Abstract
Fanconi anemia (FA) is a genetically heterogeneous chromosome instability syndrome associated with congenital abnormalities, bone marrow failure, and cancer predisposition. Eight FA proteins form a nuclear core complex, which promotes tolerance of DNA lesions in S phase, but the underlying mechanisms are still elusive. We reported recently that the FA core complex protein FANCM can translocate Holliday junctions. Here we show that FANCM promotes reversal of model replication forks via concerted displacement and annealing of the nascent and parental DNA strands. Fork reversal by FANCM also occurs when the lagging strand template is partially single-stranded and bound by RPA. The combined fork reversal and branch migration activities of FANCM lead to extensive regression of model replication forks. These observations provide evidence that FANCM can remodel replication fork structures and suggest a mechanism by which FANCM could promote DNA damage tolerance in S phase.
Keywords: fanconi anemia, replication fork
A variety of structural and chemical alterations in DNA can hinder the progression of replication forks and precipitate the formation of gross chromosomal rearrangements. These hurdles impose distinct structural constraints in the template DNA, which elicit the action of diverse lesion bypass or lesion tolerance pathways (1, 2). Covalent links between complementary DNA strands constitute a unique challenge to replicating cells, because they preclude strand separation and, hence, completely block fork progression. In mammalian cells, the repair of DNA interstrand cross-links (ICLs) is thought to take place during S phase (3). The exact mechanism of repair is unknown, but it seems to involve the interplay of different pathways, with the homologous recombination machinery, translesion DNA polymerases, and the Fanconi anemia (FA) pathway all being required for ICL tolerance (4).
FA is a genetically heterogeneous inherited disorder, which combines congenital abnormalities, bone marrow failure, and a marked cancer predisposition (5–8). FA cells are prone to spontaneous and damage-induced chromosomal aberrations and are notoriously hypersensitive to DNA interstrand cross-linking agents. FA proteins can be classified into three groups (8). Group I includes FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM. These eight FA proteins form a nuclear core complex (9–11) whose integrity is required for the conjugation of a ubiquitin moiety to the group II proteins, FANCI and FANCD2 (12, 13). Group III consists of FANCD1 (BRCA2), FANCN (PALB2), and FANCJ (BRIP1), which do not play a role in FANCD2 monoubiquitination. BRCA2 regulates formation of RAD51 nucleoprotein filaments during homologous recombination (14, 15), PALB2 is necessary for the correct association of BRCA2 with chromatin (16), and BRIP1 is a BRCA1-associated DNA helicase that contributes to homologous recombination and cross-link repair (17, 18).
The FA core complex protein FANCM can specifically bind to model replication forks and Holliday junctions and move their junction points in an ATPase-dependent manner (19). Because ATP hydrolysis by FANCM is dispensable for the monoubiquitination of FANCD2 in vivo (20), it is unclear whether translocation of FANCM on DNA is essential for the FA pathway. The ATPase-mutant protein K117R FANCM, however, fails to complement the sensitivity of FANCM-depleted cells to ICLs (20), indicating that FANCM's translocase activity is required for DNA damage tolerance.
Whereas FANCM can migrate Holliday junctions in vitro, it is dispensable for the repair of I-SceI induced breaks by homologous recombination in DT40 cells (21). It is thus unlikely that FANCM promotes branch migration of Holliday junctions during recombinational repair of double-strand breaks. On the other hand, four-way junctions not only emerge from an exchange of strands between homologous duplexes, but can also arise from the regression of stalled replication forks (22). Because FANCM binds equally well to model replication forks as to Holliday junctions (19) and FA proteins are associated with the response to replication stress, a more likely possibility is that FANCM remodels the branch point of stalled replication forks.
Using plasmid-based DNA replication structures, we show that FANCM can convert a replication fork into a four-way junction. The fork remodeling activity of FANCM is discussed in the context of the replication stress response.
Results
FANCM-Mediated Dissociation of Displacement (D)-Loop Structures.
