Abstract
Hypermodifications near the anticodon of tRNA are fundamental for the efficiency and fidelity of protein synthesis. Dimethylallyltransferase (DMATase) catalyzes transfer of a dimethylallyl moiety from dimethylallyl pyrophosphate to N6 of A37 in certain tRNAs. Here we present the crystal structures of Saccharomyces cerevisiae DMATase–tRNACys complex in four distinct forms, which provide snapshots of the RNA modification reaction catalyzed by DMATase. The structures reveal that the enzyme recognizes the tRNA substrate through indirect sequence readout. The targeted nucleotide A37 flips out from the anticodon loop of tRNA and flips into a channel in DMATase, where it meets its reaction partner di methylallyl pyrophosphate, which enters the channel from the opposite end. Structural changes accompanying the transfer reaction taking place in the crystal result in disengagement of DMATase–tRNA interaction near the reaction center. In addition, structural comparison of DMATase in the complex with unliganded bacterial DMATase provides a molecular basis of ordered substrate binding by DMATase.
Keywords: RNA modification, substrate specificity, x-ray crystallography
Among >100 modified nucleotides discovered to date in RNA, several exhibit extensive modifications (hypermodifications). These hypermodified nucleotides are clustered near the anticodon of tRNA, either at the wobble position (position 34) or at position 37. The hypermodifications are important for efficiency and fidelity of protein translation by the ribosome (1, 2).
When the third anticodon of a tRNA is A, which forms a weak base pair with the first codon U in mRNA, its 3′-adjacent nucleotide A37 is hypermodified to 2-methylthio-N6-isopentenyladenosine (ms2i6A). The first step of the modification reactions involves the transfer of a DMA moiety from dimethylallyl pyrophosphate (DMAPP) to N6 of A37 in tRNA substrate, carried out by dimethylallyltransferase (DMATase) [supporting information (SI) Fig. S1] (3–5). Studies by Moore and Poulter (6) using recombinant Escherichia coli DMATase demonstrated ordered substrate binding by the enzyme, with binding of the accepting substrate tRNA first, followed by the donating substrate DMAPP. A conserved sequence of AAA at nucleotides 36–38 of tRNA was required for efficient modification by DMATase (7, 8). Site-directed mutagenesis of E. coli DMATase also provided insight into possible roles of some conserved amino acids in tRNA binding and catalysis (9). More recently, we reported the crystal structure of a bacterial DMATase (10). The structure revealed that the core domain of DMATase contains a central channel, where we proposed the transfer reaction might occur (10). However, our previous studies did not reveal the structure of a domain of ≈85 aa (named the insertion domain) despite the presence of the insertion domain within crystals (10). In addition to the core and insertion domains in the bacterial DMATase, the eukaryotic enzyme contains an extra ≈80 aa at its C terminus (named the C-terminal addition) whose function is unknown.
To provide structural information of the insertion domain and the C-terminal addition, and to reveal the molecular basis of tRNA substrate recognition and the reaction mechanism, we crystallized Saccharomyces cerevisiae DMATase in complex with tRNACys in the presence of pyrophosphate and solved its structure at 2.95-Å resolution. Furthermore, soaking crystals of the DMATase–tRNA binary complex with dimethylallyl thio-pyrophosphate (DMASPP, a DMAPP analog) and DMAPP allowed us to obtain structures of the reaction intermediate as well as the enzyme bound to the tRNA product. Together, these structures provided us snapshots of the DMATase-catalyzed reaction along the reaction pathway.
Results
Overall Structure of the DMATase-tRNA-Pyrophosphate Complex.
