Abstract
Pyruvate formate-lyase activating enzyme generates a stable and catalytically essential glycyl radical on G734 of pyruvate formate-lyase via the direct, stereospecific abstraction of a hydrogen atom from pyruvate formate-lyase. The activase performs this remarkable feat by using an iron-sulfur cluster and S-adenosylmethionine (AdoMet), thus placing it among the AdoMet radical superfamily of enzymes. We report here structures of the substrate-free and substrate-bound forms of pyruvate formate-lyase-activating enzyme, the first structures of an AdoMet radical activase. To obtain the substrate-bound structure, we have used a peptide substrate, the 7-mer RVSGYAV, which contains the sequence surrounding G734. Our structures provide fundamental insights into the interactions between the activase and the G734 loop of pyruvate formate-lyase and provide a structural basis for direct and stereospecific H atom abstraction from the buried G734 of pyruvate formate-lyase.
Keywords: crystallography, metalloprotein, radical chemistry, S-adenosylmethionine, iron–sulfur cluster
Pyruvate formate-lyase-activating enzyme (PFL-AE) is one of the first-discovered members of the “AdoMet radical” or “Radical SAM” superfamily, which is characterized by both the presence of a conserved CX3CX2C sequence motif that coordinates an essential 4Fe-4S cluster and by the use of S-adenosylmethionine (AdoMet or SAM) for 5′-deoxyadenosyl radical (5′-dA·) generation (1–3). The electron required for reductive cleavage of AdoMet to form 5′-dA· comes from the 4Fe-4S cluster, which is reduced in Escherichia coli by flavodoxin (4). AdoMet radical enzymes act on a wide variety of biomolecules in numerous pathways. For example, biotin synthase (BioB) (5), coproporphyrinogen III oxidase (HemN) (6), and molybdenum cofactor biosynthetic enzyme A (MoaA) (7) are involved in vitamin/cofactor biosynthesis; lysine 2,3-aminomutase (LAM) facilitates the fermentation of lysine and the biosynthesis of β-lysyl antibiotics (8); spore photoproduct lyase repairs UV-induced DNA damage (9); and class III ribonucleotide reductase (RNR) activase, like PFL-AE, catalyzes the formation of glycyl radicals (10). The structural basis by which AdoMet radical enzymes are able to react with such a diverse group of substrates, including dethiobiotin, DNA, and proteins, is a key question in the field. The structures of PFL-AE from E. coli described here provide views of a member of the AdoMet radical superfamily for which the substrate is an enzyme and thus structural insight into activation of a glycyl radical enzyme (GRE).
Because of their oxygen-sensitive nature, GREs (11–14) are proposed to have evolved before the appearance of oxygen in the atmosphere, and some speculate that class III RNR may be a truly ancient enzyme (15) because the ancestral RNR was likely involved in the conversion of RNA- to DNA-based life (16). Unlike RNR, which converts ribonucleotides to deoxyribonucleotides, PFL is a central metabolic enzyme that converts pyruvate and CoA to acetyl-CoA and formate (17). Found in both prokaryotes and eukaryotes, PFL provides the sole source of acetyl-CoA for the Krebs cycle under fermentative conditions. X-ray analysis reveals a common fold for GRE family members as a 10-stranded β barrel, with two sets of five-stranded sheets running antiparallel to each other and a buried glycine residue at the tip of the second of two β finger motifs (18–23). In one of the more fascinating activation reactions in biology, PFL-AE (a monomer of 28 kDa) stereospecifically abstracts a hydrogen atom from residue G734 of PFL (homodimer of 170 kDa), generating the catalytically relevant glycyl radical species (24). Because the crystal structure of free PFL in its inactive form shows G734 buried 8 Å from the protein surface (18), the direct abstraction of a hydrogen atom from this residue is difficult to conceptualize and is believed to involve a conformational change (19, 21, 22). Once established, the glycyl radical is surprisingly stable (25), catalyzing multiple turnovers via a putative active site thiyl radical. Although stable under anaerobic conditions, the glycyl radical is susceptible to destruction by oxygen, which results in irreversible cleavage of the polypeptide and inactivation of the enzyme (26). In this circumstance, some organisms produce a short protein with high homology to the last 59 residues of PFL (including G734), called YfiD in E. coli. After activation by PFL-AE, YfiD can associate with cleaved PFL, replacing its lost radical-containing domain and yielding a working heteroenzyme complex (27). In this manner, YfiD acts as a “spare part,” restoring activity of oxygen-cleaved PFL with minimal energy expenditure. In vitro, peptides of as few as seven residues, which contain the sequence surrounding G734 (24), are substrates for PFL-AE, albeit with less activity than for full PFL (with Km = 0.22 mM and Vmax = 11 nmol/min·mg, compared with 1.4 μM and 54 nmol/min·mg for PFL). To provide insight into the mechanism by which the AdoMet radical activases catalyze formation of glycyl radicals on substrate proteins, we solved the structure of PFL-AE in both the substrate-free (AE) and substrate-bound (pept-AE) forms. Our results, along with previous biochemical and structural studies on PFL, support the proposal that a small domain of PFL harboring G734 undergoes a conformational change (19, 21, 22), thereby positioning G734 in close proximity to the AdoMet bound at the active site of PFL-AE.