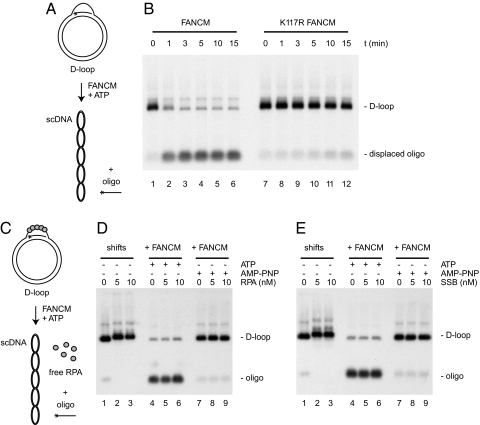
The first evidence of FANCM having a translocase activity was obtained by using a triple-stranded DNA structure (11). FANCM was found to catalyze ATP-dependent displacement of an oligonucleotide wound around B-form duplex DNA via Hoogsteen base pairing. Because Hoogsteen base pairs are qualitatively distinct from Watson–Crick base pairs, we first wanted to address whether FANCM was also able to disrupt Watson–Crick base pairing during translocation on duplex DNA. To this end, we generated a D-loop using RecA to assimilate an 80-mer oligonucleotide into supercoiled DNA (Fig. 1A).
Fig. 1.
D-loop dissociation catalyzed by FANCM. (A) Experimental scheme. Asterisks denote 32P label. Dissociation of D-loops results in the release of the labeled oligonucleotide. (B) Analysis of reaction products by agarose gel electrophoresis. FANCM (lanes 1–6) and K117R FANCM (lanes 7–12) were incubated with the D-loop substrate for the indicated time periods. (C) Experimental scheme. RPA binds to the single-stranded part of the D-loop. (D) Autoradiographs from agarose gels showing native RPA/D-loop complexes (lanes 1–3) and D-loop dissociation in the presence of RPA and ATP (lanes 4–6) or AMP-PNP (lanes 7–9) after deproteinization. (E) As in D, but D-loops were incubated with SSB instead of RPA.
When incubating the D-loop substrate with FANCM in the presence of ATP, the inserted oligonucleotide was rapidly displaced from the D-loop. After 3 min, ≈90% of the D-loop substrate was dismantled (Fig. 1B, lanes 1–6). As expected, D-loop dissociation depended on hydrolysis of ATP (Fig. 1D, lane 7) and on the integrity of the ATPase domain of FANCM (Fig. 1B, lanes 7–12). These results indicate that FANCM can remove DNA strands during translocation.
In vivo, the displaced strand within a D-loop will be bound by the ubiquitous single-strand DNA binding protein RPA. In such a situation, the presence of RPA could potentially occlude FANCM binding and inhibit D-loop disruption. We therefore asked whether FANCM was also able to dissociate D-loop structures in which the displaced strand is covered by RPA (Fig. 1C). To this end, we first confirmed by gel retardation analysis that D-loop molecules were bound by RPA (Fig. 1D, lanes 1–3). We then added FANCM and ATP to the preformed D-loop/RPA complexes. In the presence of RPA, FANCM was still able to remove the labeled oligonucleotide (Fig. 1D, lanes 4–6), indicating that RPA does not inhibit the binding to or the processing of D-loops by FANCM. The FANCM-mediated strand displacement from the D-loop/RPA complex did not depend on an interaction between FANCM and RPA, because D-loops bound by the bacterial single-strand DNA binding protein SSB were also efficiently disrupted by FANCM (Fig. 1E, lanes 4–6).
Fork Regression by FANCM.
