1 targets. One fundamental question that remains unanswered is whether Clb5 and Clb2 substrates are dephosphorylated with distinct windows.
Structural and kinetic data support a role for phosphatase Cdc14 in the preferential dephosphorylation of proteins modified by proline-directed kinases, such as Cdk1 (2). The regulation of Cdc14 in budding yeast is achieved through its subcellular localization. During most of the cell cycle, Cdc14 is sequestered within the nucleolus by associating with a nucleolar protein Net1/Cfi1 (3, 4). During late anaphase, the activation of the MEN pathway results in Cdc14 release from the nucleolus, which inactivates Clb2-Cdk1 (5). Before the activation of the MEN (mitotic exit network), yeast cells exhibit a brief FEAR (Cdc14 early anaphase release)-dependent release of Cdc14 during early anaphase (6). FEAR pathway promotes Cdc14 release by inactivating the protein phosphatase 2A (PP2ACdc55), which dephosphorylates Net1 and prevents Cdc14 release (7–9). Nevertheless, the biological significance of the two waves of Cdc14 release induced by FEAR and MEN remains unclear. Here, we present evidence indicating that FEAR-induced Cdc14 release during early anaphase and the degradation of S-phase cyclin Clb5 leads to the dephosphorylation of Clb5 substrates, which facilitates chromosome segregation.
Results
S and M Phase Cyclin Substrates Are Dephosphorylated by Cdc14 with Distinct Time Windows.
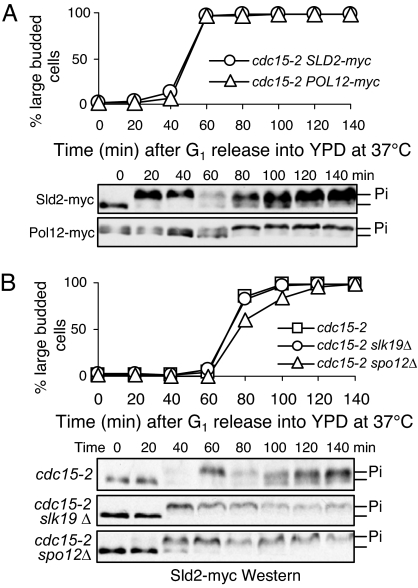
Sld2 is a Clb5-specific substrate and its phosphorylation is essential for DNA replication (1, 10). As an S-phase cyclin substrate, Sld2 phosphorylation is insensitive to the presence of Swe1, an inhibitory kinase for Clb2-Cdk1 (11, 12). In contrast, the phosphorylation of Pol12, the B subunit of DNA polymerase α, is blocked in the presence of Swe1, indicating that Pol12 might be a Clb2-Cdk1 substrate (13). Our in vitro kinase assay further confirmed that Pol12 is a preferred substrate of Clb2-Cdk1, but not Clb5-Cdk1 [supporting information (SI) Fig. S1A]. Consistently, clb5Δ strains exhibited compromised phosphorylation of Sld2, but not Pol12 (Fig. S1B). In addition, high levels of Sic1, the Cdk1 inhibitor, decreased Pol2 phosphorylation significantly (Fig. S1C). Therefore, Sld2 and Pol12 were used as the representatives of S and M-phase cyclin-specific substrates in this study. We first compared the temporal phosphorylation of Sld2 and Pol12 during cell cycle on the basis of band shift (10, 14). The phosphorylation of Sld2 occurred earlier than that of Pol12, consistent with the functional windows of Clb5 and Clb2. Interestingly, Sld2 also exhibited earlier dephosphorylation than Pol12. Sld2 became dephosphorylated 60 min after G1 release, whereas the dephosphorylation of Pol12 occurred at 120 min, demonstrating the distinct temporal windows for dephosphorylation of different Cdk substrates (Fig. 1A). Because phosphatase Cdc14 is believed to be responsible for the dephosphorylation of Cdk substrates (2), the modification of Sld2 and Pol12 was compared in WT and cdc14–1 mutant cells and their dephosphorylation was abolished in cdc14–1 cells incubated at 37°C (Fig. 1B). Altogether, S and M-phase cyclin substrates exhibit differential temporal windows of dephosphorylation, and the dephosphorylation depends on Cdc14.
