Abstract
Diapause is the classic adaptation to seasonality in arthropods, and its expression can result in extreme lifespan extension as well as enhanced resistance to environmental challenges. Little is known about the underlying evolutionary genetic architecture of diapause in any organism. Drosophila melanogaster exhibits a reproductive diapause that is variable within and among populations; the incidence of diapause increases with more temperate climates and has significant pleiotropic effects on a number of life history traits. Using quantitative trait mapping, we identified the RNA-binding protein encoding gene couch potato (cpo) as a major genetic locus determining diapause phenotype in D. melanogaster and independently confirmed this ability to impact diapause expression through genetic complementation mapping. By sequencing this gene in samples from natural populations we demonstrated through linkage association that variation for the diapause phenotype is caused by a single Lys/Ile substitution in one of the six cpo transcripts. Complementation analyses confirmed that the identified amino acid variants are functionally distinct with respect to diapause expression, and the polymorphism also shows geographic variation that closely mirrors the known latitudinal cline in diapause incidence. Our results suggest that a naturally occurring amino acid polymorphism results in the variable expression of a diapause syndrome that is associated with the seasonal persistence of this model organism in temperate habitats.
Keywords: cline, diapause, life history, mapping, tradeoff
Natural populations encounter environmental stresses that diminish individual fitness, and these stresses are variable across space and time. It is predicted that the resulting natural selection often leads to a situation in which no genotype has the highest fitness across all environments, and polymorphism is maintained. This concept is pervasive in arguments about the evolution of life history variation and associated genetic tradeoffs (1). However, the expected molecular polymorphism associated with life history tradeoffs and adaptation remains elusive. Although we would expect these phenomena to be universal, the complexities of genetic dissection of such variation suggest that the best opportunity to identify the genetic basis for life history variation lies in the study of the genetic models in their natural populations (2).
Drosophila melanogaster is a human commensal that has spread from areas of Sub-Saharan Africa to Europe and Asia, possibly over the last 5,000 to 16,000 years, and into the Western Hemisphere and Australia in the past several hundred years (3–5). This worldwide expansion from the tropics has required adaptation to the pronounced seasonality present in temperate habitats, and there are many examples of both single-gene polymorphism and quantitative trait variation that show geographic patterns associated with the transition from tropical to temperate climates in this species (6, 7). There also is good evidence that D. melanogaster overwinters at the adult stage in temperate habitats (8, 9), and that temperate populations do not merely reflect recurrent migration from more moderate climates (10, 11). This overwintering survivorship clearly presents a variety of challenges, including the need for lifespan extension well beyond that typically measured in the laboratory, as well as increased physiological tolerance of extended exposure to suboptimal conditions (12).
The best-studied adaptation to seasonality in insects is the expression of a diapause syndrome. This phenotype is analogous, and potentially in part homologous, to the dauer stage in Caenorhabditis elegans; unlike the genes and pathways underlying dauer formation, however, very little is known about the genetic basis of diapause in Drosophila. Although many aspects of insect diapause vary across taxa, the expression of diapause is associated with a suite of physiological changes that allow persistence during periods of stress exposure (12). Once thought absent in this tropical species, reproductive diapause occurs in D. melanogaster and is cued by exposure of adults to short days and low temperatures (13). The expression of this diapause is under neuroendocrine control (14) and results in lifespan extension, delayed senescence, and increased stress resistance (15). The incidence of reproductive diapause exhibits a strong latitudinal cline in eastern North American populations (16), varying from ≈30% in southern Florida to 90% in New England. Diapause incidence also varies predictably with the season (high incidence in spring, lower in fall) in temperate orchard populations in Pennsylvania and New Jersey (17). The spatial and temporal patterns of diapause variation appear to reflect a robust series of life history tradeoffs between genotypes with a high and a low propensity to express diapause (17–19). These properties of reproductive diapause in D. melanogaster offer a unique opportunity to elucidate the genetic architecture of an important fitness trait with pleiotropic effects on life histories. Similarly, the global spread of a sophisticated genetic model organism presents us with a superior opportunity to study adaptation to novel environments that has emerged over a relatively short evolutionary period, possibly as recent as 2,000 generations in the New World.
Previous analyses in a set of isogenic D. melanogaster laboratory stocks and inbred lines had demonstrated that the genetic factors affecting diapause expression were entirely associated with the third chromosome (16). In this report, we use third-chromosome recombinants in a standardized genetic background, genetic complementation, and linkage association in a natural population to identify the gene responsible for diapause variation in natural populations.