Results
PFL-AE Overall Fold.
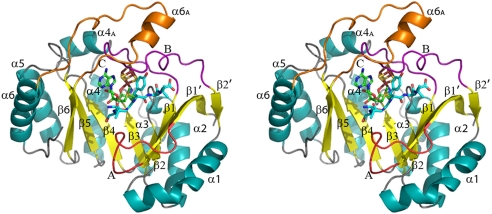
PFL-AE is a monomeric, single-domain enzyme, and with 245 aa, it is the smallest AdoMet radical enzyme of known structure [see Methods, Table 1, and supporting information (SI) Table S1]. Both the 2.25 Å resolution AE and the 2.8-Å resolution pept-AE structures show a partial TIM barrel fold (Fig. 1) with a 4Fe-4S cluster ligated by the canonical CX3CX2C motif. Although the AE crystals were grown in the presence of AdoMet, this cosubstrate is observed fully ordered only in the pept-AE structure, where it ligates the 4Fe-4S cluster as observed in other AdoMet radical enzyme structures and packs against the 7-mer RVSGYAV peptide substrate (Figs. S1 and S2). PFL-AE consists of the (β/α)6 AdoMet radical core and, unlike the other AdoMet radical enzymes, few secondary structural elements outside of the core fold are present to “close off” the opening of the partial barrel (Fig. 1). Only 65 aa of PFL-AE are not part of the core region, including β1′, loop A, loop C, and α6 (Fig. 1). The C-terminal end (top) of the partial barrel is covered by two long loops located after strands β1 (loop B) and β6 (loop C). The N-terminal end (bottom) and the lateral opening of the barrel appear highly solvent-exposed.
Table 1.
Refinement statistics
| Parameter | AE | Pept-AE |
|---|---|---|
| Resolution limits, Å* | 38.19–2.25 | 37.95–2.77 |
| (2.39–2.25) | (2.94–2.77) | |
| No. unique reflections* | 10,618 (1,428) | 8,376 (1,230) |
| No. reflections in test set* | 1,010 (147) | 438 (54) |
| Rwork, % | 22.4 | 22.9 |
| Rfree, % | 29.8 | 26.1 |
| Average B factors, Å2 | 58.3 | 80 |
| Protein† | 57.8 (1,928) | 76.9 (1,872) |
| 4Fe-4S† | 45.8 (8) | 59.7 (8) |
| AdoMet†,‡ | 61.5 (11) | 75.7 (27) |
| PEG† | 83.9 (10) | NA§ |
| Peptide† | NA§ | 104.4 (42) |
| Water† | 60.9 (60) | 81.0 (16) |
| Bond length deviation, Å | 0.008 | 0.008 |
| Bond angle deviation, ° | 1.8 | 1.5 |
*Values in parentheses refer to the high-resolution bin.
†Values in parentheses refer to number of atoms.
‡AdoMet is only partially ordered in the AE model. Only 11 AdoMet atoms were included in the AE refinement and final model.
§NA, not applicable.
Fig. 1.
Stereoview of PFL-AE with secondary structural elements assigned numerically (helices in cyan, strands in yellow). The loops after strands β1′, β1, and β6 are labeled A (red, residues 10–20), B (purple, residues 27–47), and C (orange, residues 201–225). The 4Fe-4S cluster (ruby and gold), AdoMet (green carbons), and peptide (teal carbons) are depicted in sticks with oxygens colored red and nitrogens colored blue.
AdoMet Radical Core.
