Abstract
We report the discovery and optimization of a potent inhibitor against the papain-like protease (PLpro) from the coronavirus that causes severe acute respiratory syndrome (SARS-CoV). This unique protease is not only responsible for processing the viral polyprotein into its functional units but is also capable of cleaving ubiquitin and ISG15 conjugates and plays a significant role in helping SARS-CoV evade the human immune system. We screened a structurally diverse library of 50,080 compounds for inhibitors of PLpro and discovered a noncovalent lead inhibitor with an IC50 value of 20 μM, which was improved to 600 nM via synthetic optimization. The resulting compound, GRL0617, inhibited SARS-CoV viral replication in Vero E6 cells with an EC50 of 15 μM and had no associated cytotoxicity. The X-ray structure of PLpro in complex with GRL0617 indicates that the compound has a unique mode of inhibition whereby it binds within the S4-S3 subsites of the enzyme and induces a loop closure that shuts down catalysis at the active site. These findings provide proof-of-principle that PLpro is a viable target for development of antivirals directed against SARS-CoV, and that potent noncovalent cysteine protease inhibitors can be developed with specificity directed toward pathogenic deubiquitinating enzymes without inhibiting host DUBs.
Keywords: ubiquitin-specific protease, noncovalent cysteine protease inhibitor, severe acute respiratory syndrome antiviral, X-ray structure
Proteolytic enzymes are key regulators of physiological processes in humans and are also essential for the replication of pathogenic viruses, parasites, and bacteria that cause infectious disease. Their importance in such fundamental processes has been widely recognized, and as a result, since the mid-1990s, >30 new protease inhibitors have entered the marketplace for the treatment of a wide spectrum of diseases including HIV/AIDS (1). These drugs target at least 10 structurally diverse proteases representing every class of protease (metallo, aspartic, serine, and threonine), with the exception of the cysteine proteases.
Historically, the development of cysteine protease inhibitors with drug-like properties has been plagued with a number of challenges, most notable being their toxicity and lack of specificity due to covalent modification of untargeted cysteine residues. As a result, only a small number have entered into late-phase clinical trials thus far. Despite such challenges, cysteine proteases hold significant promise as drug targets, because they are involved in many disease-related processes and as such, a number of compounds have entered into preclinical evaluation or development (2). To capitalize on the success of targeting proteases as therapeutics, especially in the antiviral drug discovery area, we report here the discovery and optimization of a noncovalent lead inhibitor of the papain-like protease (PLpro) from the coronavirus that causes severe acute respiratory syndrome (SARS), SARS-CoV.
Although the spread of SARS-CoV, which caused the pandemic of 2002–2003, was effectively halted within a few months after the initial outbreaks, the recent isolation of strains from zoonotic origins thought to be the reservoir for SARS-CoV (3, 4) accentuates the possibility of future retransmissions of SARS-CoV, or related coronaviruses, from animals to humans. The development of novel antivirals against SARS-CoV is therefore an important safeguard against future outbreaks and pandemics, but so far potent antivirals against SARS-CoV with efficacy in animal models have not yet been developed.
Because of the complex nature of SARS-CoV replication, a number of processes are considered essential to the coronaviral lifecycle and therefore provide a significant number of targets for inhibiting viral replication. An early and essential process is the cleavage of a multidomain viral polyprotein into 16 mature components termed nonstructural proteins (nsps), which assemble into complexes to execute viral RNA synthesis (reviewed in refs. 5 and 6). Two cysteine proteases that reside within the polyprotein, a PLpro and a 3C-like protease (3CLpro), catalyze their own release and that of the other nsps from the polyprotein, thereby initiating virus-mediated RNA replication (Fig. 1A). Despite numerous biochemical, structural, and inhibitor-development studies directed at 3CLpro (reviewed in ref. 7), potent antivirals that directly target 3CLpro have yet to be developed. In contrast, structural and functional studies directed at PLpro are far less numerous but have established important roles for PLpro beyond viral peptide cleavage including deubiquitination, deISGylation, and involvement in virus evasion of the innate immune response (8–12). Recent studies have also shown that an enzyme homologous to PLpro from the human coronavirus 229E, PLP2, is essential for viral replication (13).
Fig. 1.
High-throughput screening using a ubiquitin-like peptide substrate identifies a PLpro inhibitor. (A) Schematic of the SARS-CoV polyprotein, with cleavage sites of the 2 proteases, PLpro (red) and 3CLpro (blue), indicated by color-coded arrows and dashed lines. Nsp numbering is indicated below the polyprotein. (B) The sequences of the 3 PLpro polyprotein cleavage sites are aligned with the peptide substrate used for high-throughput screening. Conserved residues are shown in bold. (C) Results from screening 50,080 compounds in duplicate for inhibition of PLpro activity. The replicate plot shows the percentage inhibition of PLpro by each compound. The structure of the lead compound 7724772 is shown, and its activity is plotted in red. The hit zone for the assay (>35% inhibition) is indicated by a white box.