Results
A set of 15 third-chromosome SNP markers was used to initially map the reproductive diapause phenotype in 250 recombinant inbred lines (RILs) generated from recombination between the parental lines w;6326;VT46 and w;6326;6326. These lines were selected because the inbred line VT46 does not exhibit diapause, whereas the isogenic line 6326 expresses diapause (16) and also forms a reference genome in other studies (20). Using these markers, multi-interval mapping (21) places a single identified quantitative trait locus (QTL) between markers at cytological band positions 90D1 and 92E8 (Fig. 1). A subset of 20 RILs possessed informative recombination events within the identified interval; the placement of an additional eight SNP markers in these lines provided a higher-resolution map (Fig. 2). This refined analysis eliminated a number of genes in the initial 248-kb region and identified a final candidate gene as couch potato (cpo).
Fig. 1.
Likelihood plot for the QTL analysis of reproductive diapause. The likelihood ratio, as calculated by multiple-interval mapping for ordinal traits (21) and implemented in QTL Cartographer v.2.5 (Category Trait Mapping, Forward Model), is plotted as a function of location in cM units on chromosome 3. The positions of the markers used in the analysis are indicated by arrows, and they correspond to cytological positions 61C, 64E, 65D, 68C, 75F, 83B, 85D, 87E, 89A, 90A, 90D, 92E, 94D, 98B, and 99A. The two markers that flank the identified QTL are indicated.
Fig. 2.
SNP genotypes across the right arm of the third chromosome and diapause incidence in the 20 RILs that were recombinant in the interval to which diapause mapped. The cytological position of each SNP marker is given (Left, top) and ranged from band 83B to 99A. Four SNPs were placed in the cpo gene and are listed as cpo1–4. The variation in diapause phenotype clearly maps only to the interval between SNP markers cpo3 and cpo4; this corresponds to the 3′ end of the cpo locus containing all of the coding sequence in exon 5.
To independently test the hypothesis that variation in diapause expression is associated with the cpo gene, we carried out four sets of genetic complementation studies using cpo P element and piggyBac transposon-derived constructs in a standardized genetic background. These analyses used the alleles cpoBG02810 (22) and cpoP3 (the latter is a precise excision of the P{w+GT} element in cpoBG02810), as well as the homozygous viable cpo hypomorphs cpov3, cpocp1, and cpocp2 (23). A second set of crosses used FLP-FRT site-specific recombination (20) of piggyBac elements to create duplications and deletions of both the cpo gene region (90C6–90E1) and the 5′ flanking region outside cpo (90B7–90C1). These studies all confirm that genetic modifications of the cpo gene alone cause pronounced and repeatable effects on diapause expression [supporting information (SI) Text and Fig. S1].
Since cpo dosage appears to influence diapause, we measured the cpo transcript levels in the progenitor VT46 (RIL line 201) and 6326 (RIL line 107) lines. The results of the RT-PCR that includes dp110 and Gapdh controls showed that 6326 third instar larvae and adults possess ≈15% of the level of cpo transcript as VT46 (SI Text and Fig. S2).
We next determined whether diapause phenotype in natural populations was associated with molecular polymorphism within the cpo locus. The cpo gene spans 84 kb and encodes six discrete transcripts. The major coding region (exon 5 of the cpo-RA transcript) is 449 aa separated by 37.5 kb from a scattered 3′ set of seven small exons encoding another 286 aa (depending on the transcript and annotation). Preliminary sequence analysis of exon 5 in a diverse set of lines identified extensive silent and amino acid polymorphism, as well as two major haplotypes. The central portion of the exon also contains several short tracts of polyglutamine repeats that vary in copy number. We began our linkage disequilibrium association analysis by examining this major exon and its variation.
We sequenced 3.5 kb spanning exon 5 from 35 third chromosomes from a single natural population (Davis Peach Farm; DPF) that were placed into the standardized genetic background (w;6326) used throughout this study. The DPF third-chromosome lines were then assayed for diapause expression under the standard induction conditions. The association between diapause phenotype and each of 192 polymorphic sites over the 3.5-kb region is depicted in Fig. 3. The only polymorphic sites that were significantly associated with diapause phenotype are located in the 3′ end of cpo exon 5, and these reflect the aforementioned major haplotypes. Two of these polymorphisms are nonconservative amino acid changes. Amino acid position 356 possesses an alanine-to-valine substitution; high-diapause lines tend to possess the valine codon that is derived with respect to other Drosophila species. However, the strongest association was observed for residue 462. High-diapause alleles, without exception, were characterized by the radical, charge-changing substitution of a lysine for isoleucine.