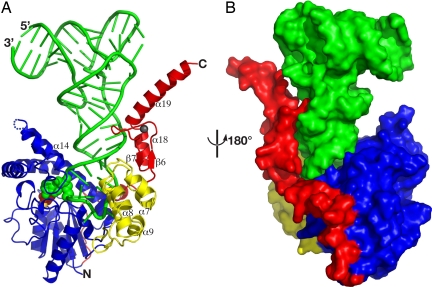
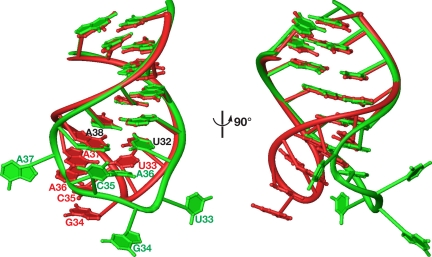
The overall structure of the complex can be schematically described as the letter L (tRNA) inversely docked into the deep cleft of the letter U (DMATase) (Fig. 1). One side of the U-shaped DMATase is composed of the core domain (Fig. 1, blue), and the opposite side is the combination of the insertion domain and part of the C-terminal addition (Fig. 1, yellow and red, respectively). The structure of the core domain of DMATase in the complex is very similar to that of the previously reported bacterial enzyme (Fig. S6) (10). The rmsd of Cα of the two structures is 1.8 Å. The insertion domain, whose structure was not observed in our previous studies of the bacterial enzyme (10), is a five-helix bundle (Fig. 1A, yellow). It connects to the core domain through two loops (Fig. 1A and Fig. S5). The C-terminal addition, unique to the eukaryotic enzyme (11, 12), can be further divided into two parts. The C terminus is a zinc finger composed of two short β-strands and two α-helices (Fig. 1A). The zinc finger is connected to the core domain by a linker of ≈30 aa composed of two short helices and loops (Fig. S5) traversing along the core domain and placing the zinc finger ≈50 Å away (Fig. 1B). A Dali structural homology search using the structure of the insertion domain or the C-terminal zinc finger failed to reveal any known proteins that are structurally similar to these two domains (13), suggesting that both the insertion domain and the C-terminal zinc finger are new RNA-binding motifs. Contrasted to little structural change of the core domain, substantial conformational change occurred in the anticodon loop of tRNA upon association of tRNA with DMATase (Fig. 2). In addition to substantial positional shift of the tRNA backbones, four nucleotides, U33, G34, C35, and A37, flip out from the anticodon loop of tRNA (Fig. 2).
Fig. 1.
Structure of the DMATase–tRNA–pyrophosphate complex. (A) Ribbon diagram. Green, tRNA; blue, DMATase core domain; yellow, DMATase insertion domain; red, DMATase C-terminal addition. The components in the reaction channel of DMATase, including the targeted A37, the pyrophosphate, and the magnesium ion, are highlighted in spheres and colored green, red, and gold, respectively. The zinc ion of the C-terminal zinc finger is depicted as a small sphere in dark gray. Part of the secondary structure relevant to Fig. 4 is labeled for clarity. (B) Surface representation of the structure viewed from the opposite side.
Fig. 2.
Conformational change of tRNA upon its association with DMATase. Shown is the superposition of the anticodon stem-loop of S. cerevisiae tRNACys in the DMATase–tRNA complex (green) with the corresponding nucleotides of E. coli tRNACys (red, Protein Data Bank ID code 1U0B). The nucleotide sequences of the anticodon stem-loops (27–43) of these two tRNAs are identical.
Interaction between tRNA and DMATase is extensive and is mainly concentrated at the anticodon stem-loop (nucleotides 27–43). Approximately 50% of the surface area of the anticodon stem-loop is buried within DMATase. The deep tRNA-binding cleft in DMATase is strongly positively charged (Fig. S7).
Molecular Recognition of the Conserved A36, A37, and A38 of tRNA.
Specific interactions between DMATase and tRNA, as well as within tRNA itself, provide insight into the molecular basis of tRNA substrate recognition (7, 8).