Fig. 1.
Cdk1 substrates exhibit differential temporal dephosphorylation. (A) Cell cycle-regulated phosphorylation and dephosphorylation of Pol12 and Sld2. Cells with indicated genotypes were synchronized in G1 phase with α-factor and then released into YPD medium at 30°C. Cells were collected every 20 min to prepare protein samples. The protein phosphorylation was determined after Western blotting with anti-myc antibody on the basis of band shift. (B) The dephosphorylation of Sld2 and Pol12 depends on Cdc14. G1-synchronized cells with indicated genotype were released into YPD at 37°C. The phosphorylation of Sld2 and Pol12 was determined as described in A.
Because Sld2 exhibits earlier dephosphorylation, we reasoned that other Clb5 substrates might be dephosphorylated earlier as well. Ase1, Sli15, the other known Clb5-specific substrates (1), also exhibited earlier dephosphorylation than Pol12, the Clb2 substrate (Fig. S2). Although there is no evidence indicating that Ask1 is a Clb5-Cdk1 substrate, its phosphorylation during S-phase suggests that it could be phosphorylated by Clb5-Cdk1 (15). The dephosphorylation of Ask1 also occurred much earlier than Pol12, suggesting that Clb5 substrates are dephosphorylated earlier than Clb2 substrates.
The Different Role of MEN and FEAR in the Dephosphorylation of Sld2 and Pol12.
FEAR and MEN promote Cdc14 release during early and late anaphase, respectively. Because Cdc14 dephosphorylates Cdk substrates, we reasoned that cells lacking FEAR and MEN pathways should show defective dephosphorylation of Cdk substrates. Therefore, the dephosphorylation kinetics of Sld2 and Pol12 were first examined in cdc15–2 mutants, wherein the MEN is inactive when grown at 37°C (16). As shown in Fig. 1B, Sld2 and Pol12 became dephosphorylated after their phosphorylation in synchronized WT cells incubated at 37°C, but the dephosphorylation of Pol12 was totally blocked in cdc15–2 mutant. In contrast, Sld2 could still be dephosphorylated in cdc15–2 cells, even though the dephosphorylation was incomplete (Fig. 2A). As FEAR induces a brief Cdc14 release in cdc15–2 mutant cells (6), active FEAR might be responsible for Sld2 dephosphorylation. To test this idea, we examined Sld2 dephosphorylation in cells lacking both FEAR and MEN. Strikingly, deletion of either SLK19 or SPO12, which encode FEAR components (6), abolished Sld2 dephosphorylation completely in cdc15–2-arrested cells (Fig. 2B). Because Ask1 and Sli15 are likely Clb5 substrates, their dephosphorylation was also compared in cdc15–2 and cdc15–2 slk19Δ mutants. Like Sld2, Ask1 and Sli15 could be dephosphorylated in cdc15–2-arrested cells and deletion of SLK19 abolished the dephosphorylation (Fig. S3), indicating that FEAR-induced Cdc14 release promotes the dephosphorylation of Clb5 substrates but not those of Clb2. We therefore conclude that FEAR and MEN-induced sequential Cdc14 release contributes to the differential temporal dephosphorylation of S and M-phase Cdk substrates.
Fig. 2.
The dephosphorylation of Clb5 and Clb2 substrates in MEN and FEAR mutants. (A) The dephosphorylation of Sld2 and Pol12 in cdc15–2 mutants. G1-arrested cdc15–2 SLD2-myc and cdc15–2 POL12-myc cells were released into YPD medium and incubated at 37°C. Cells were collected at 20 min intervals for the preparation of protein samples. Budding index and the immunoblots of Sld2 and Pol12 are shown. (B) The dephosphorylation of Sld2 in cdc15–2 mutant cells depends on FEAR. G1-synchronized cdc15–2, cdc15–2 slk19Δ, and cdc15–2 spo12Δ mutants with myc-tagged Sld2 were released into YPD medium at 37°C. Cells were harvested every 20 min for the preparation of protein samples. The phosphorylation of Sld2 was examined after Western blotting.