Fig. 3.
The association between diapause phenotype and each of 192 identified polymorphisms in the 3.5-kb region encompassing cpo exon 5. Each point represents the transformed P value resulting from a nominal logistic regression of nucleotide state on diapause incidence in the 35 extracted third chromosomes from the DPF population. The dashed line indicates significance threshold based on Bonferroni adjustment for multiple testing. The four sites (two synonymous and two nonsynonymous substitutions) that are significantly associated with diapause phenotype span 322 bp and are in significant linkage disequilibrium with one another (D ranges from 0.162 to 0.238, P < 0.0001 for each). The amino acid polymorphism at residue 462 is present in only the smallest cpo transcript (cpo-RH).
A series of nested regressions were used to evaluate the relative contribution of the four significantly associated nucleotide polymorphisms. When the nucleotide state at residue 462 was taken into consideration, no single other site accounted for any additional variance in diapause phenotype in the sequenced population [site 2069: F1,34 = 0.83, P > 0.368; site 2145 (amino acid position 356): F1,34 = 0.083, P > 0.775; site 2163: F1,34 = 1.53, P > 0.226]. In contrast, adding residue 462 did explain a significant additional variance in regressions for all other SNP sites. Thus, although the linkage group in the 3′ portion of exon 5 is significantly associated with diapause phenotype, this is driven exclusively by the polymorphic Lys/Ile change and disequilibrium between this and other polymorphic sites.
We have shown that diapause incidence varies with latitude in the eastern United States (16). Therefore, any putative quantitative trait nucleotide for diapause would be predicted to also exhibit a latitudinal cline in allele frequency, and this should extend to any polymorphisms that are in significant linkage disequilibrium with the causal site or sites. To address this expectation, we used restriction enzymes to screen five cpo SNPs (in cpo exons 1, 5, and 6 that span ≈70 kb) from 11 natural populations across the latitudinal gradient in the eastern United States (Fig. 4). Between exons 1, 5, and 6 there is only weak linkage disequilibrium among the SNPs, and within exon 5 the strength of association among sites becomes nonsignificant at distances of 1–2 kb, which is typical of other studies in D. melanogaster (24–27). The SNP showing the strongest association with latitude is the BsiE1 site in exon 5 (R2 = 0.95; F = 82.97, P < 1.2 × 10−5). This corresponds to the Ala/Val nonsynonymous polymorphism at residue 356, and the derived allele (Val) increases in frequency with latitude. This pattern is consistent with the observation that alleles increasing in frequency with spread of D. melanogaster into temperate regions are most often derived (7). Diapause incidence was previously observed to segregate as an autosomal dominant and varied from 33–80% in these populations (16). This relationship predicts an associated allele frequency cline in cpo of 0.18 to 0.55; the observed linear allele frequency cline of 0.21 to 0.53 is strikingly similar.
Fig. 4.
Variation in allele frequency for the five assayed cpo SNPs as a function of latitude of the populations. (A) Latitudinal clines for four reference SNPs in exons 1 (diamonds), 5 (circles), and 6 (5′ SNP, triangles; 3′ SNP, squares). The SNP in the 3′ end of exon 6 exhibits a significant cline in frequency (F1,10 = 11.82, P < 0.007, R2 = 0.568), whereas patterns of geographic allele frequency variation are homogeneous for the other three markers. (B) The SNP corresponding to the polymorphism at amino acid residue 356 exhibits an allele frequency cline (F1,10 = 82.97, P < 0.00001, R2 = 0.92), and it is the derived allele (Val) that increases positively with latitude. This polymorphism is in strong linkage disequilibrium with the amino acid polymorphism at residue 462 (D = 0.216, χ2 = 37.99, P < 0.0001). This suggests that the cpo462Ile/cpo462Lys polymorphism also varies significantly with geography. This is further supported by direct sequencing of cpo alleles in third chromosomes from the northernmost and southernmost populations sampled. The frequency of the derived cpo462Lys allele was 0.15 in the southern (n = 23 sequences) and 0.61 in the northern (n = 24 sequences) population (data not shown).