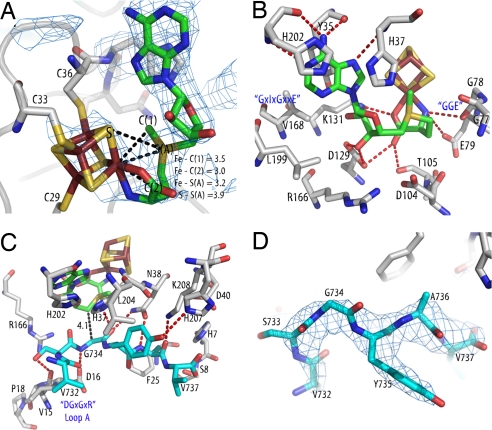
The AdoMet radical core fold of PFL-AE consists of a “splayed” sheet similar to those observed in structures of other AdoMet radical family members (5–8, 28, 29) (see Table S2 for rmsd values). The active sites of these enzymes are located within the lateral opening and are made up mainly of residues originating from β strands and loops at the top of the core's sheet (Fig. 1). The 4Fe-4S cluster of PFL-AE is located at the top of the partial barrel (Fig. 1), with the unique iron coordinated by AdoMet (Fig. 2A) through the amino nitrogen (2.2 Å) and carboxyl oxygen (2.2 Å). The distances observed between AdoMet and the cluster (Fig. 2A) are in agreement with the spectroscopic data reported for this enzyme (30–32) and are similar to those seen in the other AdoMet radical structures (5–8). These close interactions between AdoMet and the cluster are consistent with the theory that innersphere electron transfer from the 4Fe-4S cluster of PFL-AE to AdoMet precedes generation of 5′-dA· (30). Several residues that form the AdoMet-binding site comprise motifs conserved among all of the AdoMet radical enzymes, such as the GGE motif (G77, G78, and E79 in PFL-AE) and the GxIxGxxE motif (PFL-AE residues V168, V170, G172, and E175) (33). As discussed in ref. 33, these two motifs bind the AdoMet methionine directly and serve to stabilize the binding site for the adenine moiety, respectively (Fig. 2B). Finally, two residues (D104 and R166) do not make specific contacts with either AdoMet or the peptide substrate, but they are both completely conserved and located in close proximity to the C5′ atom of AdoMet (Fig. 2B), suggesting a potential role in catalysis. Additional interactions with AdoMet (Fig. 2B) are similar to those observed in other AdoMet radical enzymes, although the identity of the residues is often not conserved (5–8, 28, 29), a difference caused in part by the large number of contacts made by backbone atoms. One interesting variation is the presence of two histidines, H37 and H202, which pack against the AdoMet adenine ring. As discussed below, these residues are likely important for the ability of PFL-AE to bind a protein substrate.
Fig. 2.
Substrate and cofactor binding. Colors are as in Fig. 1, with protein side chain carbons in gray. Conserved motifs (33) are labeled in blue. Composite omit maps are shown as a blue mesh and are contoured at 1σ. Hydrogen bond lengths and other distances are represented as dashed lines. (A) Detail of cluster–AdoMet interaction with composite omit map contoured around the AdoMet. Distances of interest between the unique iron of the 4Fe-4S cluster and AdoMet atoms are shown. (B) AdoMet–protein interactions. (C) Peptide–protein interactions. (D) Omit map contoured around the peptide.
Peptide Binding.
The peptide (RVSGYAV) binds in a bent conformation (Fig. 2 C and D) across the lateral opening of the partial barrel, interacting primarily with residues from loops A–C (Fig. 1). Six of the seven peptide residues are visible in the structure, with the first peptide residue, R731, disordered because of a nearby crystal lattice contact. Binding of the peptide seals the active site from solvent, burying the cluster and AdoMet and providing a protected environment for radical generation (Fig. S2). The binding of peptide also appears to stabilize AdoMet binding to PFL-AE. In the absence of substrate, both previous spectroscopic studies (30–32, 34) and our crystallographic experiments show ligation of the cluster's unique iron by the amino acid moiety of AdoMet. However, in our substrate-free model we observe poor-quality electron density for the remaining AdoMet atoms, indicating disorder (Figs. S1a and S3). In contrast, in the presence of peptide, electron density improves significantly, and the entire molecule of AdoMet can be refined (Fig. 2 and Figs. S1b and S3). Stabilization of AdoMet in this site upon substrate binding is attractive from a biological perspective because it provides a potential mechanism to prevent uncoupling between AdoMet cleavage and glycyl radical generation: only in the presence of substrate would AdoMet favor a stable catalytic conformation. Although the AdoMet is buried in the pept-AE structure, a large surface area including the peptide itself and the N-terminal sides of some β strands remains solvent-exposed. Presumably, these surfaces will be buried in the full PFL–PFL-AE complex.