Results and Discussion
Identification of a SARS-CoV PLpro Inhibitor.
The numerous functions and requisite roles of PLpro in viral replication and pathogenesis suggest that PLpro may serve as an attractive target for antiviral drugs. Therefore, to identify potential inhibitors of PLpro, we developed a sensitive fluorescence-based high-throughput screen based on our initial work, which showed that PLpro is more catalytically active toward ubiquitin-derived substrates relative to polyprotein-based peptide substrates (11). We used a commercially available peptide substrate representing the 5 C-terminal residues of ubiquitin derivatized with a C-terminal 7-amido-4-methylcoumarin (AMC) fluorogenic reporter group (Fig. 1B). To validate the assay, we performed a prescreen of 10,000 diverse compounds in the absence of reducing agent to assess the reactivity of PLpro's active site cysteine with electrophiles common to many diverse compound libraries. The vast majority of hits displaying >60% inhibition were determined either to be known electrophiles or to exhibit no inhibitory activity in the presence of reducing agent during follow-up analysis (data not shown). Although the majority of cysteine protease inhibitors described in the literature act covalently, the inherent electrophilic nature of these compounds often leads to nonspecific reactivity with unintended nucleophiles, resulting in adverse side effects (14). In the interest of discovering and developing only noncovalent inhibitors against PLpro, 5 mM DTT was incorporated into all subsequent primary high-throughput screens.
A primary screen of 50,080 diverse, lead-like, and drug-like compounds was performed in 384-well plates, in duplicate, which resulted in a Z′-factor of 0.8 [supporting information (SI) Fig. S1). Only a small number of compounds, 17 total (0.04%), were found to have >35% inhibitory activity toward PLpro (Fig. 1C). These 17 compounds were subjected to a series of confirmatory and secondary assays to test for interference of AMC fluorescence, dose-dependent inhibition of PLpro, and inhibition of the enzyme in the presence of Triton-X, a test to eliminate promiscuous inhibitors (15). Of the original 17 hits, 9 compounds were found to interfere with the fluorescence of the AMC reporter group, and of the remaining 8 compounds, only 2 reproducibly inhibited PLpro in a dose-dependent manner, both in the absence and presence of Triton-X. Compound 7724772, a racemic mix of 2-methyl-N-[1-(2-naphthyl)ethyl]benzamide (Fig. 1C), inhibited PLpro with an IC50 value of 20.1 ± 1.1 μM and was chosen for further development (Fig. 2A).
Fig. 2.
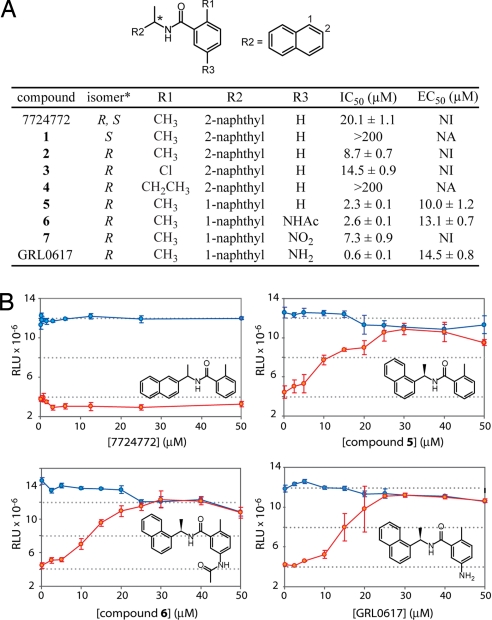
PLpro inhibitors follow a structure–activity relationship and have antiviral activity against SARS coronavirus. (A) The structure–activity relationship of synthetic compounds based on the HTS lead compound 7724772 is shown in a table format. An asterisk indicates the position of the chiral carbon in the chemical structure at top. IC50 values represent inhibitory activity of PLpro in vitro. EC50 values represent antiviral activity of the compounds against SARS-CoV. NI, no inhibition; NA, not assayed. (B) SARS-CoV infected (red points) and mock-infected (blue points) Vero E6 cells were incubated in the presence of inhibitor compounds 7724772, 5, 6, or GRL0167 at the concentrations indicated for 48 h. Cell viability was measured 48 h after infection by using the CellTiter-Glo Luminescent Cell Viability Assay (Promega), and output was expressed as RLU. The error bars represent the standard deviation between triplicate samples. Structures of the tested inhibitors are included as Insets.