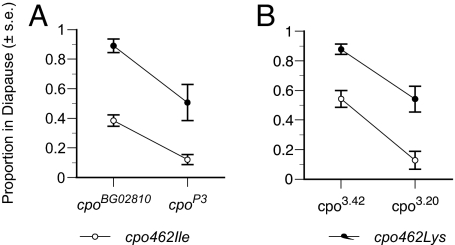
To further evaluate whether the two identified amino acid variants have differential effects on patterns of diapause expression, we used 12 cpo462Ile and 12 cpo462Lys alleles in two sets of complementation analyses (Fig. 5). It should be noted that the only nucleotide position that was identical among allelic replicates was the candidate site (residue 462). The cpo hypomorph, cpoBG02810, was associated with higher diapause expression than the wild-type revertant, cpoP3, created by the precise excision of the P element in cpoBG02810 (χ2 = 31.93, P < 0.0001). In this background, there was a pronounced difference in the level of diapause expression between cpo462Ile and cpo462Lys (χ2 = 74.51, P < 0.0001). The complementation of these alleles, as clearly indicated by a lack of a significant interaction term (χ2 = 0.0071, P > 0.93), demonstrated that the magnitude of the difference between the two natural cpo alleles was similar across all genetic backgrounds. Identical results were obtained in complementation studies using the cpo region piggyBac-derived deletion (cpo3.42) and duplication (cpo3.20). Again, the manipulation of the cpo locus predictably affected diapause expression (χ2 = 64.18, P < 0.0001). Patterns of diapause expression were distinct between cpo462Ile and cpo462Lys alleles (χ2 = 61.73, P < 0.0001), and this difference was maintained in both the cpo deletion and duplication backgrounds (interaction term χ2 = 0.93, P > 0.76).
Fig. 5.
Complementation analyses of 24 cpo alleles (12 cpo462Ile, 12 cpo462Lys) derived from the DPF population. (A) Diapause incidence in F1 progeny from crosses to the cpo hypomorphic allele cpoBG02810 and the wild-type revertant (cpoP3) created by the precise excision. Patterns of diapause expression exhibited codominance and stepwise effects of allelic substitution. Genotypes possessing two high-diapause alleles (cpoBG02810 or cpo462Lys) exhibited a strong diapause phenotype; the substitution of a low-diapause allele (cpoP3 or cpo462Ile) resulted in intermediate diapause expression, and substituting a second low-diapause allele resulted in a low-diapause phenotype. In both genetic backgrounds, the cpo462Lys allele is associated with a higher diapause expression than the cpo462Ile allele. (B) Patterns of diapause incidence in F1 progeny from crosses to the cpo deletion (cpo3.42) and duplication (cpo3.20) were qualitatively identical. The observation that the duplication of the cpo allele from the high-diapause parental strain (stock 6326) results in a reduction in diapause incidence again indicates that diapause phenotype may be governed by cpo expression.
Discussion
Our study shows that the variation in reproductive diapause expression is associated with the cpo gene and, moreover, a single linkage group in the 3′ end of cpo exon 5. The most likely cause is an isoleucine-to-lysine mutation in residue 462 that also shows a latitudinal cline consistent with that previously observed for diapause incidence. This does not rule out other genes contributing to the variation in diapause in natural populations, as these analyses were conducted in a standardized genetic background where the second and X chromosomes were isogenic. The manipulations of levels of cpo expression by hypomorphic alleles and gene duplications/deletions support the hypothesis that variation for diapause phenotype in D. melanogaster is associated with cpo expression level. This hypothesis was further supported by initial RT-PCR expression analyses of the parental lines used to map diapause to cpo. Given these results, it is interesting that the candidate site is an amino acid polymorphism in the coding region of a single transcript: the change in primary protein sequence may be of direct functional significance and/or affect some aspect of dosage. At present, there are no data to test these hypotheses.