FEAR Mutants Are More Sensitive to High Levels of Clb5 Protein.
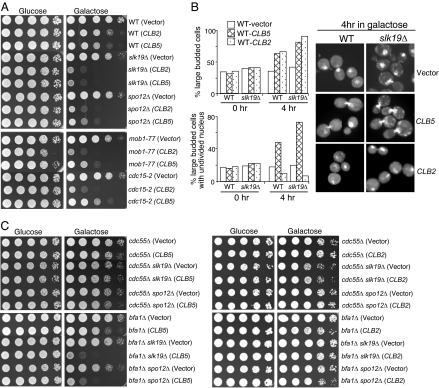
If the function of FEAR is to promote the dephosphorylation of Clb5 substrates, then FEAR may function as a Clb5-Cdk1 terminator. Therefore, high levels of Clb5 protein might be toxic to FEAR mutants. Indeed, slk19Δ or spo12Δ FEAR mutant cells containing PGAL-CLB5 plasmids grew poorly on galactose plate when Clb5 was overproduced (Fig. 3A). Overexpression of CLB2 also showed toxicity to FEAR mutants, presumably due to the mitotic exit defects (6). However, overexpression of CLB5 and CLB2 exhibited different toxicity toward MEN mutants. Overexpression of CLB2 caused lethality in MEN mutant strains mob1–77 and cdc15–2, because the function of MEN pathway is to inactivate Clb2-Cdk1. In contrast, CLB5 overexpression showed much less or no toxicity to mob1–77 and cdc15–2 mutant cells (Fig. 3A), suggesting that FEAR mutant cells are more sensitive to CLB5 overexpression than MEN mutants cells.
Fig. 3.
The comparison of the toxicity of high levels of Clb2 and Clb5 to FEAR and MEN mutant cells. (A) Overexpression of CLB5 and CLB2 shows differential toxicity to FEAR and MEN mutants. Stationary phase cells with vector, PGAL-CLB2 or PGAL-CLB5 plasmids were 10-fold diluted and spotted onto URA dropout plates containing either glucose or galactose. The plates were scanned after 3-day incubation at room temperature. (B) CLB5 overexpression leads to nuclear division defects. WT and slk19Δ cells with either vectors or PGAL-CLB2, PGAL-CLB5 plasmids were incubated in raffinose medium to midlog phase. Galactose was added into the medium to a final concentration of 2% and cells were collected at 0 and 4 h for the examination of budding index and nuclear division after DAPI staining. The percentage of large budded cells and cells with unseparated nucleus and the nuclear morphology in some representative cells are shown. (C) Hyperactive FEAR, but not MEN, suppresses the toxicity of CLB5 overexpression. Stationary phase cultures with indicated genotypes were tenfold diluted and spotted onto URA dropout plates containing glucose or galactose. The plates were scanned after 3 day incubation at 30°C.
To address why Clb5 and Clb2 are toxic to FEAR mutants, we compared the cell cycle progression of WT and FEAR mutant cells overexpressing CLB2 or CLB5. High levels of either Clb2 or Clb5 in WT cells led to the accumulation of large budded cells and deletion of SLK19 exaggerated the phenotype. The DAPI staining revealed that CLB2 overexpression increased the population of large budded cells with divided nuclei. In contrast, CLB5 overexpression led to the increase of large budded with an undivided nucleus in both WT and alk19Δ cells (Fig. 3B). spo12Δ mutants responded to the overexpression of CLB5 or CLB2 in a similar manner as slk19Δ did (data not shown). Therefore, high levels of Clb5 and Clb2 are toxic to FEAR mutants for distinct mechanisms. High level of Clb2 blocks the cell cycle in telophase in FEAR mutants, whereas CLB5 overexpression interferes with chromosome segregation.