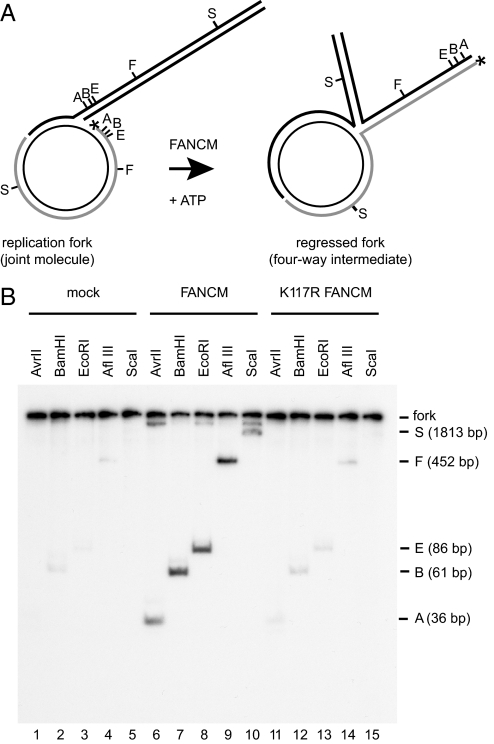
D-loops can be viewed either as structural mimics of recombination intermediates or as hemireplicated molecules in which the 3′-end of the invading strand is analogous to the leading strand of a replication fork. Hence, the ability of FANCM to disrupt D-loops might also reflect a role for FANCM in displacing nascent strands at replication forks. In addition to being able to disrupt D-loops, we have shown previously (19) that FANCM can translocate the branch point of synthetic three- and four-way junctions. We therefore examined the possibility that the coupling of these two activities of FANCM could promote replication fork regression. To investigate this, we constructed a joint molecule, which mimics a model replication fork, by annealing an open circular plasmid and a homologous linear duplex bearing complementary single-stranded gaps (Fig. 2A) (23, 24). The gapped circular plasmid is labeled at its 5′-end. In a fork-reversal reaction, the label is transferred into a fourth arm extruded at the branch point of the replication fork, and the extent of fork regression can be evaluated by restriction analysis. In our experimental conditions, spontaneous fork reversal was negligible (Fig. 2B, lanes 1–5). After incubation of the model replication fork with FANCM and ATP, digestion of reaction products with restriction enzymes released labeled duplex fragments of predefined lengths, which indicate fork regression (Fig. 2B, lanes 6–10). Fork-regression products were barely above background levels when the substrate was incubated with the ATPase mutant protein K117R FANCM, indicating that fork regression depends on ATP hydrolysis by FANCM (Fig. 2B, lanes 11–15).
Fig. 2.
FANCM can promote fork regression. (A) Experimental scheme. Fork reversal results in the transfer of the 32P label (asterisk) from the gapped molecule to the protruding arm of the four-way junction. Extent of fork regression can be estimated by restriction analysis with AvrII (A), BamHI (B), EcoRI (E), AflIII (F), and ScaI (S). (B) Autoradiographs from polyacrylamide gels showing fork regression in the absence of proteins (lanes 1–5), in the presence of FANCM (lanes 6–10), and in the presence of K117R FANCM (lanes 11–15).
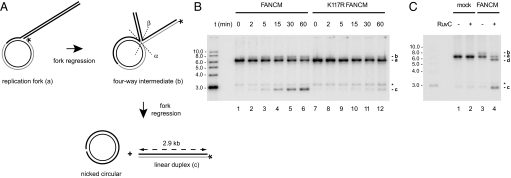
To test whether FANCM could bring the fork-regression reaction to completion on this substrate, reaction products were resolved by electrophoresis on agarose gels in the absence of restriction digestion. Complete fork regression is expected to produce linear duplex and nicked circular DNA, whereby the labeled strand is transferred from the gapped circular molecule to the 2.9-kb linear duplex DNA (Fig. 3A). In the presence of FANCM and ATP, the linear duplex product was detected already after 5 min and accumulated during a 1-h incubation period (Fig. 3B, lanes 1–6). In contrast, the ATPase mutant protein K117R FANCM displayed a greatly diminished fork-regression activity (Fig. 3B, lanes 7–12). Fork regression by FANCM was concentration dependent, whereas the fork-regression rate did not augment with increasing concentrations of K117R FANCM, nor did it exceed the level of spontaneous branch migration [supporting information (SI) Fig. S1].
Fig. 3.
Extensive fork regression catalyzed by FANCM. (A) Experimental scheme. Complete fork regression results in the formation of linear duplex and nicked circular DNA molecules. Asterisks indicate 32P label. Dashed lines show the two possible orientations of RuvC-mediated Holliday-junction cleavage, which result in the formation of nicked circular and labeled linear duplex DNA of 2.9 kb (cleavage in the α orientation) and labeled linear duplex DNA of 5.7 kb (cleavage in the β orientation), respectively. (B) Analysis of fork-regression products by agarose gel electrophoresis. FANCM (lanes 1–6) and K117R FANCM (lanes 7–12) were incubated with the replication-fork substrate for the indicated periods of time. The different labeled species are (top down) the four-way junction intermediate arising during the reaction (b), the original replication fork (a), the labeled gap molecule (asterisk; background in all lanes, because of incomplete annealing during fork generation), and the linear duplex molecule (c), the end product of fork regression. (C) Analysis of RuvC-mediated resolution of the Holliday junction-like intermediate formed by FANCM (b; lane 3) into linear duplex DNA of 2.9 kb (c), and linear duplex DNA of 5.7 kb (d), respectively (lane 4).