The peptide substrate in the pept-AE structure corresponds to the seven-residue consensus sequence around G734 of PFL, RVSGYAV. This motif is completely conserved in PFL. There is, however, poor consensus at this site among all glycyl radical-containing enzymes, with only the residues corresponding to R731 and G734 being completely conserved (Fig. S4). Specific residues on PFL-AE required for or involved in the activation reaction have not been examined in detail, although it is known that the enzyme is not active toward a peptide corresponding to the similar class III RNR consensus sequence, RVCGYLG, which substitutes a Cys for S733, a Leu for A736, and a Gly for V737 (24).
Despite some observed specificity toward the PFL glycine loop sequence (24), contacts are made mainly to peptide substrate backbone atoms (Fig. 2C). In contrast, all of the contacts to the peptide are made by side chains of PFL-AE, each of which is highly conserved and includes a DGxGxR motif located on loop A (Figs. 1 and 2C). The key interactions between PFL-AE and the peptide appear to serve three critical purposes: (i) orienting the glycine in the active site, (ii) controlling the overall peptide conformation, and (iii) imparting selectivity. Three specific residues ensure the proper orientation of G734 with respect to AdoMet in the active site: D16 of the DGxGxR motif and fully conserved N38 hydrogen bond to the glycine amino and carboxyl groups, whereas H37 reaches across the AdoMet adenine ring to help orient the residue (Fig. 2C). These specific interactions ideally position G734 for hydrogen atom abstraction, with an AdoMet C5′ to G734 Cα distance of 4.1 Å (Fig. 2C), similar to those observed in BioB (3.9 and 4.1 Å) (5) and LAM (3.8 Å) (8). The peptide conformation is dictated by the van der Waals interactions with loop A and hydrogen bonding interactions provided by K208 and N38 to A736. Finally, selectivity appears to arise from interactions with F25 at residue A736 (F25 should prevent binding of a larger amino acid) and with L204 and H207 at Y735 (through packing and hydrogen bonding interactions) (Fig. 2C). Because the other interactions observed in this structure are to peptide backbone atoms, specificity at these other positions is likely governed by the effect of side chain identity on the conformation of the peptide backbone itself. Unfortunately, crystal packing prevents modeling of R731, leaving its role in the activation reaction and the reason for its conservation unclear, although others have proposed that it is involved in the stabilization of the glycine loop structure in the native state of a GRE (19). The pept-AE interactions observed here are not electrostatic in nature, consistent with the observation that the activation reaction is not affected by increasing the salt concentration of the activation reaction buffer (35).
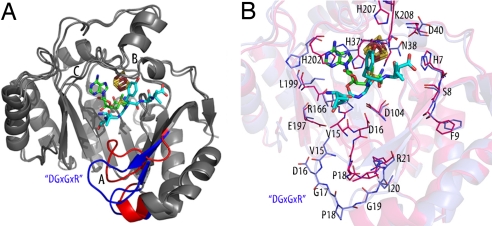
Comparison of the AE and pept-AE structures (rmsd = 0.774 Å) allows us to investigate structural rearrangements associated with AdoMet and peptide binding (Fig. 3). The major conformational change upon binding of the peptide occurs in loop A (Fig. S5), which harbors the activase-specific DGxGxR motif (D16G17xG19xR21 in PFL-AE) (Fig. 3A). As the loop swings up toward the active site to form contacts with the peptide, the largest movement occurs in residue D16, which is displaced ≈10 Å at the Cβ atom (Fig. 3B). R21 of the DGxGxR motif anchors the beginning of strand β1 and may stabilize the β sheet during this motion. This movement is likely essential to the activation reaction, in enabling a structural change in PFL and/or orienting the glycine loop in the active site. Several other side chain rearrangements or reordering occur and appear important in forming the AdoMet binding site (H37, L199, H202) and binding peptide (N38, H207, K208).
Fig. 3.
Conformational change of PFL-AE upon substrate binding. Colors are as in Fig. 1 unless otherwise noted. (A) The AE and pept-AE models were superimposed and colored in gray, with areas undergoing a conformational change highlighted (blue, AE; red, pept-AE). Loops A–C are labeled. (B) Close-up view of the active site showing side-chain rearrangements upon substrate binding. AE carbon atoms are colored blue, pept-AE carbon atoms red, oxygen red, and nitrogen blue.
Docking Studies.