Because the FEAR controls Cdc14 release by inhibiting PP2ACdc55 (9), the FEAR pathway is hyperactive in cdc55Δ mutants, wherein the B-regulatory subunit of PP2A is deleted (7, 8). To further confirm that CLB5 overexpression is toxic to FEAR mutants, we examined the growth of FEAR mutants overexpressing CLB5 in the absence of Cdc55. When Clb5 is overproduced, cdc55Δ mutant cells grew better than WT cells, and the poor growth phenotype of slk19Δ and spo12Δ cells induced by CLB5 overexpression was completely suppressed by cdc55Δ. In contrast, deletion of BFA1, which encodes a negative regulator of MEN, was unable to do so. Unlike Clb5, deletion of either CDC55 or BFA1 was able to suppress the toxicity caused by Clb2 overproduction (Fig. 3C). Therefore, CLB2 overexpression is toxic to both FEAR and MEN mutants, but high levels of Clb5 are only toxic to FEAR mutants, supporting the opposite roles of Clb5 and FEAR in cell cycle regulation.
Released Cdc14 Induced by FEAR or MEN Does Not Show Substrate Specificity.
We next asked why active FEAR only induces the dephosphorylation of Clb5 substrates, but not those of Clb2. To test the possibility that FEAR-induced Cdc14 exhibits specificity toward Clb5 substrates, the dephosphorylation of Sld2 was examined in cells with hyperactive FEAR or MEN. Nocodazole is a microtubule poison that arrests cells at metaphase with nucleolar localized Cdc14. Deletion of CDC55 results in Cdc14 release in nocodazole-treated cells due to the activation of FEAR (7, 8). Cells lacking Bfa1, the negative regulator of MEN, also show released Cdc14 in the presence of nocodazole (17). In nocodazole-arrested cells, Sld2 exhibited hyperphosphorylated forms, but deletion of CDC55 or BFA1 led to Sld2 dephosphorylation (Fig. 4A), indicating that either FEAR or MEN-induced Cdc14 release is able to dephosphorylate Clb5 substrates.
Fig. 4.
Low levels of Clb5 contribute to the dephosphorylation of Clb5-specific substrates during early anaphase. (A) The dephosphorylation of Sld2 in nocodazole-treated wild-type, cdc55Δ, and bfa1Δ mutants. The G1-arrested cells were released into YPD medium containing 20 μg/ml nocodazole and incubated at 30°C. Cells were collected at 20 min intervals for the preparation of protein samples. The budding index and Sld2 protein modification are shown. (B) cdc15–2 and cdc15–2 spo12Δ cells exhibit low levels of Clb5, but high levels of Clb2. The G1-arrested cells with indicated genotypes were released into YPD medium at 37°C. Cells were harvested every 20 min for protein preparation. Clb5-HA and Clb2-HA levels were determined after Western blotting and the protein levels of Pgk1 are shown as loading controls. (C) The outline of the experimental procedure for D and E. (D) CLB5 overexpression results in decreased Sld2 dephosphorylation. The cdc15–2 SLD2-myc cells with either a vector or a PGAL-CLB5 plasmid were treated as described in C. Here, the modification of Sld2 after Western blotting is shown. (E) Overexpression of SIC1 leads to the dephosphorylation of Clb2 substrate Pol12 in cdc15–2 mutants. The cdc15–2 POL12-myc and cdc15–2 slk19Δ POL12-myc cells with either a vector or a PGAL-SIC1 plasmid were treated as described in C. The modification of Pol12-myc protein is shown after Western blotting. (F) Stabilized Clb5 decreases Sld2 dephosphorylation in cdc15–2-arrested cells. The G1-synchronized cells with indicated genotypes were released into YPD medium at 37°C. α-factor was added back to WT and clb5-dbΔ cell cultures after the majority of the cells became budded to block the second round of cell cycle. Cells were harvested to analyze the phosphorylation of Sld2.
Differential Clb5 and Clb2 Levels Contribute to the Distinct Dephosphorylation Windows of Cdk1 Substrates.