The appearance of linear duplex DNA was accompanied by a new smeary signal above the replication-fork structure, which likely reflects the formation of a four-way intermediate (Fig. 3 B and C, lane 3). To formally prove this, the bacterial Holliday junction resolvase RuvC was added to the reaction mixtures (Fig. 3C, lane 4). Indeed, addition of RuvC led to the disappearance of the smeary band and to the formation of the two expected Holliday junction resolution products: linear duplex DNA of 2.9 kb (cleavage along the α axis) and 5.7 kb (cleavage along the β axis). Resolution products were not detected in the absence of FANCM (Fig. 3C, lane 2). Taken together, these results indicate that FANCM can convert replication forks into four-way junctions, and carry out extensive fork regression.
Remodeling of RPA-Bound Replication Forks by FANCM.
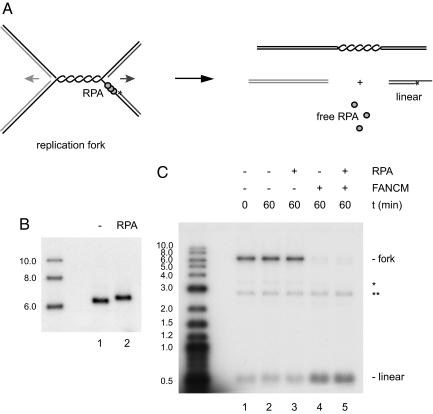
Whereas FANCM binds preferentially to branched DNA structures with at least two duplex arms (19), replication intermediates in vivo are expected to contain significant stretches of single-stranded DNA regions in the vicinity of the branch point, which will be bound by RPA. To mimic more closely the in vivo situation, we constructed a model replication fork that contains a 114-nt gap ahead of the lagging strand. To this end, two open circular plasmids, identical in sequence but containing single-stranded gaps of different sizes, were annealed, treated with topoisomerase I, and then digested by AflIII (Fig. S2). Fork regression results in the transfer of the radioactive label from the replication-fork structure to a 0.5-kb linear duplex molecule (Fig. 4A). Incubation of the substrate with FANCM in the presence of ATP led to efficient fork regression (Fig. 4C, lane 4). Thus, FANCM can reverse replication forks that contain either a 14-nt gap ahead of the leading strand (Figs. 2 and 3) or a 114-nt gap ahead of the lagging strand (Fig. 4), indicating that symmetry of the replication fork is not required for FANCM to promote fork reversal.
Fig. 4.
Regression of an RPA-covered replication fork. (A) Experimental scheme. RPA binds to the 114-nt gap on the lagging strand template of the replication fork. Fork regression results in the transfer of the 32P label (asterisk) from the replication fork to the 0.5-kb linear duplex product. (B) Binding of RPA to the replication fork was verified by agarose gel-shift analysis. (C) Analysis of fork regression by agarose gel electrophoresis. FANCM and the replication-fork substrate were incubated for the indicated time periods without (lane 4) and with (lane 5) preincubation of the replication fork with RPA. As a control, the substrate was incubated without FANCM in the absence (lanes 1 and 2) or presence (lane 3) of RPA. Asterisks indicate background of labeled gapped DNA (*) and labeled linear DNA (**).
We next went on to examine whether RPA bound to the lagging-strand gap might inhibit the ability of FANCM to translocate the branch point of the replication fork and promote fork regression. As expected, analysis by gel retardation showed that the model replication fork was efficiently bound by RPA (Fig. 4B). When the fork containing the 114-nt gap ahead of the lagging strand was preincubated with RPA, FANCM was still able to promote fork regression (Fig. 4C, lane 5), suggesting that FANCM can displace RPA from the lagging strand of a model replication fork during fork reversal.