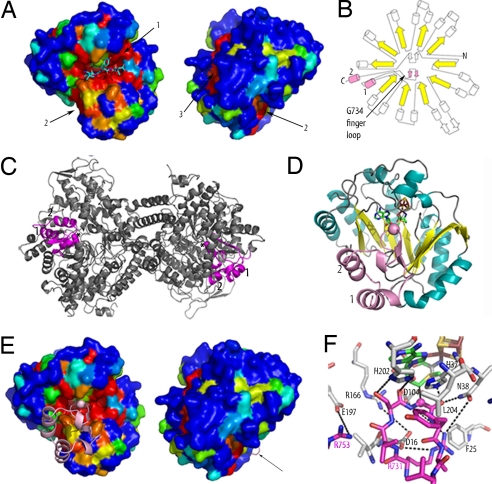
Areas of high conservation on the PFL-AE surface that could be involved in protein–protein interactions, such as with PFL or flavodoxin, were identified (see SI Materials and Methods). These include: (i) the active site, (ii) the N-terminal side of the β sheet, and (iii) behind loop B (Fig. 4A). The preservation observed at the active site (region 1) was expected and is more extensive than just the area involved in binding the peptide. Regions 1 and 2 are adjacent to one another, suggesting that the two areas may together form the PFL-binding surface. The location of region 3 proximal to the 4Fe-4S cluster is ideal for interactions with flavodoxin.
Fig. 4.
Docking studies of PFL-AE. Colors are as in Fig. 1 unless specified. (A) Surface representation of the pept-AE colored by sequence conservation as calculated by ESPript (43), showing the same view of the pept-AE as in Fig. 1 and rotated 180° from that orientation. Sequence conservation is represented in rainbow colors, with areas of 100% conservation in red, and 0% conservation in blue. The three conserved regions are numbered. (B) Topology diagram of PFL showing 10-stranded α/β barrel (strands in yellow) with RD highlighted in pink and helices denoted 1 (PFL residues 712–720) and 2 (744–752). An N-terminal domain is omitted for clarity. (C) Ribbon representation of the PFL dimer in gray with RD in magenta and G734 in spacefill. (D) Best docking model output by ZDOCK, with Cα of G734 displayed in spacefill and RD colored as in B. (E) Docking model, with RD displayed as in D. The pept-AE is colored as in A and is shown in the same orientation. An arrow indicates a loop from RD. (F) Detail of active site of docking model colored as in D, with side chains of interest shown in sticks. RD side chains are labeled in magenta, pept-AE side chains in black. Dashed lines indicate possible side-chain interactions between RD and AE.
A theoretical model of the mode of interaction between PFL and PFL-AE was generated by manual and computational docking studies (see Methods and Fig. S6 a–c). The best docking model was obtained by using the pept-AE structure and a fragment cut from the PFL model [Protein Data Bank (PDB) ID code 2PFL] with high homology to YfiD (the small protein capable of undergoing activation by PFL-AE) (27). This region of PFL, which was implicated as being involved in the activation reaction on the basis of high structural homology to the corresponding region of the GRE glycerol dehydratase (22), will be referred to as a “radical domain” (RD) and encompasses PFL residues 712–759 (Fig. 4 B and C). In the docking model, the glycine loop of RD points into the active site of AE, positioned similarly to the peptide observed bound in the experimental maps (Fig. 4 D–F), although increased conformational flexibility allows the peptide to bind in a more extended conformation toward its C terminus (Fig. S6d). This difference in conformations could also be in part because RD was treated as a rigid body in the docking studies and not allowed to adjust to its new binding environment. In addition, based on differences in Km values between the peptide substrates and PFL, one would not expect identical binding interactions. Despite these caveats, the distance predicted between AdoMet and RD G734 Cα (4.6 Å) in the docking model agrees reasonably well with that observed between the AdoMet and G734 Cα of the bound peptide (4.1 Å). The interactions between H37, N38, or L204 and RD are similar to those observed in the pept-AE structure, although the bidentate interaction of N38 and the contacts between F25, K208, and the peptide are lost. Residues from PFL-AE and RD in the docking model are also poised to make several additional interactions (Fig. 4F), including RD residue R731 with PFL-AE residues E10 (the side chain of which is disordered in this structure), D16, N38, and D104. In addition, R753 of RD could interact with E197 or the C terminus, highly conserved PFL-AE residues N38 and H202 are positioned to interact with RD glycine loop and RD Y735 stacks against PFL-AE L204, which is always hydrophobic (Fig. 4F). Of the residues involved in these possible interactions, E10, D16, N38, and H202 are notable in that they are fully conserved among the PFL activases, and E197 is always either glutamate or aspartate. RD also contacts conserved surface regions 1 and 2 (Fig. 4E), providing a reasonable explanation for their preservation across the activases.