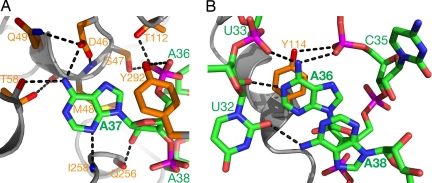
The targeted A37 flips out from the anticodon stem-loop of tRNA and flips into a channel in the core domain of DMATase (Fig. 1A, green spheres). Recognition of A37 is achieved through its extensive interactions with several conserved amino acid residues in DMATase (Fig. 3A). The conserved DSMQ motif in DMATase (residues 46–49) plays a major role in A37 recognition. The side chain of Asp-46, whose position is secured through hydrogen bonding with the side chain of Gln-49 and main-chain amide of Ser-47, forms a hydrogen bond with N6 of A37; the side chain of Ser-47 donates a hydrogen bond to the phosphate group of A37; and the side chain of Met-48 stacks on the base of A37 (Fig. 3A). In addition to the DSMQ motif, other DMATase residues (Thr-58, Thr-112, Gln-256, Ile-258, and Tyr-292) are also involved in A37 binding (Fig. 3A).
Fig. 3.
Molecular recognition of the conserved AAA sequence at nucleotides 36–38 of the tRNA substrate. (A) Recognition of the targeted A37 by DMATase. The main chain of DMATase is represented as gray ribbons. The side chains of DMATase and tRNA are depicted as sticks. Hydrogen bonds are depicted as dotted lines in black. The carbon atoms of DMATase and tRNA are colored orange and green, respectively. Hetero-atoms are colored individually, with nitrogen in blue, oxygen in red, phosphorus in magenta, and sulfur in yellow. The oxygen atom of the phosphate group of A37 that forms hydrogen bonds with the side chains of Ser-47 and Thr-112 is hidden by the OH group of Tyr-292. (B) Recognition of A36 and A38 through specific interactions within tRNA itself.
Surprisingly, no specific interaction between DMATase and A36 or A38 was observed. Instead, recognition of A36 and A38 is achieved through specific interactions within tRNA itself (Fig. 3B). N6 of A36 donates two hydrogen bonds to the phosphate groups of U33 and C35, and N1 of A36 accepts a hydrogen bond from O2′ of U32 (Fig. 3B). The position of the base of A36 is further secured by stacking with the side chain of Tyr-114 as well as the base of A38. N6 of A38 donates a hydrogen bond to O2 of U32, a position that is termed a universal hydrogen bond acceptor (Fig. 3B). The position of the base of A38 is secured by its stacking with the base of A36 and the base of U39 (data not shown).
Interactions Between DMATase and tRNA.
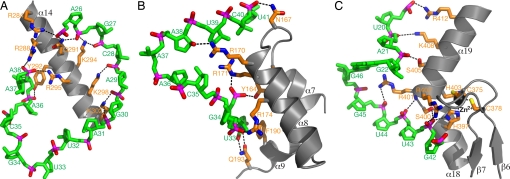
In addition to A37 recognition, the core domain has contacts with tRNA, and these are primarily concentrated at helix 14, which sits in the major groove of the anticodon stem-loop (Fig. 1A). A series of basic residues facing RNA (Arg-284, Arg-288, Gln-291, Lys-294, Arg-295, Lys-298, and Lys-302) interact with phosphate groups of the nucleotides in the anticodon stem-loop (Fig. 4A). Tyr-292, which is involved in A37 recognition (Fig. 3A), also resides on helix 14 (Fig. 4A).
Fig. 4.
Major contacts between DMATase and tRNA. (A) Interaction of helix 14 from the core domain with tRNA. DMATase is depicted and colored as in Fig. 3. tRNA is in green except phosphate groups, which are highlighted in red (oxygen) and magenta (phosphorus). With the exception of U33, G22, and G46, the bases of tRNA are omitted for clarity. (B) Interaction of the insertion domain with tRNA. The base of U33 is specifically recognized by the side chain of Gln-193 through two hydrogen bonds. (C) Interaction of the C-terminal zinc finger with tRNA. The zinc ion is depicted as a small sphere in dark gray.