Discussion
The experiments described here show that FANCM can convert a replication-fork structure into a four-way junction, which implies the concerted displacement and annealing of nascent and parental DNA strands. Importantly, fork reversal also occurs on a substrate that mimics a naturally occurring replication fork with a gap ahead of the lagging strand, indicating that FANCM can displace nascent strands from replication forks without the requirement of symmetry at the branch point. This feature is also reflected in the ability of FANCM to displace the inserted oligonucleotide from a D-loop, a structure that can be regarded as a hemireplicated molecule. Moreover, FANCM can regress an asymmetric fork that is bound by the single-strand DNA binding protein RPA, as would be the case in vivo, suggesting that FANCM can displace RPA during fork remodeling. Finally, we find that the ability of FANCM to promote reversal of replication forks combines with its branch migration activity and results in extensive fork regression.
In E. coli, it has been clearly established that under certain circumstances, such as impairment of helicase or replisome function, stalled replication forks can regress to form a Holliday-junction-like structure (25–27). RecG seems to play a crucial role in this context (28, 29). In contrast to prokaryotes, the occurrence of fork regression in eukaryotes is under debate. Studies in S. cerevisiae demonstrate that extensive fork regression upon hydroxyurea treatment occurs more frequently in checkpoint-deficient than in wild-type cells (30, 31), suggesting that at least extensive fork regression is a pathological event, which is usually prevented by a functional checkpoint.
Several lines of evidence suggest that during replication of damaged DNA, the FA core complex promotes DNA damage tolerance via mutagenic processes, thereby preventing gross chromosomal rearrangements (32, 33). First, deletions represent the most prevalent class of spontaneous mutations in FA cells, whereas base substitutions usually predominate in cells derived from healthy donors (34, 35). Second, the knockdown of fancg in hamster CHO cells leads to an increase in the ratio of spontaneous deletions versus base substitutions (36). Third, integrity of the FA pathway is required for normal mutagenic tolerance to photoactivated psoralens (37, 38) and UVC (39). Fourth, the human FA core complex promotes assembly of the translesion DNA polymerase Rev1 into nuclear foci, independently of FANCD2 and PCNA monoubiquitination (39).
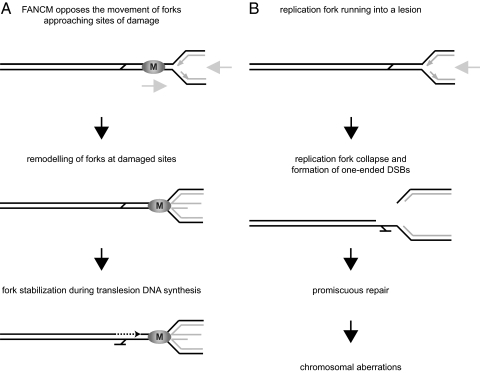
The data presented in this study show that FANCM has the ability to reverse replication forks in vitro. In a cellular context, however, where replication forks are covered by the replisome, FANCM might antagonize the movement of replication forks, rather than actually promote fork reversal. In a speculative scenario (Fig. 5), FANCM could counteract the advancement of replication forks and prevent them from running into lesions and collapsing. Under certain circumstances, e.g., when the replication fork is stalled at a DNA ICL, the action of FANCM might eventually result in fork reversal. Because the replisome is likely to obstruct access to the damaged template DNA, the remodeling of replication structures might be required to dislodge the replisome from the junction point, thereby allowing DNA caretaker proteins to signal, repair, and/or bypass the lesion. An analogous situation occurs when chemical alterations in DNA block RNA polymerase II elongation. The Swi/Snf family ATPase CSB (Rad26 in yeast) is required to remove stalled RNA polymerase II molecules to allow repair by the nucleotide excision repair machinery (40, 41).
Fig. 5.
Model for the role of FANCM in the response to replication stress. (A) In this model, FANCM antagonizes the movement of replication forks advancing toward sites of damage, which might result in fork reversal once the replication fork is stalled. Remodeling of forks by FANCM would stabilize the stalled replication fork and provide time and space for the lesion site to be repaired. (B) Replication forks running into damaged sites may lead to replication-fork collapse and the formation of one-ended DSBs. These are susceptible to promiscuous repair events and are potentially the cause of chromosomal aberrations.
The remodeling of replication forks by FANCM could thus promote the action of translesion polymerases in the context of linear duplex DNA. In contrast, in the absence of FANCM where stalled forks would not be regressed, attempts to repair lesions at the junction point of a stalled replication fork could result in the collapse of the replication fork and the formation of a one-ended double-strand break. One-ended double-strand breaks, however, are potentially dangerous to a cell because they carry the risk of promiscuous recombination events, which can lead to chromosomal aberrations, a hallmark of FA cells.