Discussion
The AdoMet radical superfamily is capable of catalyzing some of the most difficult chemical reactions known in biology via the breaking of C–H bonds to form substrate radicals (2, 3). The structures described here illustrate the surprising plasticity of the core fold and extend our understanding of the activation reactions of the GRE family. The AdoMet radical structures (5–8, 28, 29) show that, in general, the size of the specific enzyme tends to increase as its substrate gets smaller, a point emphasized by the small size and conspicuous lack of noncore secondary structural elements in PFL-AE. As the most compact of the structurally characterized AdoMet radical enzymes, with 245 residues compared with next-smallest MoaA 340 residues, PFL-AE defines the minimal machinery required for generation of 5′-dA·. The majority of interactions between PFL-AE and its substrate occur outside of the 180-residue AdoMet radical core because less than one-half of the contacts between PFL-AE and the peptide substrate originate from within the core.
The mechanism of activation of PFL and other GREs is an open question, despite the six independent structures (18–23) of inactive GREs have revealed their 10-stranded β barrel folds. Although the radical-harboring G734 of PFL is buried 8 Å from the surface of the protein (18), hydrogen atom abstraction by 5′-dA· to form the glycyl radical occurs directly and stereospecifically (24). To allow access of the activase to the catalytic glycine, a dramatic conformational change within PFL would have to occur. This ATP-independent process could proceed in two ways, the first through the action of the activase via conformational changes induced by its binding. Alternatively, PFL itself could be conformationally flexible, at times adopting a more open conformation that exposes G734, but which is less thermodynamically stable than the conformation observed in the crystal structures.
The findings presented here provide important insight into the possible modes of interaction in the full physiological complex, although they do not answer the question of how the necessary conformational changes occur. Because the peptide used in these studies is known to stimulate AdoMet cleavage by PFL-AE (24), albeit with lower activity, the pept-AE structure is likely relevant to the physiological reaction. The location of peptide binding and the AdoMet C5′ to G734 Cα distance are consistent with the substrate-binding sites observed in the other AdoMet radical enzyme structures (5, 8). Additionally, the torsional angles of G734 as modeled in this structure are similar to both those seen in the PFL structure (18, 21) and those predicted for the PFL glycyl radical by quantum-chemical studies of the EPR parameters of the PFL radical (36). The pept-AE structure illustrates one possible mode of interaction between PFL and the activase in which the glycine loop extends across the surface of the PFL-AE active site. The unexpected motion undergone by loop A to bind substrate is likely a feature conserved among all of the activases, as evidenced by high conservation of the DGxGxR motif and by the conservation of the residues that come into contact with the motif after it binds substrate. This loop could play a major role in the conformational change of PFL by shifting the glycine loop into the PFL-AE active site, and it is most certainly essential to the proper orientation of G734.
Through sequence comparisons and docking studies, we were able to identify a fragment of PFL, residues 712–759, that we propose to be the main unit that interacts with PFL-AE in the full complex. This 48-residue fragment forms a small domain that connects to the β barrel of PFL through a short loop. Modeling suggests that rotation of this RD about a single hinge point allows the glycine loop to move out of the barrel with minimal steric clashes, making it accessible to the activase (Fig. S7). Our current view of the PFL–PFL-AE complex formation is that PFL-AE binding either helps position a RD outside of the PFL barrel or captures a conformationally flexible RD in its “out” conformation, either of which would involve motion of PFL-AE loop A to allow proper binding and orientation of G734. The PFL-AE–RD docking model is attractive in terms of the positioning of G734 with respect to the AdoMet, explaining two regions of PFL-AE surface conservation and providing a role for conservation of residues D16, H37, and N38 in RD binding. After radical generation, PFL would once again undergo a conformational change, likely with the help of PFL-AE to shield the radical, returning the activated glycine loop to the active site. After generating a radical in one monomer of the PFL dimer, PFL-AE's task is complete. Allosteric effects are thought to prevent activation of the second monomer, explaining the observed half-of-the-sites reactivity (18).
The PFL system is responsible for a fundamental, metabolically essential chemical transformation and is believed to be representative of one of Nature's most ancient enzymes. Our PFL-AE structures identify the regions and residues involved in activation and complex formation with PFL and provide a much needed structural framework for future experiments on this system.