The insertion domain, located on the opposite side of helix 14 (Fig. 1A, yellow), interacts with the nucleotides in the anticodon stem-loop of tRNA that helix 14 is not able to reach (Fig. 4B). Several amino acids on helices 7 and 8 as well as on the intervening loop (Tyr-164, Asn-167, Arg-170, Arg-171, and Arg-174) make contacts with phosphate groups of the nucleotides in the anticodon stem-loop. Of particular interest is the formation of the two hydrogen bonds between the side chain of Gln-193 and the base of U33 (Fig. 4B). Because Gln-193 is strictly conserved in DMATase, and virtually all tRNAs contain U at position 33, this interaction is specific and is likely the favorable interaction that makes the flipping out of U33 energetically accessible.
Sitting on top of the insertion domain is the C-terminal zinc finger (Fig. 1A, red), which is involved in interactions with nucleotides at the top part of anticodon stem (nucleotides 42–44) as well as part of the D loop (nucleotides 20–22) (Fig. 4C). The overall structure of the zinc finger can be interpreted as an uninterrupted long α-helix that is forced to make a kink by coordination of His-397 and His-403 with a zinc ion. This results in a zinc finger whose shape is complementary to the structure of tRNA near the junction of anticodon stem and D loop (Figs. 1A and 4C). As such, the side chains of some basic residues on helix 19 (Arg-401, Arg-402, Lys-408, and Arg-412) are able to reach phosphate groups of the nucleotides in D loop (Fig. 4C). Residues in the kinked region make close contact with nucleotides at the top of anticodon stem (side chain of Ser-400 to the phosphate group of U43; main chain amide of Arg-401 to the phosphate group of U44). Of particular interest is the possibly dual role played by the side chain of His-397. The imidazole ring of His-397 appears to serve as a ligand for the zinc ion on one side and to donate a hydrogen bond to the phosphate group of U43 on the other. In our structure, the imidazole ring of His-397 is not perfectly aligned to be capable of both zinc coordination and hydrogen bonding with RNA. The distance between NE2 and the zinc ion is 2.5 Å, and the angle of the zinc ion with the plane of the imidazole ring of His-397 is 140°. On the other hand, the distance between O1P of U43 and ND1 of His-397 is 2.6 Å, and the angle of O1P with the imidazole ring is 170°. However, if the imidazole ring is rotated slightly, the hydrogen bond would be in the same distance with a perfect angle of 180°, and the distance and the angle for the zinc coordination would be improved to 2.4 Å and 145°, respectively. The imperfect stereo chemistry and distances of the described interactions may be a consequence of the modest 2.95-Å resolution of our structure.
Snapshots of the DMATase-Catalyzed Reaction.
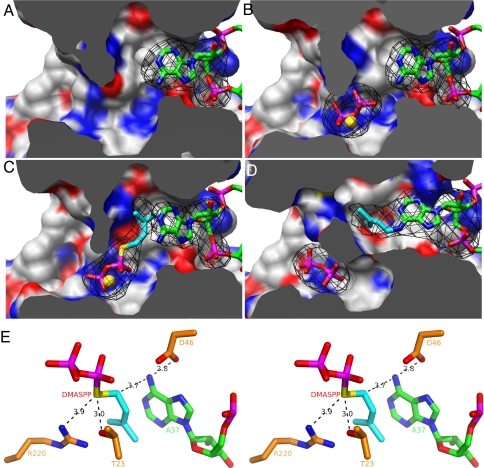
Through improvement in sample preparation and further optimization of crystallization conditions, we were able to obtain a 3.1-Å-resolution structure of DMATase–tRNA binary complex in the absence of pyrophosphate. More importantly, this allowed us to probe the mechanism involved in the transfer reaction through various soaking experiments. Together, a total of four structures provided us snapshots of the DMATase-catalyzed reaction (Fig. 5).
Fig. 5.