Because compound 7724772 contains a stereogenic center adjacent to the carboxamide moiety, both the (S) enantiomer, 1, and the (R) enantiomer, 2, were synthesized to determine whether PLpro exhibits any stereospecificity. At 100 μM, the (S) enantiomer was found to have only slight inhibitory activity (14%), whereas the (R) enantiomer inhibited PLpro activity >90% with an IC50 value of 8.7 ± 0.7 μM (Fig. 2A). The stereochemical preference for the (R) over the (S) enantiomer established 2 as a lead compound and is consistent for a protein that utilizes the 4-location model for stereospecific recognition (16). Therefore, compound 2 served as the basis for further rounds of synthetic optimization.
Synthetic Optimization of Lead Increases Potency.
The preference for a methyl group over hydrogen at the R1 position of 7724772 was found to be required for potency based on a structure–activity analysis of the primary high-throughput screen (HTS) data and on the follow-up studies of the primary screen. Therefore, other substituents at the R1 position were explored. The substitution of a chlorine atom at R1, 3, resulted in a 2-fold decrease in inhibitory potency (IC50 = 14.5 ± 0.9 μM) compared with 2, and the substitution of a larger ethyl group, 4, almost abolished activity (IC50 > 100 μM), suggesting that the optimum size of a substituent at the R1 position is a methyl group. Compound 2 was further optimized by probing the effect of changing the orientation of the relatively bulky naphthalene group (R2 in Fig. 2A). Replacing the 2-naphthyl group of 2 with a 1-naphthyl to form compound 5 resulted in a 4-fold increase in inhibitory potency (IC50 = 2.3 μM). Finally, the addition of a second functional group to the R3 position of the ortho-methyl benzene ring of 5 was explored. Addition of an NHAc group, 6, was found to have little effect on the IC50 value (2.6 ± 0.1 μM) compared with 5, whereas the addition of a nitro group, 7, decreased activity nearly 3-fold. In contrast, the addition of an amino group at the same position increased the inhibitory potency almost 4-fold (IC50 = 0.6 ± 0.1 μM), suggesting that an additional hydrogen bond may be formed in the enzyme–inhibitor complex. The resulting compound, designated GRL0617, is the most potent inhibitor in the series (Fig. 2A) and was therefore subjected to more extensive mechanistic and structural analyses.
Mechanism of Inhibition of Most Potent Lead-GRL0617.
To characterize the mechanism of inhibition by the GRL0617 class of compounds, kinetic and biochemical studies of the enzyme–inhibitor complexes were performed. A kinetic study of PLpro activity, in which the concentration of its optimal substrate, ISG15-AMC, was varied relative to fixed concentrations of inhibitor, reveals that GRL0617 is a potent competitive inhibitor of PLpro with a Ki value of 0.49 ± 0.08 μM (Fig. S2A). Progress curve analysis also suggested that the inhibitor is noncovalent. To further probe for noncovalent inhibition of PLpro by GRL0617, PLpro was incubated for 1 h with 12 μM GRL0617 (>20-fold the Ki value), and the resulting complex was dialyzed to allow the inhibitor to diffuse away and therefore restore enzymatic activity (Fig. S2B). Approximately 25% of the PLpro activity was recovered after 3 h of dialysis compared with a recovery of 100% for the enzyme without inhibitor. The inability to fully recover PLpro activity after 3 h could be either a result of a slow off-rate of the inhibitor from the PLpro-GRL0617 complex or a result of covalent modification of the active-site cysteine by a direct reaction with the inhibitor or by indirect oxidation. Because GRL0617 has no apparent thiol-reactive groups, the inability to recover enzymatic activity is likely a result of both mechanisms, a slow off-rate of inhibitor and oxidation of the cysteine, despite the use of reducing agents throughout all studies. Evidence for cysteine oxidation is revealed in the structural studies described below.
SARS-CoV Antiviral Activity.
To probe the antiviral activity of the PLpro inhibitors, several compounds were assayed for their ability to rescue cell culture from SARS-CoV infection. The viability of virus-infected Vero E6 cells as a function of inhibitor concentration was measured relative to mock-infected cells by using a luminescence assay, which allows for the evaluation of both inhibitor efficacy and cytotoxicity (Fig. 2B). Most significantly, GRL0617 and compounds 5 and 6 display significant antiviral activity with EC50 values ranging from 10 to 15 μM without toxicity up to the highest concentration tested (Fig. 2 A and B). Notably, the increasing antiviral potency correlates with the in vitro inhibition of PLpro, suggesting that the compounds work directly on the enzyme in cells.
Structural Basis for Potent Inhibition of SARS-CoV PLpro Revealed by X-Ray Crystallography.