The couch potato gene was first identified in a screen for genes expressed in sensory organ precursor cells during peripheral nervous development (23). It was shown subsequently to produce a nuclear protein and encodes an RNA-binding domain that is expressed in the peripheral and central nervous system of embryos, larvae, and adults, and in such tissues as the midgut, glia, and salivary glands (28). Also, cpo is highly expressed in the ring gland (29), the primary endocrine structure in Drosophila; this is particularly interesting because diapause is under neuroendocrine control. We also identified a series of cpo ecdysone response elements, suggesting that the effects of cpo on diapause in D. melanogaster may be mediated by ecdysteroids (30). Loss-of-function cpo mutations are lethal, and partial loss-of-function mutations generate a variety of behavioral phenotypes (23) and neurological abnormalities (31). The three predicted ORFs contain a nuclear localization sequence, polyglutamine OPA repeat regions, and an RRM-RNA recognition motif in five of the six transcripts (28). We see that the RRM region is very highly conserved across the 12 sequenced Drosophila genomes, but the OPA repeats show extensive copy number variation across taxa. Of particular note is that this alternate splicing predicts that the Ile/Lys amino acid polymorphism in residue 462 lies in the only protein lacking the RNA-binding domain. Our RT-PCR was not designed to target this transcript specifically.
Two recent studies have also investigated the genetic architecture for the reproductive diapause trait in D. melanogaster. Williams et al. (32) linked natural variation in reproductive diapause to the insulin-regulated phosphatidylinositol 3-kinase gene, dp110, also found close to cpo on the third chromosome at band 92F3. This association with diapause was further supported by manipulations of dp110 using transgene constructions. However, no naturally occurring amino acid sequence variation was observed in the dp110 gene, and no significant differences in dp110 expression were detected between the high- and low-diapause lines. In C. elegans the homologue of dp110, age-1, is associated with dauer formation, and this concurrence suggests a common role for this gene in countering environmental stress across species. The identification of dp110 as a diapause gene also suggests that aspects of diapause in Drosophila may be regulated by insulin signaling, and that C. elegans dauer and D. melanogaster diapause may be more than analogous phenotypes (33). Insulin signaling also has been shown to affect diapause expression in the mosquito Culex pipiens (34).
Diapause induction was also studied in association with allelic variation in the gene timeless (tim) (35), which encodes a light-responsive component of the circadian clock (36). As diapause expression is dependent on photoperiod, the authors hypothesized that this gene might impact reproductive diapause. In particular, a newly derived allele, ls-tim, increases in frequency across Europe, raising a possible association between temperate habitats, diapause incidence, and tim allele frequencies. Tauber et al. (36) showed that in three populations there was a significant relationship between the proportion expressing diapause, the testing photoperiod, and homozygous genotypes for the timeless alleles s-tim and ls-tim. It will be interesting to determine whether a similar association exists for the timeless alleles and diapause in North American populations, and in particular whether there is a latitudinal cline, as would be predicted.
In D. melanogaster the genetic variance associated with reproductive diapause has been shown to have profound pleiotropic effects on a number of other fitness-related phenotypes. These include lifespan, rates of senescence, fecundity profiles, development time, lipid content, and resistance to a variety of stressors (16–19). The absence of diapause induction in Drosophila simulans and African populations of D. melanogaster (19) suggests that this trait is of recent evolutionary origin (14) or is very rare in these populations. Our analysis identified a single gene and single nucleotide polymorphism that explains the observed variance in diapause expression in natural populations. However, it is likely that the diapause trait is polygenic in D. melanogaster, and that more genes will be discovered that modify its expression and variation. Nevertheless, the combined quantitative mapping analysis, complementation studies, detailed evaluation of molecular variation, and the predicted association of geographic variation are compelling in singling out cpo. A number of important questions remain. These concern the function of cpo, and in particular that of the splicing product cpo-RH that replaces the putative RNA-binding domain with a highly basic lysine/arginine rich terminus of 41 aa. Whether this product in turn indirectly impacts many downstream genes or acts more directly remains to be deciphered to explain the many pleiotropic effects that variation in this gene has on life history traits.
Materials and Methods
Stocks.
All lines and cpo alleles were placed into a common genetic background of w;6326 using marker-assisted introgression (37). The complementation analyses used cpoBG02810 (22) and cpoP3 (a precise excision of the P{w+GT} element in cpoBG02810). Mobilization of the P element used standard crosses to a male stock carrying a transposase source; excisions were recovered in males, extracted, and the background replaced. Excision was confirmed by PCR fragment analysis and sequencing of the gene region. The other cpo hypomorphs used (cpov3, cpocp1, and cpocp2) have been described (23). Line w;6326;VT46 (low-diapause parental genotype) is derived from an inbred line, VT46, collected in 1997 from Whiting, Vt (16). Line w;6326;6326 (high-diapause parental genotype) is a derivative of Bloomington line 6326 with the white-marked chromosome from Bloomington stock 2475.