Clb5 is degraded before anaphase entry, whereas Clb2 levels are high until late anaphase. In cdc15–2-arrested cells, Clb2 protein levels were high, but little Clb5 protein was detected (Fig. 4B). We reasoned that the low levels of Clb5 in cdc15–2 cells could shift the equilibrium toward a hypophosphorylated state for Clb5 substrates after FEAR-induced Cdc14 release. If that is the case, high levels of Clb5 are expected to decrease the dephosphorylation of Clb5 substrates. To test this possibility, cdc15–2 SLD2-myc cells with either a vector or a PGAL-CLB5 plasmid were first arrested with nocodazole at 25°C and then released into galactose medium and incubated at 37°C (Fig. 4C). As expected, Sld2 dephosphorylation was decreased in cells overexpressing CLB5 (Fig. 4D). Nevertheless, a transient dephosphorylation was still observed after release for 20 min, indicating that the briefly released Cdc14 overwhelmed Clb5-Cdk1.
If the low levels of Clb5 contribute to the dephosphorylation of Clb5-specific substrates during early anaphase, inactive Clb2-Cdk1 should also allow the dephosphorylation of Clb2 substrates in a similar manner. To test this idea, we used the same protocol to examine the dephosphorylation of Clb2 substrate Pol12 in cdc15–2 cells overproducing Sic1, the Cdk inhibitor in budding yeast. High levels of Sic1 resulted in the dephosphorylation of Pol12 and this process was dependent on FEAR, as deletion of SLK19 totally abolished Pol12 dephosphorylation (Fig. 4E). We also examined the Sld2 dephosphorylation in cells expressing Clb5-dbΔ (destruction box deleted) from its endogenous promoter, as deletion of the destruction box stabilizes Clb5 (18). Stabilized Clb5 clearly impaired Sld2 dephosphorylation in cdc15–2 mutants (Fig. 4F). Therefore, the combination of the unavailability of Clb5 protein and FEAR-induced Cdc14 release during early anaphase contributes to the dephosphorylation of Clb5 substrates, because the substrates are unable to be rephosphorylated once they are dephosphorylated.
FEAR-Induced Cdc14 Release Is Required for Anaphase Progression.
What is the biological significance of the dephosphorylation of Clb5-Cdk1 substrates during early anaphase? The dephosphorylation of Ase1 and Fin1, two Clb5-Cdk1 substrates, has been shown to promote spindle stabilization and elongation (19, 20). The reversal of Clb5-Cdk1-imposed phosphorylation might be critical for the normal spindle behavior during anaphase. Hence, the spindle morphology in cdc15–2-arrested telophase cells was examined in the presence and absence of FEAR. After G1 release into YPD medium at 37 °C for 150 min, 82% of cdc15–2 single mutant cells exhibited fully elongated spindles, but the numbers were decreased to 37% and 39% in cdc15–2 slk19Δ and cdc15–2 spo12Δ cells, respectively (Fig. 5B). Although deletion of either SLK19 or SPO12 resulted in broken and short spindles in cdc15–2 background, cdc15–2 slk19Δ cells showed more severe broken spindle phenotype, presumably due to the direct role of Slk19 in spindle stability (Fig. 5 C and D) (19, 21). These observations suggest that FEAR may facilitate spindle elongation by antagonizing Clb5-Cdk1 function.
Fig. 5.
FEAR pathway facilitates anaphase progression. G1-arrested cdc15–2, slk19Δ cdc15–2 and spo12Δ cdc15–2 cells with GFP-tagged TUB1 were released into YPD medium at 37°C. Cells were removed every 30 min for the examination of spindle morphology by fluorescence microscopy. The budding index and the percentage of cells with elongated spindles are shown in A and B. (C) The percentage of cells with broken spindle at the indicated time points is shown. (D) Some representative cells at 150 min after G1 release are shown. The percentage of cells with broken or fully elongated spindles is a result of three experiments.