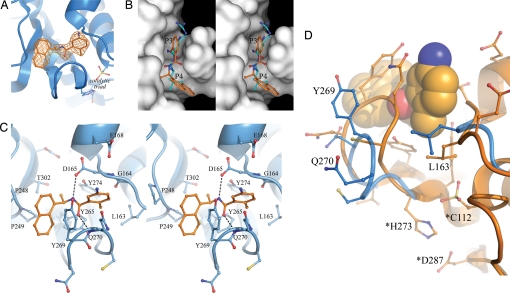
To better understand the molecular basis for inhibition of PLpro by GRL0617 and the structure–activity relationship described above, we determined the X-ray structure of the PLpro-GRL0617 complex to a resolution of 2.5 Å (Table S1). The structure reveals unambiguous electron density for the inhibitor, which binds in a cleft leading to the active site (Fig. 3A). The inhibitor is well-removed from the catalytic triad and instead bound within the S3 and S4 subsites of PLpro (Fig. 3B). The interaction between GRL0617 and PLpro is stabilized through a pair of hydrogen bonds and a series of hydrophobic interactions stemming from residues lining the pocket. Specifically, the amide group of the inhibitor forms hydrogen bonds with the side chain of D165 and the backbone nitrogen of Q270 (Fig. 3C). D165 is highly conserved among the ubiquitin-specific protease (USP) family of deubiquitinating enzymes (17) and among most coronaviral papain-like proteases (11, 18). Several structural studies of USPs have revealed that this aspartic acid residue hydrogen bonds with the backbone of ubiquitin molecules at the P4 position, an interaction presumed to be important for ligand stabilization (19–21).
Fig. 3.
Inhibitor GRL0617 binds within the S3 and S4 subsites of PLpro and induces loop closure over the inhibitor. (A) An Fo-Fc electron density omit map, generated with the inhibitor excluded from the phase calculation, is shown contoured at 4σ (orange mesh). The inhibitor is shown as orange sticks. The location of the catalytic triad is indicated, as is the oxidized active site cysteine residue. (B) Stereoview of PLpro (white surface) bound to the inhibitor (orange), which lies within a narrow groove proposed to bind to the P3 and P4 positions of substrate (cyan). The structure of the C-terminal residues of ubiquitin, which were modeled into the PLpro apoenzyme active site (10) is shown superimposed on the inhibitor-bound complex. For clarity, only the P3 (Arg) and P4 (Leu) residues of substrate are shown. (C) Stereoview of the hydrogen bonds and hydrophobic interactions between the inhibitor (orange) and PLpro (blue). Hydrogen bonds are indicated by dashed lines. (D) A ribbon diagram of apo PLpro (blue) superimposed on the inhibitor-bound enzyme (orange). The inhibitor is shown as spheres representing van der Waals radii. Catalytic triad residues are designated by asterisks.
Aside from the 2 aforementioned hydrogen bonds, the majority of contacts between PLpro and inhibitor GRL0617 are hydrophobic in nature. The 1-naphthyl group is partly solvent-exposed but forms hydrophobic interactions with the aromatic rings of Y265 and Y269 and with the side chains of P248 and P249 (Fig. 3C). These residues line the pocket and accommodate the leucine at the P4 position of PLpro substrates (10) (Fig. 3B). The (R)-methyl group, attached to the stereocenter of the inhibitor, points directly into the interior of the protein between Y265 and T302, where it is accommodated by a cavity that is mostly polar in nature. The positions of 3 bound water molecules in this cleft suggest the potential for extending the (R)-methyl group further into the pocket by the addition of polar substituents.
The di-substituted benzene ring at the opposite end of the inhibitor occupies the putative P3 position of bound substrate. The benzene ring stacks against the aliphatic portions of G164, D165 and Q270, whereas the ortho-methyl substituent at the R1 position points into the floor of the cavity, which is lined by the side chains of Y265, Y274 and L163 (Fig. 3C). The other ring substituent, -NH2 at the R3 position of GRL0617, extends from the opening of the cleft where it is surrounded by a series of polar groups, including the side chain oxygens of Q270 and E168 and the hydroxyl of Y269, any of which could serve as a hydrogen bond acceptor (Fig. 3 C and D).
Comparison of the unbound and inhibitor-bound structures reveals 2 significant conformational differences, both presumed to be induced by inhibitor binding. In the apoenzyme structure, a highly mobile loop hinged by 2 glycine residues (G267-G272) is positioned in different conformations in each of the 3 monomers of the asymmetric unit (10). Movements of homologous loops in the deubiquitinating enzymes USP14 and HAUSP upon substrate binding have been observed (19, 20). With PLpro, inhibitor-binding induces closure of this loop such that it clamps the inhibitor to the body of the protein (Fig. 3D). The side chains of Y269 and Q270 become well-defined and reorient to close over the inhibitor, whereas the mainchain of the loop moves to within hydrogen bonding distance of the carbonyl at the center of the inhibitor. Additional movements are observed upon inhibitor binding whereby the side chain of L163 moves to cradle the ortho-methyl of the benzene ring while simultaneously blocking access to the catalytic triad. The plasticity of this region, especially the G267-G272 loop, which is a highly variable region both in length and sequence among papain-like proteases, may account for the range of substrates recognized by these enzymes.