To create site-specific duplications and deletions across the cpo gene region, piggyBac FLP-FRT-facilitated recombination (20) was carried out between insertions PBac{WH}CG7357[f00521] and PBac{WH}CG7785[f06154]. More than 100 third recombinant chromosome lines were recovered over the TM3 balancer chromosome, screened, and characterized. The manipulation of the cpo gene region resulted in a 214-kb duplication (designated cpo3.20) and deletion (cpo3.42) that span polytene bands 90C6 to 90E1. This region also covers four other genes (CG31246, tinc, Rim, and Dnase2). The selected gene deletion (cpo3.42) is homozygous, lethal, and is clearly visible in polytene preparations. The selected gene duplication (cpo3.20) was confirmed by PCR fragment analysis (20). Polytene chromosome preparations showed cpo3.20 as a duplication. Duplications and deletions of the 5′ region immediately flanking cpo were created by piggyBac FLP-FRT-facilitated recombination between insertions PBac{WH}CG14325[f03448] and PBac{WH}CG31249[f07289]. This region covers 58.14 kb and includes eight genes (alt, ald, CG31360, CG31249, CG31251, CG7655, CG7523, and CG14322) and 10 tRNAs. Allele cpo1.36 was confirmed by PCR and fragment analysis as a genomic duplication and was homozygous viable; allele cpo1.45 was confirmed as a deletion that was homozygous lethal. All third-chromosome lines were placed into the common w;6326 genetic background.
Mapping.
To map diapause QTL, a single F1 female from a cross between the high-diapause (w;6326;6326) and low-diapause (w;6326;VT46) parental lines was mated to w;6326;TM3/Dr males, and more than 250 recombinant male progeny were recovered. Each w;6326;TM3/+RIL male was mated to the w;6326;TM3/Dr balancer line to directly establish RILs for the third chromosome. All of the lines had identical X and second chromosomes. Using restriction site polymorphisms designed from Hoskins et al. (38) and from direct sequencing, 15 SNP sites were used initially to map 275 recombination events in 201 homozygous fertile lines. All lines were phenotyped for diapause, and we recovered 112 high-diapause and 79 low-diapause RILs. Each RIL was discretely partitioned into a high- or low-diapause class (see methods below). Thus, the phenotype data were binary in nature and were analyzed by multiple-interval mapping for ordinal traits (21). This method is a maximum likelihood-based approach that uses multiple marker intervals to determine significance for models that contain variable numbers of putative QTLs. Data were analyzed in Windows QTL Cartographer 2.5. Subsequently, an additional eight SNP markers were designed within the interval between cytological bands 90D1 and 92E8 by direct sequencing of parental lines.
Diapause Phenotyping.
The diapause induction phenotype of the RILs was tested in the homozygous condition (+RIL/+RIL), as well as over the TM3 balancer (+RIL/TM3), which expresses a low-diapause phenotype (16). These results indicated codominance of high- and low-diapause alleles: +HD/+HD flies were nonvitellogenic, but +HD/TM3 flies contained stage 8 oocytes and a mean of 1.2 stage 14 oocytes per set of ovaries. In contrast, +LD/+LD flies, as well as +LD/TM3 flies, were strongly vitellogenic, with an average of 14.3 stage 14 oocytes per ovary set. For purposes of mapping analysis, all lines were scored in the homozygous state. All lines and crosses that were phenotyped were maintained at low density in vial cultures on standard cornmeal-molasses medium at 25°C, 12L:12D. Females were collected within 2 h of eclosion from replicate vial cultures and placed at 11°C, 10L:14D, in Percival I36VL incubators. These females were dissected 4 weeks later, and the developmental status of the ovaries was assessed according to King (39). A female was scored as diapausing if egg development was arrested before vitellogenesis (before stage 8); a female was scored as nondiapausing if vitellogenin was observed in either ovary (stage 8 or later). At least 100 females were scored from 10 replicate cultures for each RIL and line cross. A bimodal distribution was observed: the incidence of diapause in a given RIL was either high (>70%) or low (<25%), with no intermediate frequencies observed. Thus, the trait was treated as binary and each RIL scored as a high- or low-diapause genotype.
Allele Frequency Clines.