We further examined the chromosome segregation in cdc15–2 single and cdc15–2 slk19Δ, cdc15–2 spo12Δ double mutants by visualizing GFP-tagged Mtw1, a kinetochore protein (22). More than 80% of cdc15–2-arrested cells showed fully separated kinetochore clusters after G1 release for 150 min, i.e., the two GFP dots had migrated to the ends of two daughter cells, but the number was reduced to ≈50% in the double mutants, indicating the role of FEAR in chromosome segregation. We also examined chromosome separation at centromeric and telomeric regions by visualizing Nuf2-Cherry and GFP-marked telomere on chromosome V (Tel-GFP) (23–25). Although no kinetochore separation delay was observed in cdc15–2 slk19Δ and cdc15–2 spo12Δ cells, we noticed a telomere separation delay in the double mutants, consistent with the previous reports (Fig. S4) (24, 26). Interestingly, the double mutant cells showed more frequent asynchronous separation of telomeres and kinetochores, as indicated by a single Tel-GFP dot and two separated Nuf2-Cherry dots (Fig. S4). Collectively, our data suggest that FEAR-induced Cdc14 release during early anaphase promotes spindle elongation by reversing Clb5-Cdk1-imposed phosphorylation, which facilitates spindle elongation and the subsequent chromosome segregation, especially at telomeric regions.
Discussion
Cyclin-dependent substrate specificity ensures the unique temporal phosphorylation window for a given Cdk substrate, which is important to the correct order of cell cycle events, but it remains unclear whether protein phosphatase plays a role in this regulation. Here, we show evidence indicating that the FEAR and MEN-induced waves of Cdc14 release provide the molecular basis for the sequential dephosphorylation of Clb5 and Clb2 substrates. Our data also suggest that the differential dephosphorylation windows of Clb5 and Clb2 substrates are not due to the substrate specificity of Cdc14 phosphatase. Instead, the low Clb5 protein levels contribute to the dephosphorylation of Clb5-specific substrates during early anaphase, because the substrates cannot be rephosphorylated once dephosphorylated by Cdc14. Therefore, in addition to the tightly regulated Cdk activity, the two waves of Cdc14 release induced by FEAR and MEN are also critical for the temporal phosphorylation of Cdk substrates, which limits the function of a given Cdk substrate within a specific window during the cell cycle.
Why do S-phase Cdk substrates need to be dephosphorylated during early anaphase? Among the Clb5-specific substrates, the phosphorylation of Sld2 has been proven to be essential for DNA replication (10), but it remains unclear whether the dephosphorylation of Sld2 is necessary to terminate DNA replication and reset the machinery for the following S-phase. Clb5-specific substrates also include some microtubule-associated proteins, such as Ase1 and Fin1 (1). As the dephosphorylation of Ase1 and Fin1 facilitates spindle stabilization and elongation (19, 20), the phosphorylation of these proteins by Clb5-Cdk1 may inhibit anaphase. Therefore, Clb5-Cdk1 not only promotes DNA replication, but also functions as a negative regulator of anaphase. FEAR-induced Cdc14 release relieves the inhibition imposed by Clb5-Cdk1.
Mammalian cells express Cdc14A and Cdc14B, the homologues of budding yeast Cdc14. PRC1, the human homologue of Ase1, also becomes dephosphorylated during metaphase to anaphase transition and this modification is essential for the spindle midzone formation (27). Therefore, budding yeast and mammalian cells may share a conserved mechanism to stimulate anaphase through the dephosphorylation of some Cdk substrates. In summary, our work reveals a new layer of regulation of the transition from metaphase to anaphase in budding yeast. FEAR-induced Cdc14 release during early anaphase reverses Clb5-Cdk1-imposed protein phosphorylation, which facilitates spindle elongation and the subsequent chromosome segregation.
Materials and Methods
Yeast Strains and Plasmids.
All of the yeast strains are derivatives of Y300 isogenic to W303 and are listed in Table S1. The clb5-dbΔ mutant was confirmed with PCR. Epitope-tagging of endogenous genes was carried out using a PCR-based method. The YPD medium was used for the growth of yeast strains except for those carrying plasmids. Cells harboring plasmids were grown in URA dropout medium before G1 release. For Clb5 and Sic1 induction, YEP-galactose was used. Nocodazole treatment, α-factor treatment, and protein analysis were performed as previously described (13).
Microscopy.