Methods
AE and pept-AE were purified as described (30, 37) with 1 mM DTT in all buffers and crystallized anaerobically by the vapor-diffusion technique (see SI Materials and Methods). Multiwavelength anomalous diffraction data were collected at the iron edge on beamlines 9-2 and 9-1 at Stanford Synchrotron Radiation Laboratory (SSRL) and beamline 5.0.2 at Advanced Light Source (ALS). Sites were identified with SOLVE (38) and refined in Sharp (39), yielding interpretable density maps to 3.7 Å (AE) and 2.77 Å (pept-AE) resolution. Model building was carried out in XtalView (40), and the model was refined in CNS (41), yielding Rwork (Rfree) of 22.4% (29.8%) to 2.25-Å resolution (AE) and 22.9% (26.1%) to 2.77-Å resolution (pept-AE). Ramachandran diagrams were as follows: for the final AE model, 87.8% of residues were in the allowed regions, 11.3% were generously allowed, and 0.9% additionally allowed, with none disallowed. For the final pept-AE model, 84.3% of residues were in the allowed regions, 15.3% generously allowed, and 0.5% additionally allowed. The AE and pept-AE models were docked, both manually and with the docking algorithm ZDOCK (42), with a fragment of the PFL model (18). The resulting docking complexes were evaluated based on the position of PFL G734 in the AE active site, possible hydrogen-bonding contacts, interaction of RD with conserved AE residues, and agreement with biochemical data.
Supplementary Material
Acknowledgments.
We thank T. Doukov and C. Smith for help during data collection. Data were collected at the Advanced Light Source (ALS) and Stanford Synchrotron Radiation Laboratory (SSRL). Support for the synchrotron sources is provided by the U.S. Department of Energy (ALS, SSRL) and by the National Institutes of Health (SSRL). This work was supported by the National Science Foundation (C.L.D.), the Massachusetts Institute of Technology Center for Environmental Health Sciences (C.L.D.), the National Institutes of Health (J.B.B.), and the William Asbjornsen Albert Fellowship (J.L.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3C8F and 3CB8).
This article contains supporting information online at www.pnas.org/cgi/content/full/0806640105/DCSupplemental.
References
- 1.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang SC, Frey PA. S-Adenosylmethionine as an oxidant: The radical SAM superfamily. Trends Biochem Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Cheek J, Broderick JB. Adenosylmethionine-dependent iron-sulfur enzymes: Versatile clusters in a radical new role. J Biol Inorg Chem. 2001;6:209–226. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- 4.Blaschkowski HP, Neuer G, Ludwig-Festl M, Knappe J. Routes of flavodoxin and ferredoxin reduction in Escherichia coli: CoA-acylating pyruvate:flavodoxin and NADPH:flavodoxin oxidoreductases participating in the activation of pyruvate formate-lyase. Eur J Biochem. 1982;123:563–569. [PubMed] [Google Scholar]
- 5.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 2003;22:6214–6224. doi: 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci USA. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lepore BW, Ruzicka FJ, Frey PA, Ringe D. The x-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale. Proc Natl Acad Sci USA. 2005;102:13819–13824. doi: 10.1073/pnas.0505726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebeil R, et al. Spore photoproduct lyase from Bacillus subtilis spores is a novel iron-sulfur DNA repair enzyme which shares features with proteins such as class III anaerobic ribonucleotide reductases and pyruvate-formate lyases. J Bacteriol. 1998;180:4879–4885. doi: 10.1128/jb.180.18.4879-4885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamarit J, Mulliez E, Meier C, Trautwein A, Fontecave M. The anaerobic ribonucleotide reductase from Escherichia coli: The small protein is an activating enzyme containing a [4Fe-4S]2+ center. J Biol Chem. 1999;274:31291–31296. doi: 10.1074/jbc.274.44.31291. [DOI] [PubMed] [Google Scholar]
- 11.Selmer T, Pierik AJ, Heider J. New glycyl radical enzymes catalysing key metabolic steps in anaerobic bacteria. Biol Chem. 2005;386:981–988. doi: 10.1515/BC.2005.114. [DOI] [PubMed] [Google Scholar]