Eleven populations from the US East Coast were included in this study. Ten were collected by Brian Verrelli in 1997 and have been described previously (7). The DPF population was collected in 2005 in Mount Sinai, NY, by Thomas Merritt. Isofemale lines were immediately established in the field from these populations, and the third chromosomes were extracted using a TM3/Dr stock. A subset (n = 46 for DPF, n = 48 for all other populations) of homozygous, viable, third-chromosome lines was used in the restriction fragment length polymorphism assay. The primer pairs and restriction enzymes are: exon 1 reference SNP, TspRI: 5′-GTCAAAGCGGGGAAAATATAGC-3′ and 5′-AAATGTGTGGTAAAACCTCTGCG-3′; exon 5 SNPs, AfeI: 5′-ACAGCAACACCAGTGCAGGAG-3′ and 5′-TCCATGCTCTGCGAAAGTCC-3′, and BsiEI (residue 356 Ala/Val): 5′-ACAGCAACACCAGTGCAGGAG-3′ and 5′-TCCATGCTCTGCGAAAGTCC-3′; and both exon 6 SNPs, DdeI: 5′-CGCTCAAAAGTAACGCTCGC-3′ and 5′-CTCACCGATGCAGTTTTGCC-3′.
Supplementary Material
Acknowledgments.
We thank J. True, A. Paaby, and A. Roddy for commenting on earlier versions of the manuscript, and J. Plotkin for suggestions regarding linkage association analyses. H. Bellen (Baylor College of Medicine, Houston, TX) kindly provided the cpo mutant allelic series. We would like to acknowledge both the Bloomington and Exelixis Stock Centers for providing critical transposon insertion lines. A portion of this work is presented in a PhD dissertation by C.-T.Z. to the Graduate Program in Ecology and Evolution, Stony Brook University. This study was supported by National Science Foundation Collaborative Grants DEB-0542859 (to P.S.S.) and DEB-0543050 (to W.F.E.), and a junior investigator award from the American Federation for Aging Research (to P.S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805485105/DCSupplemental.
References
- 1.Zera AJ, Harshman LG. The physiology of life history trade-offs in animals. Annu Rev Ecol Syst. 2001;32:95–126. [Google Scholar]
- 2.Feder ME, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nat Rev Genet. 2003;4:651–657. doi: 10.1038/nrg1128. [DOI] [PubMed] [Google Scholar]
- 3.Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudry E, Derome N, Huet M, Veuille M. Contrasted polymorphism patterns in a large sample of populations from the evolutionary genetics model Drosophila simulans. Genetics. 2006;173:759–767. doi: 10.1534/genetics.105.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- 6.Berry A, Kreitman M. Molecular analysis of an allozyme cline: Alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics. 1993;134:869–893. doi: 10.1093/genetics/134.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sezgin E, et al. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics. 2004;168:923–931. doi: 10.1534/genetics.104.027649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitrovski P, Hoffmann AA. Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: Evidence for clinal variation under semi-natural conditions. Proc Biol Sci. 2001;268:2163–2168. doi: 10.1098/rspb.2001.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouletreau-Merle J, Fouillet P, Varaldi J. Divergent strategies in low temperature environment for the sibling species Drosophila melanogaster and D. simulans: overwintering in extension border areas of France and comparison with African populations. Evol Ecol. 2003;17:523–548. [Google Scholar]
- 10.Ives PT. Genetic changes in American populations of Drosophila melanogaster. Proc Natl Acad Sci USA. 1954;40:87–92. doi: 10.1073/pnas.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ives PT. Further genetic studies on the South Amherst population of Drosophila melanogaster. Evolution. 1970;24:507–518. doi: 10.1111/j.1558-5646.1970.tb01785.x. [DOI] [PubMed] [Google Scholar]
- 12.Denlinger DL. Regulation of diapause. Annu Rev Entomol. 2002;47:93–122. doi: 10.1146/annurev.ento.47.091201.145137. [DOI] [PubMed] [Google Scholar]
- 13.Saunders DS, Henrich VC, Gilbert LI. Induction of diapause in Drosophila melanogaster: Photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc Natl Acad Sci USA. 1989;86:3748–3752. doi: 10.1073/pnas.86.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders DS, Gilbert LI. Regulation of ovarian diapause in Drosophila melanogaster by photoperiod and moderately low temperature. J Insect Physiol. 1990;36:195–200. [Google Scholar]
- 15.Tatar M, Chien SA, Priest NK. Negligible senescence during reproductive dormancy in Drosophila melanogaster. Am Nat. 2001;158:248–258. doi: 10.1086/321320. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt PS, Matzkin L, Ippolito M, Eanes WF. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution. 2005;59:1721–1732. [PubMed] [Google Scholar]