Snapshots of the DMATase-catalyzed reaction. (A) Structure of DMATase–tRNA binary complex. DMATase is depicted as protein surface, with carbon in white, nitrogen in blue, and oxygen in red. tRNA is depicted and colored as in Fig. 3. The structure of DMATase is clipped across the reaction channel to reveal contents in the channel. (B) Structure of DMATase–tRNA–PP ternary complex. The pyrophosphate is in sticks, and Mg2+ is depicted as a sphere in gold. (C) Structure of DMATase–tRNA–DMASPP ternary complex. The bridging sulfur and the DMA moiety in DMASPP are colored yellow and cyan, respectively. (D) Structure of DMATase–tRNA(P)–PP ternary complex. A37 of the tRNA substrate in A–C, i6A37 of the tRNA product in D, and pyrophosphate, DMASPP, and Mg2+ in A–D are covered with simulated annealing omit maps colored black. The maps are contoured at 5.0 σ. (E) Stereo view of the final model of DMATase–tRNA–DMASPP ternary complex near the active site showing the catalytic apparatus and substrates. The distances between atoms relevant to mechanistic discussions are labeled in black.
The structure of DMATase-tRNA-PP ternary complex revealed that, although the targeted A37 of tRNA occupies the entrance of one side of the channel (Fig. 5A), the pyrophosphate and its coordinated Mg2+ ion are located in the middle of the channel (Fig. 5B). Recognition of pyrophosphate is mainly achieved by the N-terminal conserved P-loop (GxTGxGKS sequence motif from residues 21 to 28), as well as the conserved Arg-220 (Fig. S8A). The overall structure of DMATase–tRNA binary complex is very similar to the structure of DMATase–tRNA–PP ternary complex, with an rmsd of 0.3 Å. Nevertheless, small but noticeable structural changes can be observed for those amino acids involved in pyrophosphate binding. For example, both the position of TGxG motif (residues 23–26) within the P-loop and the side chain of Arg-220 have shifted ≈1.3 Å as a result of pyrophosphate binding. The number may be statistically significant considering that the overall rmsd is 0.3 Å.
In crystals of the DMATase–tRNA binary complex soaked with DMASPP, DMASPP occupied the channel (Fig. 5C). The DMA moiety was found in a hydrophobic pocket adjacent to and stacked with the base of A37 (Fig. 5C). The two terminal methyl groups pointed in the direction of the side chains of Ile-258 and Met-224 (Fig. 6A). Because the only difference between DMASPP and the real substrate DMAPP is substitution of the bridging oxygen in DMAPP by a sulfur in DMASPP (Fig. 5C, yellow), the relative configuration of the targeted A37 and DMASPP shown in Fig. 5C no doubt represents a close mimic of the reaction intermediate of the DMATase-catalyzed reaction.
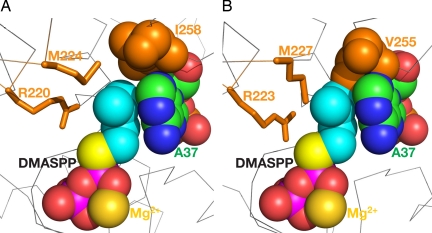
Fig. 6.
Location of the DMA moiety and molecular basis of ordered substrate binding by DMATase. (A) Structure of DMATase–tRNA–DMASPP ternary complex in the reaction channel. DMASPP, Mg2+, A37 of tRNA, and Ile-258 of DMATase are depicted as spheres to highlight their relative positions when correctly sized. (B) Docking DMASPP, Mg2+, and A37 from A into the structure of the bacterial DMATase. The equivalent residues of DMATase shown in A are highlighted.
Remarkably, crystals of the DMATase–tRNA binary complex soaked with DMAPP resulted in the transfer reaction taking place within the crystals (Fig. 5D). During data collection at the synchrotron, whereas six crystals soaked with DMASPP diffracted in the range of 3.1–3.3 Å, a resolution comparable to the unsoaked crystals, six crystals soaked with DMAPP diffracted in the range of 3.6–3.9 Å. Because these crystals were mounted from the same crystallization drop and soaked under identical conditions except for soaking with DMASPP or DMAPP, the reduction of diffraction resolution most likely resulted from the chemical reaction of DMAPP with tRNA substrate in the crystals. Indeed, structural change resulting from the transfer reaction can be observed both globally and locally. The rmsd of the structures of the reacted complex vs. any one of the unreacted complexes is 1.4 Å. This is a 1.1-Å increase when compared with the rmsd of the structures of two unreacted ones (0.3 Å). As the transfer reaction proceeds, some amino acids originally interacting with A37 become disengaged within the reaction channel. And both reaction products, i6A37 of tRNA and pyrophosphate, are on their way out (Fig. 5D; compare their positions to corresponding counterparts in Fig. 5B). Most residues involved in binding to both A37 and DMAPP are no longer within a distance to contribute such binding energy (Fig. S9). As such, the shape of the channel in DMATase has changed substantially (Fig. 5D, compare with Fig. 5 A–C).