In contrast to the motions observed outside of the catalytic center, the residues of the catalytic triad of PLpro (C112, H273, D287) undergo limited movement between the bound and unbound conformations. Interestingly, a significant amount of residual electron density surrounding the sulfur atom of the catalytic cysteine was observed. Modeling and refinement of this density against a fully oxidized sulfur atom was consistent with a sulfonic acid moiety, vs. sulfinic or sulfenic acids (Fig. S3). This observation likely explains the inability of PLpro to regain full activity after incubation with inhibitor over extended periods of time. The presence of reducing agents in solution most likely helps to maintain the active site cysteine of PLpro in a reduced state. However, upon inhibitor binding, loop closure may restrict access to the cysteine by reducing agents but still allow for oxidation, thereby generating an inactive enzyme. A similar mechanism has been proposed for protein tyrosine phosphatase 1B inhibitors (22).
Inhibitor Specificity.
Structural and functional studies have revealed that PLpro is homologous to human deubiquitinating enzymes and is capable of cleaving ubiquitin and ubiquitin-like modifiers such as ISG15 (9–12, 23). Because there are >50 putative deubiquitinating enzymes in humans that are also cysteine proteases (24), it is important that any inhibitors being developed be selective for PLpro. To test the selectivity of the lead inhibitor against PLpro, the inhibitory activities of GRL0617 against a series of cysteine proteases, including the human deubiquitinating enzymes HAUSP, USP18, UCH-L1, UCH-L3, and a papain-like protease (PLP2) from the human coronavirus NL63, were tested (Fig. 4A). Although the tested enzymes share similar active site architectures to PLpro, it is significant that none of these DUB-like enzymes were inhibited by GRL0617. Structural alignment of PLpro with 1 of its closest structural neighbors, HAUSP, reveals that at least 2 residues of HAUSP, F409 and K420, sterically clash with the inhibitor-binding site (Fig. 4B). Based on a structural alignment of 54 human ubiquitin-specific proteases (USPs), these 2 residues are >80% identical among family members (17, 21), suggesting that GRL0617 is unlikely to inhibit other human USPs.
Fig. 4.
Inhibitor GRL0617 is selective for SARS-CoV PLpro. (A) IC50 values of compounds 5, 6, and GRL0617 are listed for PLpro and 5 other papain-like proteases. (B) Stereo representation of HAUSP (Protein Data Bank ID no. 1NBF, with ubiquitin ligand removed) superimposed with the PLpro-inhibitor complex. HAUSP is shown in green, PLpro in blue, and the inhibitor molecule in orange. HAUSP active site residues that clash with the inhibitor are labeled. (C) An α-HA Western blot of lysed Vero E6 cells treated with HA-Ub-VS in the presence of N-ethyl-maleimide (NEM) or varying concentrations of GRL0617. (D) An α-HA Western blot of lysed Vero E6 cells mixed with 0.1 μg of PLpro and HA-Ub-VS, both in the absence and presence of 40 μM GRL0617.
To further explore the specificity of inhibitor GRL0617 for PLpro over human DUBs, we probed the ability of human deubiquitinating enzymes from cellular lysates to be modified by the active-site-directed probe HA-Ub-vinyl sulfone (VS) in the presence and absence of GRL0617. HA-Ub-VS and similar Ub-derived covalent modifiers have been invaluable tools in identifying and studying various novel cellular and viral deubiquitinating enzymes (25–27). Numerous cellular DUBs become modified when treated with HA-Ub-VS and can therefore be visualized by Western blot analysis by using an anti-HA antibody. If these DUBs are potentially inhibited by compound GRL0617, concurrent treatment with the compound should reduce the extent of VS modification. When lysed cells were treated with HA-Ub-VS in the absence and presence of GRL0617, no change was noted in the immunoblot pattern (Fig. 4C). When PLpro was added to the lysate, it too underwent modification by the HA-Ub-VS, but unlike the cellular DUBs, its modification by the VS was almost completely eliminated in the presence of GRL0617S (Fig. 4D).