- 17.Schmidt PS, Conde DR. Environmental heterogeneity and the maintenance of genetic variation for reproductive diapause in Drosophila melanogaster. Evolution. 2006;60:1602–1611. [PubMed] [Google Scholar]
- 18.Schmidt PS, Paaby AB. Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution. 2008;62:1204–1215. doi: 10.1111/j.1558-5646.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt PS, Paaby AB, Heschel MS. Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution. 2005;59:2616–2625. [PubMed] [Google Scholar]
- 20.Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Wang S, Zeng ZB. Multiple-interval mapping for ordinal traits. Genetics. 2006;173:1649–1663. doi: 10.1534/genetics.105.054619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellen HJ, et al. The BDGP gene disruption project: Single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellen HJ, et al. The Drosophila couch potato gene: An essential gene required for normal adult behavior. Genetics. 1992;131:365–375. doi: 10.1093/genetics/131.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyashita NT, Aguade M, Langley CH. Linkage disequilibrium in the white locus region of Drosophila melanogaster. Genet Res. 1993;62:101–109. doi: 10.1017/s0016672300031694. [DOI] [PubMed] [Google Scholar]
- 25.Long AD, Lyman RF, Langley CH, Mackay TF. Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics. 1998;149:999–1017. doi: 10.1093/genetics/149.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langley CH, Lazzaro BP, Phillips W, Heikkinen E, Braverman JM. Linkage disequilibria and the site frequency spectra in the su(s) and su(w(a)) regions of the Drosophila melanogaster X chromosome. Genetics. 2000;156:1837–1852. doi: 10.1093/genetics/156.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macdonald SJ, Pastinen T, Long AD. The effect of polymorphisms in the enhancer of split gene complex on bristle number variation in a large wild-caught cohort of Drosophila melanogaster. Genetics. 2005;171:1741–1756. doi: 10.1534/genetics.105.045344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellen HJ, Kooyer S, D'Evelyn D, Pearlman J. The Drosophila couch potato protein is expressed in nuclei of peripheral neuronal precursors and shows homology to RNA-binding proteins. Genes Dev. 1992;6:2125–2136. doi: 10.1101/gad.6.11.2125. [DOI] [PubMed] [Google Scholar]
- 29.Harvie PD, Filippova M, Bryant PJ. Genes expressed in the ring gland, the major endocrine organ of Drosophila melanogaster. Genetics. 1998;149:217–231. doi: 10.1093/genetics/149.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard DS, et al. Vitellogenesis in diapausing and mutant Drosophila melanogaster: Further evidence for the relative roles of ecdysteroids and juvenile hormones. J Insect Physiol. 2001;47:905–913. [Google Scholar]
- 31.Glasscock E, Tanouye MA. Drosophila couch potato mutants exhibit complex neurological abnormalities including epilepsy phenotypes. Genetics. 2005;169:2137–2149. doi: 10.1534/genetics.104.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams KD, et al. Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proc Natl Acad Sci USA. 2006;103:15911–15915. doi: 10.1073/pnas.0604592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatar M, Yin C. Slow aging during insect reproductive diapause: Why butterflies, grasshoppers and flies are like worms. Exp Gerontol. 2001;36:723–738. doi: 10.1016/s0531-5565(00)00238-2. [DOI] [PubMed] [Google Scholar]
- 34.Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc Natl Acad Sci USA. 2008;105:6777–6781. doi: 10.1073/pnas.0802067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandrelli F, et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science. 2007;316:1898–1900. doi: 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- 36.Tauber E, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–1898. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- 37.Merritt TJ, Sezgin E, Zhu CT, Eanes WF. Triglyceride pools, flight and activity variation at the Gpdh locus in Drosophila melanogaster. Genetics. 2006;172:293–304. doi: 10.1534/genetics.105.047035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoskins RA, et al. Single nucleotide polymorphism markers for genetic mapping in Drosophila melanogaster. Genome Res. 2001;11:1100–1113. doi: 10.1101/gr.178001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King RC. Ovarian Development in Drosophila melanogaster. New York: Academic; 1970. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.