Discussion
Insight obtained from four structures described here, combined with previous biochemical studies, allowed us to address several fundamental issues of DMATase-catalyzed RNA modification reaction, including tRNA substrate recognition, the reaction mechanism, and ordered substrate binding.
Studies based on E. coli DMATase indicated a strict requirement of the conserved nucleotide sequence A36-A37-A38 in tRNA substrate (7, 8). Therefore, we expected specific interactions between certain conserved amino acids in DMATase and A36, A37, and A38 in tRNA. However, our finding revealed that, although the targeted A37 is specifically recognized by the enzyme (Fig. 3A), there is no specific interaction among A36, A38, and DMATase (Fig. 3B). Instead, A36 and A38 are recognized through specific but non-base-pair interactions within tRNA itself (Fig. 3B). Formation of the specific RNA configuration shown in Fig. 3B, which allows A36 and A38 to be recognized, results from the flipping out of several nucleotides in the anticodon loop, A37 and U33 in particular (Fig. 2). Because A37 interacts with the core domain and U33 interacts with the insertion domain, DMATase accomplishes recognition of A36 and A38 through an indirect readout mechanism.
The snapshots presented in Fig. 5 allowed us to propose the following mechanism of DMATase-catalyzed RNA modification reaction. Binding of tRNA substrate by DMATase results in the targeted A37 flipped into the reaction channel in DMATase (Fig. 5A). Relevant to the catalysis is the hydrogen bonding between the side chain of Asp-46 and N6 of A37 (Figs. 3A and 5E). Flipping of A37 into the channel expands the channel near Ile-258, providing sufficient room for DMAPP to enter the channel from the opposite end (Fig. 5C). Important for catalysis is the hydrogen bonding between the side chain of Thr-23 and the bridging oxygen in DMAPP (shown as a sulfur atom in Fig. 5E and Fig. S8B). Nucleophilic attack of N6 of A37 on the carbon atom adjacent to the bridging oxygen in DMAPP, which is 3.7 Å away from N6 of A37 and well aligned for the reaction (Figs. 5E and 6A), results in transfer of the DMA moiety from DMAPP to N6 of A37 in tRNA (Fig. 5D). Asp-46 acts as a general base to accept a proton from N6 of A37, and the role of Thr-23 is to activate the transferring DMA moiety. Although the side chain of Arg-220 is too far away to form a hydrogen bond with the bridging sulfur of DMASPP (Fig. 5E and Fig. S8B), it forms a hydrogen bond (3.2 Å) with the pyrophosphate (Fig. S8A). Therefore, the role of Arg-220 is likely to stabilize the leaving pyrophosphate group after the reaction. Mutating the equivalent Asp-46, Thr-23, and Arg-220 to alanine in E. coli enzyme reduced the enzymatic activity by 20-, 600-, and 25-fold, respectively (9), consistent with this proposed mechanism.