Conclusions
This work validates PLpro as a candidate drug target for the development of therapeutics against SARS-CoV. We discovered and optimized the potency of a first generation of lead compounds that act as noncovalent competitive inhibitors of PLpro by binding to the S4-S3 subsites and inducing a conformational change that renders the active site nonfunctional. Although the in vivo potency of this first generation of compounds is likely not yet sufficient to make them therapeutics, they will serve as an important basis for further optimization of in vivo potency and ADMET properties. Together, these results hold significant promise for the development of noncovalent inhibitors directed against virally encoded cysteine proteases that also share significant structural homology with host-encoded deubiquitinating enzymes. A number of viral DUBs are predicted to exist (28), and in addition to SARS-CoV PLpro (11), the deubiquitinating activity and X-ray structures of herpesvirus UL36USP (29) and adenovirus proteinase (30) have been reported. Although our goal has not been to discover inhibitors of human deubiquitinating enzymes, recent studies have begun to evaluate this group of enzymes as potential anticancer drug targets (31). However, because of the high active-site sequence homology among >50 USPs, selective targeting of 1 USP vs. another remains an unresolved issue (25). Here, we demonstrate that selective noncovalent targeting of an enzyme highly homologous to human USPs can be achieved, which engenders new opportunities and potential for selectively targeting this class of enzymes.
Materials and Methods
PLpro Purification and Kinetic Assays.
Untagged native SARS-CoV PLpro (polyprotein residues 1541–1855) was expressed and purified to >99% purity, as described (11). Kinetic assay development was first optimized in a 96-well plate format to establish suitable assay conditions and incubation times. The fluorogenic peptide substrate, Arg-Leu-Arg-Gly-Gly-AMC (RLRGG-AMC), was purchased from Bachem Bioscience. PLpro activity as a function of substrate concentration was measured to determine a suitable subsaturating substrate concentration for HTS. Enzyme concentration and incubation time with substrate were optimized to yield a linear response in a 6-min time frame. BSA was included in the assay to stabilize PLpro, to prevent the adsorption of PLpro to the assay plate, and to reduce the effects of promiscuous inhibitors. Reducing agent, 5 mM DTT in this case, was included in all assays to eliminate cysteine-reactive compounds.
Primary HTS Screening.
A compound library consisting of 50,080 structurally diverse small molecules was purchased from ChemBridge and maintained as 10 mM stock solutions dissolved in DMSO and stored desiccated at −20°C. The automated primary screen was performed on a Tecan Freedom EVO 200 robot equipped with a Tecan 3 × 3 mounted 96-well dispenser and a 384-pin stainless steel pin tool (V&P Scientific) with a 100-nL capillary capacity. Fluorescence values were measured on an integrated Tecan Genios Pro microplate reader. All assays were performed in duplicate at room temperature, in black flat-bottom 384-well plates (Matrix Technologies) containing a final reaction volume of 50 μL. The assays were assembled as follows: 40 μL of 142 nM PLpro in Buffer A (50 mM Hepes, pH 7.5; 0.1 mg/ml BSA; and 5 mM DTT) was dispensed into wells and then incubated with 100 nL of 10 mM inhibitor (20 μM final concentration) for ≈5 min. Reactions were then initiated with 10 μL of 250 μM RLRGG-AMC in Buffer A, shaken vigorously for 30 s, and then incubated for 6 min. Reactions were subsequently quenched with 10 μL of 0.5 M acetic acid, shaken for 30 s, and measured for fluorescence emission intensity (excitation λ: 360 nm; emission λ: 460 nm). Each 384-well plate contained 32 positive control wells (100 nL of DMSO replacing 100 nL of inhibitor in DMSO) and 32 negative control wells (assay components lacking PLpro). Because of the low hit rate of compounds displaying significant PLpro inhibition, compounds that showed >35% inhibition were selected for further analysis. Secondary screening methods are included in SI Text.
Synthesis of GRL0617 and Related Compounds.
1H-NMR and 13C-NMR spectra were recorded on Varian Oxford 300, Bruker Avance 400 spectrometers. Optical rotations were recorded on Perkin-Elmer 341 polarimeter. Anhydrous solvent was obtained by distillation of dichloromethane from CaH2. All other solvents were reagent grade. Column chromatography was performed with Whatman 240–400 mesh silica gel under low pressure of 3–5 psi. TLC was carried out with E. Merck silica gel 60-F-254 plates. The detailed protocols for synthesis of compounds 1–7 are included in SI Text.
IC50 Value Determination.
IC50 measurements were performed by hand, in duplicate, in a 96-well plate format. Buffer, enzyme, and substrate conditions matched those of the primary screen. Reactions containing 50 μM substrate, 2% DMSO, and varying concentrations of inhibitor (0–200 μM) were initiated with the addition of enzyme. Reaction progress was monitored continuously on a Tecan Genios Pro microplate reader (excitation λ: 360 nm; emission λ: 460 nm). Data were fit to the equation : vi = vo/(1 + [I]/IC50) by using the Enzyme Kinetics module of SigmaPlot (v. 9.01 Systat) where vi is the reaction rate in the presence of inhibitor, vo is the reaction rate in the absence of inhibitor, and [I] is the inhibitor concentration.