Methods
Proteins.
FANCM, K117R FANCM, and RPA were purified as described (19, 42), RuvC was a gift from S.C. West (Cancer Research UK, South Mimms, UK), SSB was from Promega, and RecA was from NEB.
Generation of D-loops.
The D-loop substrate was essentially generated as described (43). Briefly, 25 nM 5′-end-labeled invading oligonucleotide (5′- GAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTG-3′) was coated with 5.2 μM RecA for 5 min at 37°C. Then 36 nM supercoiled pUC19 plasmid was added for 10 min at 37°C. The reaction buffer contained 25 mM Tris (pH 7.4), 10 mM MgCl2, 1 mM DTT, 2.5 mM AMP-PNP, and 100 μg/ml BSA. The DNA was deproteinized with 2 mg/ml proteinase K and 0.4% SDS for 15 min at 37°C. Reaction products were gel-purified, precipitated, and resuspended in 10 mM Tris (pH 8.0).
Binding of RPA and SSB to D-loops.
3 nM purified D-loop was incubated with 0, 5, or 10 nM RPA and SSB, respectively, in buffer A [25 mM Na2HPO4/NaH2PO4 (pH 7.0), 75 mM NaCl, 5% glycerol, 0.005% Nonidet P-40, 0.25 mM EDTA, 1 mM TCEP, 100 μg/ml BSA] for 45 min at room temperature. Native protein/D-loop complexes were either resolved on a 0.8% agarose gel or used for the D-loop dissociation assay.
D-loop Dissociation Assay.
1 nM FANCM or K117R FANCM and 1.5 nM purified D-loop were incubated at 37°C for the indicated periods of time in buffer A containing 0.5 mM MgCl2 and 1 mM ATP or AMP-PNP, as indicated. Reaction products were deproteinized as described above and resolved on a 0.8% agarose gel. For the assays in the presence of RPA or SSB, FANCM was added to the preformed protein/D-loop complexes to a final concentration of 0.25 nM. Reactions were carried out for 15 min at 37°C in the same conditions as described above and resolved on a 0.8% agarose gel.
Generation of Joint Molecule.
The substrate was created essentially as described (23, 24). Briefly, pG46 was gapped by the nicking endonuclease Nt.BbvCI to yield pG46B and labeled with T4 polynucleotide kinase. pG68 was gapped by digestion with Nb.BbvCI, yielding pG68A, and subsequently linearized with XhoI. The gapped plasmids were then annealed together for 15 min at 65°C and for 15 min at 37°C.
Fork Regression Assays with Joint Molecule.
Reactions (10 μl) were performed in buffer A and contained 2 nM protein, 0.5 nM 5′-32P labeled DNA substrate, 10 nM oligonucleotide (as competitor), 0.5 mM MgCl2, and 1 mM ATP. Reactions were carried out at 37°C for 30 min, except when otherwise stated. Reactions were deproteinized, and reaction products were resolved on a 0.8% agarose gel containing 0.5 μg/ml EtBr. Alternatively, deproteinized reaction products were subjected to phenol extraction, precipitated, resuspended in water, and digested with restriction enzymes according to the manufacturer's instructions. Digested reaction products were then separated on a 10% polyacrylamide gel. For Holliday junction resolution, 5 nM FANCM was incubated for 20 min in the same reaction conditions as described above, before 10 μl of a mix containing 200 nM RuvC, 40 mM Tris·HCl (pH 7.5), 30 mM MgCl2, 2 mM DTT, and 200 μg/ml BSA were added. Reactions were incubated for another 60 min at 37°C, deproteinized, and run on a 0.8% agarose gel containing 0.5 μg/ml EtBr.
Generation of Replication Fork with Lagging-Strand Gap.
To generate this substrate (pRF-lag), a 247 bp cassette (RF-lag100) was cloned into pUC18 as described in Fig. S2. The sequences of RF-lag100 and the oligonucleotides used for its construction are available in Tables S1 and S2.
Binding of RPA to Gapped Replication Fork.
Purified DNA substrate (0.5 nM) was incubated with 50 nM RPA in buffer A for 30 min at room temperature. RPA/DNA complexes were resolved on a 0.6% agarose gel.