Moore and Poulter (6) reported ordered substrate binding by E. coli DMATase, with binding of accepting substrate tRNA first followed by the donating substrate DMAPP. The study implies structural change required in DMATase as a result of tRNA binding to allow DMAPP to be bound. Therefore, in addition to crystallization of DMATase–tRNA complex, we have also carried out crystallization of the apo-enzyme of S. cerevisiae DMATase to address this issue. Unfortunately, crystallization of the apo-enzyme was not successful despite our extensive effort. Therefore, comparison of the structure of the eukaryotic DMATase–tRNA complex with our previously reported structure of the bacterial DMATase, although not ideal, may be informative. As shown in Fig. 6A, the DMA moiety of DMASPP, A37 of tRNA, and Ile-258 of DMATase fit well together without any steric clash. On the other hand, the equivalent residue to Ile-258, Val-255, clashes sterically with both DMASPP and A37 when tRNA and DMASPP were docked to the structure of the bacterial DMATase (Fig. 6B). Therefore, if this structural comparison is valid, binding of tRNA causes a conformational change in the loop between helices 12 and 13 centered at Ile-258, resulting in expansion of DMA binding pocket. This expansion, combined with a possible energetic contribution of DMA moiety stacking on the base of A37, allows DMAPP to be bound by DMATase after tRNA binding.
Unlike the bacterial DMATase, eukaryotic DMATase contains a C-terminal zinc finger, which stacks on top of the insertion domain and provides additional interactions with tRNA. Clearly, this domain is not required for the modification reaction because the bacterial enzyme does not have it. We suggest two possible functions for the C-terminal zinc finger in the eukaryotic organisms. One is to reinforce the stability of the insertion domain, which may be susceptible to protease degradation in eukaryotes. Alternatively, additional interactions of the C-terminal zinc finger with tRNA in the eukaryotic DMATase may enhance the substrate specificity. Resolving these issues will require additional experiments, such as enzymatic and stability tests of both the full-length and the truncated eukaryotic DMATase.
Materials and Methods
Complex Formation and Purification.
S. cerevisiae DMATase was overexpressed in E. coli, and S. cerevisiae tRNACys was in vitro transcribed. Both the protein and tRNA were purified. A detailed description of the protein and tRNA purifications may be found in SI Text. Purified DMATase and tRNAcys were mixed in a molar ratio of 1:1.2, and the resulting mixture was incubated on ice for 40 min. The mixture was then injected into a Superdex 200 size-exclusion column. Fractions containing DMATase–tRNA complex were pooled and concentrated to ≈6 mg/ml for crystallization.
Crystallization.
Crystallization was carried out by using the hanging drop vapor diffusion method at 4°C. DMATase–tRNA complex (6 mg/ml) was mixed with an equal volume of well solutions. Initially, tiny crystals were obtained with a well solution containing 100 mM Mes (pH 6.5), 25% PEG400, 100 mM NaCl, and 5 mM MgCl2. Crystals were improved by incorporating 5 mM pyrophosphate into the crystallization drop. Although the size of crystals was unchanged, on x-ray diffraction of the crystals, substitution of PEG400 with Pentaerythritol propoxylate 426 (Aldrich) as the precipitant improved the resolution. In the later stage of crystallization, we were able to obtain high-quality crystals of DMATase–tRNA complex in the absence of pyrophosphate, allowing us to carry out DMASPP and DMAPP soaking experiments. To collect data at a low temperature, crystals were soaked briefly in a cryoprotecting solution containing all of the components of the well solution supplemented with 15% (vol/vol) glycerol. The cryoprotected crystals were then mounted in a nylon loop and flash-frozen in liquid nitrogen.
Data Collection and Refinement.
Both the native and single-wavelength anomalous dispersion (SAD) data were collected at 21-ID beamlines at the Advanced Photon Source. Data were reduced with Denzo and Scalepack (14). Phase was determined and the initial electron density map was calculated based on the SAD data by using the program Phenix (15). The initial model of DMATase–tRNA–PP complex was manually built by using the programs O and Coot (16, 17). Refinements were carried out with the program CNS (18). The structural components of DMATase and tRNA of the DMATase–tRNA–PP ternary complex were used as the initial model for the structural refinements of the DMATase–tRNA binary complex (Table S1).
Soaking Experiments.