Reversibility of Inhibition.
To test the reversibility of inhibition, 50 nM PLpro was incubated with and without inhibitor (at 20-fold the inhibitor IC50 concentration) in buffer containing 0.05 mg/ml BSA; 50 mM Hepes, pH 7.5; 5 mM DTT; and 1% DMSO in a final volume of 3 mL, for 1 h at room temperature; 1.5 mL of each sample was then dialyzed against 1 L of dialysis buffer (50 mM Hepes, pH 7.5; 5 mM DTT) for 3 h at room temperature by using 10,000 molecular weight cutoff Slide-A-Lyzer dialysis cassettes (Pierce). Samples were transferred to 1 L of fresh dialysis buffer each hour. The other 1.5 mL of each sample (undialyzed samples) were excluded from dialysis but remained at room temperature for the 3-h time period. All samples were assayed for activity after the 3-h incubation in the same manner as used for IC50 measurements.
PLpro de-ISGylating Assays.
PLpro activity with ISG15-AMC (Boston Biochem) was measured in 96-well half-volume plates at 25°C in buffer containing 50 mM Hepes, pH 7.5; 0.1 mg/ml BSA; 5 mM DTT; 2% DMSO; and fixed inhibitor concentrations of 0, 0.1, 1, and 3 μM. Substrate concentration was varied from 0 to 16 μM, and release of AMC was measured in the same manner as for the IC50 measurements described above. The Ki and mode of inhibition of inhibitor GRL0617 were determined through Lineweaver–Burk analysis of the above data by using the Enzyme Kinetics module of SigmaPlot.
Inhibitor Specificity Assays.
The specificity of inhibitors 2, 5, and GRL0617 was tested against 2 human ubiquitin C-terminal hydrolases, UCH-L1 and UCH-L3, the human deubiquitinating enzyme HAUSP, the human de-ISGylating enzyme USP-18, and a coronaviral papain-like protease from HCoV NL63, PLP2. Full-length UCH-L1 and full-length UCH-L3 were purchased from Biomol International; the catalytic domain of HAUSP (residues 213–548) and full-length USP-18 were purchased from Boston Biochem; and PLP2 was purified as described (32). All kinetic assays were performed at 25°C in 50 mM Hepes, pH 7.5; 0.1 mg/ml BSA; and 5–10 mM DTT in a 96-well plate format. Enzymes were assayed in the absence and presence of 100 μM inhibitor, with 100 nM ubiquitin-AMC (Boston Biochem) as substrate (excitation λ: 360 nm; emission λ: 460 nm), with the exception of USP-18, which was assayed with 1 μM ISG15-AMC (Boston Biochem) as substrate. PLpro was assayed under the same conditions, as a control.
Electrophilic Labeling and Detection of DUB Profile in Vero E6 Cells.
For each labeling reaction, 50 μg of Vero E6 lysate was preincubated with 0–40 μM GRL0617 for 30 min in 60 μL of homogenization buffer (50 mM Tris, pH 7.5; 5 mM MgCl2; 0.5 mM EDTA; 2 mM DTT; 2 mM ATP; and 250 mM sucrose). After the incubation, 0.8 μg of the probe HA-Ub-VS (Boston Biochem) was added and then incubated for an additional 30 min at room temperature. Samples were subjected to 10% SDS/PAGE, and the proteins were then transferred to nylon membrane and immunoblotted with mouse monoclonal antibody specific to HA followed by goat anti-mouse IgG-HRP. For samples containing PLpro, 0.1 μg of purified enzyme was added to cell lysate samples and analyzed as above. The DUB profile approach was modeled from the experiments of Kattenhorn et al. (33) and detailed materials and methods of lysis, labeling, and detection are included in SI Text.
SARS-CoVAntiviral Activity Assays.
Vero E6 cells were maintained in Minimal Essential Media (MEM) (Gibco) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin (Gibco), and 10% FCS (Gemini Bio-Products). The SARS-CoV Urbani strain used in this study was provided by the Centers for Disease Control and Prevention (34). All experiments using SARS-CoV were carried out in a Biosafety Level 3 facility by using approved biosafety protocols.
Vero E6 cells were seeded onto flat-bottom, 96-well plates at a density of 9 × 103 cells per well. Cells were either mock-infected with serum-free MEM or infected with 100-fold the median tissue culture infective dose of SARS-CoV Urbani per well in 100 μL of serum-free MEM and incubated for 1 h at 37°C with 5% CO2. After the 1-h incubation period, the viral inoculum was removed and, 100 μL of MEM supplemented with 2% FCS and containing the inhibitor compound of interest at the desired concentration (serial 2-fold dilutions from 50 to 0.1 μM) was added. Cells were incubated for a period of 48 h at 37°C with 5% CO2. Each condition was set up in triplicate, and antiviral assays were performed independently on at least 2 separate occasions. Cell viability was measured 48 h after infection by using the CellTiter-Glo Luminescent Cell Viability Assay (Promega), according to the manufacturer's recommendations. Cell viability for the CellTiter-Glo Luminescent Cell Viability Assay was measured as luminescence and output expressed as relative luciferase units (RLU).