The analysis of Tub1-GFP, Mtw1-GFP, Tel-GFP, and Nuf2-Cherry in fixed cells was carried out using fluorescence microscopy. The Tub1-GFP cells were fixed in 3.7% formaldehyde for 30 min at room temperature, whereas other strains were fixed for 5 min for the examination of fluorescence signal. Three hundreds cells were counted for each sample.
Protein Kinase Assay.
The POL12 gene was cloned into pTBSG vector and the resulting plasmid was transformed into a bacteria strain for the preparation of Pol12 protein (28). To prepare Clb-Cdk1, 60 ml of WT, CLB2-HA, and CLB5-HA cells were collected and resuspended in RIPA buffer containing protease inhibitors. The cell lysates were immunoprecipitated and resuspended in 20 μl kinase buffer containing Pol12, Histone H1, or no substrate. After incubation, the reactions were ended and the samples were separated with SDS/PAGE gel. The gel was then dried and exposed to a phosphorimager.
Supplementary Material
Acknowledgments.
We thank Drs. David Gilbert, Akash Gunjan, and Stephen Elledge for reading through the manuscript. We thank Dr. Tim Cross (Florida State University, Tallahassee) for providing the protein expression system. The original strain containing Nuf2-Cherry was a generous gift from the Davis laboratory (University of Washington, Seattle). The MTW1-GFP strain was from the Biggins laboratory (Fred Hutchinson Cancer Center, Seattle) and the clb5-dbΔ strain was from the Cross laboratory (The Rockefeller University, New York). This work is supported by the Research Scholar Grant (RSG-08–104-010CCG) from American Cancer Society and the Planning Grant from FSU Council on Research and Creativity.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808719105/DCSupplemental.
References
- 1.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 2.Gray CH, Good VM, Tonks NK, Barford D. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 2003;22:3524–3535. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shou W, et al. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 4.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 5.Visintin R, et al. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 6.Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Ng TY. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:80–89. doi: 10.1091/mbc.E04-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yellman CM, Burke DJ. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:658–666. doi: 10.1091/mbc.E05-04-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 10.Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- 11.Keaton MA, et al. Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr Biol. 2007;17:1181–1189. doi: 10.1016/j.cub.2007.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu F, Aparicio OM. Swe1 regulation and transcriptional control restrict the activity of mitotic cyclins toward replication proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:8910–8915. doi: 10.1073/pnas.0406987102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Wang Y. The function and regulation of budding yeast Swe1 in response to interrupted DNA synthesis. Mol Biol Cell. 2006;17:2746–2756. doi: 10.1091/mbc.E05-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foiani M, Liberi G, Lucchini G, Plevani P. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol Cell Biol. 1995;15:883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Elledge SJ. The DASH complex component Ask1 is a cell cycle-regulated Cdk substrate in Saccharomyces cerevisiae. Cell Cycle. 2003;2:143–148. [PubMed] [Google Scholar]
- 16.Bardin AJ, Boselli MG, Amon A. Mitotic exit regulation through distinct domains within the protein kinase Cdc15. Mol Cell Biol. 2003;23:5018–5030. doi: 10.1128/MCB.23.14.5018-5030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Hu F, Elledge SJ. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson MD, Gray S, Yuste-Rojas M, Cross FR. Testing cyclin specificity in the exit from mitosis. Mol Cell Biol. 2000;20:4483–4493. doi: 10.1128/mcb.20.13.4483-4493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: Separase cleavage and localization of Slk19. Nat Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang X, Wang Y. Pds1/Esp1-dependent and -independent sister chromatid separation in mutants defective for protein phosphatase 2A. Proc Natl Acad Sci USA. 2006;103:16290–16295. doi: 10.1073/pnas.0607856103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- 25.Shimogawa MM, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- 27.Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci USA. 2006;103:6196–6201. doi: 10.1073/pnas.0506926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin H, et al. Construction of a series of vectors for high throughput cloning and expression screening of membrane proteins from mycobacterium tuberculosis. BMC Biotechnol. 2008;8:51. doi: 10.1186/1472-6750-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.