Six crystals of DMATase–tRNA binary complex, mounted from a single crystallization drop, were incubated with 20 μl of the crystallization well solution containing either 1 mM DMASPP or 1 mM DMAPP (three crystals for each) for 1 h at 4°C. The same experiment was repeated under the same conditions except the incubation time was 20 h instead of 1 h. The soaked crystals were cryoprotected and mounted as described for the unsoaked crystals, and several datasets were collected at the synchrotron. As in the case of structural determination of DMATase–tRNA binary complex, the structural components of DMATase and tRNA of the DMATase–tRNA–PP ternary complex were used as the initial model for the structures of DMATase–tRNA–DMASSP and DMATase–tRNA(P)–PP ternary complexes (Table S1). Figures were prepared with PyMol (Figs. 1, 3, 4, 5E, and 6) (19), Ribbons (Fig. 2) (20), and Chimera (Fig. 5 A–D) (21).
Supplementary Material
Acknowledgments.
We thank S. Harrison, D. Söll, C. Wraight, D. Kranz, D. Shapiro, R. Switzer, J. Gerlt, and S. Nair for comments on the manuscript and the staff of beamline 21ID at the Advanced Photon Source (J. Brunzelle and K. Brister) for their assistance during data collection. This work was supported by National Institutes of Health Grant CA90954.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3EPH, 3EPJ, 3EPK, and 3EPL).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805680105/DCSupplemental.
References
- 1.Curran JF. Modified nucleosides in translation. In: Grosjean H, Benne B, editors. Modification and Editing of RNA. Washington, DC: Am Soc Microbiol; 1998. pp. 493–516. [Google Scholar]
- 2.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartz JK, Kline LK, Soll D. N6-(Delta 2-isopentenyl)adenosine: Biosynthesis in vitro in transfer RNA by an enzyme purified from Escherichia coli. Biochem Biophys Res Commun. 1970;40:1481–1487. doi: 10.1016/0006-291x(70)90035-5. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbaum N, Gefter ML. Delta 2-isopentenylpyrophosphate: Transfer ribonucleic acid 2-isopentenyltransferase from Escherichia coli. Purification and properties of the enzyme. J Biol Chem. 1972;247:5675–5680. [PubMed] [Google Scholar]
- 5.Caillet J, Droogmans L. Molecular cloning of the Escherichia coli miaA gene involved in the formation of delta 2-isopentenyl adenosine in tRNA. J Bacteriol. 1988;170:4147–4152. doi: 10.1128/jb.170.9.4147-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore JA, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: A binding mechanism for recombinant enzyme. Biochemistry. 1997;36:604–614. doi: 10.1021/bi962225l. [DOI] [PubMed] [Google Scholar]
- 7.Motorin Y, Bec G, Tewari R, Grosjean H. Transfer RNA recognition by the Escherichia coli delta2-isopentenyl-pyrophosphate:tRNA delta2-isopentenyl transferase: Dependence on the anticodon arm structure. RNA. 1997;3:721–733. [PMC free article] [PubMed] [Google Scholar]
- 8.Soderberg T, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: Essential elements for recognition of tRNA substrates within the anticodon stem-loop. Biochemistry. 2000;39:6546–6553. doi: 10.1021/bi992775u. [DOI] [PubMed] [Google Scholar]
- 9.Soderberg T, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: Site-directed mutagenesis of highly conserved residues. Biochemistry. 2001;40:1734–1740. doi: 10.1021/bi002149t. [DOI] [PubMed] [Google Scholar]
- 10.Xie W, Zhou C, Huang RH. Structure of tRNA dimethylallyltransferase: RNA modification through a channel. J Mol Biol. 2007;367:872–881. doi: 10.1016/j.jmb.2007.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dihanich ME, et al. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golovko A, Hjalm G, Sitbon F, Nicander B. Cloning of a human tRNA isopentenyl transferase. Gene. 2000;258:85–93. doi: 10.1016/s0378-1119(00)00421-2. [DOI] [PubMed] [Google Scholar]
- 13.Holm L, Sander C. Protein structure comparison by allignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Methods in Enzymology. Vol 277. San Diego: Academic; 1997. Processing of x-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 15.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 16.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 17.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 18.Brünger AT, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.DeLano WL. The PyMOL Molecular Graphic System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 20.Carson M. Ribbon models of macromolecules. J Mol Graphics. 1987;5:103–106. [Google Scholar]
- 21.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.