Crystallization, X-Ray Data Collection, and Structure Refinement.
The complex of inhibitor GRL0617 with PLpro was crystallized by vapor diffusion in a sitting-drop format after a 16-h incubation of 8 mg/ml PLpro (in 20 mM Tris, pH 7.5; 10 mM DTT) with 2 mM inhibitor at 4°C. Immediately before crystallization, the sample was clarified by centrifugation. A 1-μL volume of the enzyme-inhibitor solution was then mixed with an equal volume of well solution containing 1 M LiCl; 0.1 M Mes, pH 6.0; and 30% PEG 6,000 and equilibrated against well solution at 20°C. Before data collection, crystals were soaked in a cryosolution containing well solution, 400 μM inhibitor, and 16% glycerol. Details of X-ray data collection and structure refinement are included in SI Text. Final X-ray data collection and refinement statistics are given in Table S1.
Supplementary Material
Acknowledgments.
We thank S. Forrester for help with crystallization condition screening, Y. M. Baez for assistance with kinetic specificity assays, B. D. Santarsiero for crystal structure data collection support, D. C. Mulhearn for computational modeling support, and T. E. O'Brien for assistance with statistical analysis of antiviral activity. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by Public Health Service Research Grant P01 AI060915 (“Development of Novel Protease Inhibitors as SARS Therapeutics”).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates for the crystal structure of the PLpro-inhibitor complex have been deposited with the Research Collaboratory for Structural Bioinformatics Protein Data Bank (accession nos. 3E9S and RCSB049054).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805240105/DCSupplemental.
References
- 1.Turk B. Targeting proteases: Successes, failures and future prospects. Nat Rev Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 2.Leung-Toung R, Li W, Tam TF, Karimian K. Thiol-dependent enzymes and their inhibitors: A review. Curr Med Chem. 2002;9:979–1002. doi: 10.2174/0929867024606704. [DOI] [PubMed] [Google Scholar]
- 3.Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 4.Lau SK, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziebuhr J. The coronavirus replicase. Curr Top Microbiol Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziebuhr J. In: Nidoviruses. Perlman S, Gallagher T, Snijder EJ, editors. Washington, DC: ASM Press; 2008. pp. 65–82. [Google Scholar]
- 7.Yang H, Bartlam M, Rao Z. Drug design targeting the main protease, the Achilles' heel of coronaviruses. Curr Pharm Des. 2006;12:4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- 8.Devaraj SG, et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindner HA, et al. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratia K, et al. Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barretto N, et al. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulea T, Lindner HA, Purisima EO, Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J Virol. 2005;79:4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziebuhr J, et al. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J Virol. 2007;81:3922–3932. doi: 10.1128/JVI.02091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung D, Abbenante G, Fairlie DP. Protease inhibitors: Current status and future prospects. J Med Chem. 2000;43:305–341. doi: 10.1021/jm990412m. [DOI] [PubMed] [Google Scholar]
- 15.Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat Protoc. 2006;1:550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mesecar AD, Koshland DE., Jr A new model for protein stereospecificity. Nature. 2000;403:614–615. doi: 10.1038/35001144. [DOI] [PubMed] [Google Scholar]
- 17.Quesada V, et al. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Commun. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 18.Sulea T, Lindner HA, Purisima EO, Menard R. Binding site-based classification of coronaviral papain-like proteases. Proteins. 2006;62:760–775. doi: 10.1002/prot.20802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu M, et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu M, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 21.Renatus M, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure(London) 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 23.Lindner HA, et al. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daviet L, Colland F. Targeting ubiquitin specific proteases for drug discovery. Biochimie. 2008;90:270–283. doi: 10.1016/j.biochi.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 26.Borodovsky A, et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 27.Borodovsky A, et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindner HA. Deubiquitination in virus infection. Virology. 2007;362:245–256. doi: 10.1016/j.virol.2006.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlieker C, et al. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol Cell. 2007;25:677–687. doi: 10.1016/j.molcel.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGrath WJ, Ding J, Didwania A, Sweet RM, Mangel WF. Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: Insights on a new fold. Biochim Biophys Acta. 2003;1648:1–11. doi: 10.1016/s1570-9639(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson B, Marblestone JG, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191–199. doi: 10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, et al. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.