Fork Regression Assays with Gapped Replication Fork.
5′-32P labeled DNA substrate (0.5 nM) was preincubated with 0 or 50 nM RPA in buffer A for 30 min at room temperature; 2 nM FANCM, 0.5 mM MgCl2, and 1 mM ATP were added in the same reaction buffer as above. Reactions were incubated at 37°C for 0 or 60 min and subsequently de-proteinized. Reaction products were resolved on a 0.8% agarose gel.
Supplementary Material
Acknowledgments.
We thank Stephen C. West for reagents and all of the laboratory members for critical reading of the manuscript. This work was supported by the Swiss National Science Foundation (PP00A-102891 and PP00A-118991). L.W. is funded by Cancer Research U.K.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804777105/DCSupplemental.
References
- 1.Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amsterdam) 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 2.Lambert S, Froget B, Carr AM. Arrested replication fork processing: Interplay between checkpoints and recombination. DNA Repair (Amsterdam) 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 3.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niedzwiedz W, et al. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: New faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 7.Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25:5875–5884. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- 8.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 9.Meetei AR, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 10.Meetei AR, et al. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- 11.Meetei AR, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 13.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 15.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 16.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 18.Litman R, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 20.Xue Y, Li Y, Guo R, Ling C, Wang W. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum Mol Gen. 2008;17:1641–1652. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosedale G, et al. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 22.Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 23.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 24.Blastyak A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores MJ, Bierne H, Ehrlich SD, Michel B. Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J. 2001;20:619–629. doi: 10.1093/emboj/20.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grompone G, Ehrlich D, Michel B. Cells defective for replication restart undergo replication fork reversal. EMBO Rep. 2004;5:607–612. doi: 10.1038/sj.embor.7400167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 29.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 30.Lopes M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 31.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 32.Hinz JM, Nham PB, Salazar EP, Thompson LH. The Fanconi anemia pathway limits the severity of mutagenesis. DNA Repair (Amsterdam) 2006;5:875–884. doi: 10.1016/j.dnarep.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 33.Thompson LH, Hinz JM, Yamada NA, Jones NJ. How Fanconi anemia proteins promote the four Rs: Replication, recombination, repair, and recovery. Environ Mol Mutagen. 2005;45:128–142. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- 34.Papadopoulo D, Guillouf C, Mohrenweiser H, Moustacchi E. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc Natl Acad Sci USA. 1990;87:8383–8387. doi: 10.1073/pnas.87.21.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laquerbe A, Sala-Trepat M, Vives C, Escarceller M, Papadopoulo D. Molecular spectra of HPRT deletion mutations in circulating T-lymphocytes in Fanconi anemia patients. Mutat Res. 1999;431:341–350. doi: 10.1016/s0027-5107(99)00177-3. [DOI] [PubMed] [Google Scholar]
- 36.Hinz JM, Nham PB, Urbin SS, Jones IM, Thompson LH. Disparate contributions of the Fanconi anemia pathway and homologous recombination in preventing spontaneous mutagenesis. Nucleic Acids Res. 2007;35:3733–3740. doi: 10.1093/nar/gkm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papadopoulo D, Laquerbe A, Guillouf C, Moustacchi E. Molecular spectrum of mutations induced at the HPRT locus by a cross-linking agent in human cell lines with different repair capacities. Mutat Res. 1993;294:167–177. doi: 10.1016/0921-8777(93)90025-c. [DOI] [PubMed] [Google Scholar]
- 38.Guillouf C, Laquerbe A, Moustacchi E, Papadopoulo D. Mutagenic processing of psoralen monoadducts differ in normal and Fanconi anemia cells. Mutagenesis. 1993;8:355–361. doi: 10.1093/mutage/8.4.355. [DOI] [PubMed] [Google Scholar]
- 39.Mirchandani KD, McCaffrey RM, D'Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amsterdam) 2008 doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tijsterman M, Brouwer J. Rad26, the yeast homolog of the cockayne syndrome B gene product, counteracts inhibition of DNA repair due to RNA polymerase II transcription. J Biol Chem. 1999;274:1199–1202. doi: 10.1074/jbc.274.3.1199. [DOI] [PubMed] [Google Scholar]
- 41.Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- 42.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: Expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 43.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.