- 12.Raynaud C, Sarcabal P, Meynial-Salles I, Croux C, Soucaille P. Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc Natl Acad Sci USA. 2003;100:5010–5015. doi: 10.1073/pnas.0734105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leuthner B, et al. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: A new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol Microbiol. 1998;28:615–628. doi: 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 14.Yu L, Blaser M, Andrei PI, Pierik AJ, Selmer T. 4-Hydroxyphenylacetate decarboxylases: Properties of a novel subclass of glycyl radical enzyme systems. Biochemistry. 2006;45:9584–9592. doi: 10.1021/bi060840b. [DOI] [PubMed] [Google Scholar]
- 15.Reichard P. The evolution of ribonucleotide reduction. Trends Biochem Sci. 1997;22:81–85. doi: 10.1016/s0968-0004(97)01003-7. [DOI] [PubMed] [Google Scholar]
- 16.Stubbe J. Ribonucleotide reductases: The link between an RNA and a DNA world? Curr Opin Struct Biol. 2000;10:731–736. doi: 10.1016/s0959-440x(00)00153-6. [DOI] [PubMed] [Google Scholar]
- 17.Knappe J, Wagner AF. Glycyl free radical in pyruvate formate-lyase: Synthesis, structure characteristics, and involvement in catalysis. Methods Enzymol. 1995;258:343–362. doi: 10.1016/0076-6879(95)58055-7. [DOI] [PubMed] [Google Scholar]
- 18.Becker A, et al. Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat Struct Biol. 1999;6:969–975. doi: 10.1038/13341. [DOI] [PubMed] [Google Scholar]
- 19.Logan DT, Andersson J, Sjoberg BM, Nordlund P. A glycyl radical site in the crystal structure of a class III ribonucleotide reductase. Science. 1999;283:1499–1504. doi: 10.1126/science.283.5407.1499. [DOI] [PubMed] [Google Scholar]
- 20.Leppanen VM, et al. Pyruvate formate-lyase is structurally homologous to type I ribonucleotide reductase. Structure. 1999;7:733–744. doi: 10.1016/S0969-2126(99)80098-7. [DOI] [PubMed] [Google Scholar]
- 21.Lehtio L, Leppanen VM, Kozarich JW, Goldman A. Structure of Escherichia coli pyruvate formate-lyase with pyruvate. Acta Crystallogr D. 2002;58:2209–2212. doi: 10.1107/s0907444902016402. [DOI] [PubMed] [Google Scholar]
- 22.O'Brien JR, et al. Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: Preliminary biochemical and structural characterization. Biochemistry. 2004;43:4635–4645. doi: 10.1021/bi035930k. [DOI] [PubMed] [Google Scholar]
- 23.Lehtio L, Grossmann JG, Kokona B, Fairman R, Goldman A. Crystal structure of a glycyl radical enzyme from Archaeoglobus fulgidus. J Mol Biol. 2006;357:221–235. doi: 10.1016/j.jmb.2005.12.049. [DOI] [PubMed] [Google Scholar]
- 24.Frey M, Rothe M, Wagner AF, Knappe J. Adenosylmethionine-dependent synthesis of the glycyl radical in pyruvate formate-lyase by abstraction of the glycine C-2 pro-S hydrogen atom: Studies of [2H]glycine-substituted enzyme and peptides homologous to the glycine 734 site. J Biol Chem. 1994;269:12432–12437. [PubMed] [Google Scholar]
- 25.Knappe J, Neugebauer FA, Blaschkowski HP, Ganzler M. Post-translational activation introduces a free radical into pyruvate formate-lyase. Proc Natl Acad Sci USA. 1984;81:1332–1335. doi: 10.1073/pnas.81.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner AF, Frey M, Neugebauer FA, Schafer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner AF, et al. YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem Biophys Res Commun. 2001;285:456–462. doi: 10.1006/bbrc.2001.5186. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki Y, et al. Crystal structure of the radical SAM enzyme-catalyzing tricyclic modified base formation in tRNA. J Mol Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Goto-Ito S, et al. Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis. Acta Crystallogr D. 2007;63:1059–1068. doi: 10.1107/S0907444907040668. [DOI] [PubMed] [Google Scholar]
- 30.Walsby CJ, et al. Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe-4S]+ cluster of pyruvate formate-lyase activating enzyme. J Am Chem Soc. 2002;124:3143–3151. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 31.Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. An anchoring role for FeS clusters: Chelation of the amino acid moiety of S-adenosylmethionine to the unique iron site of the [4Fe-4S] cluster of pyruvate formate-lyase-activating enzyme. J Am Chem Soc. 2002;124:11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 32.Walsby CJ, et al. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “radical-Sam” protein superfamily. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 33.Nicolet Y, Drennan CL. AdoMet radical proteins, from structure to evolution: Alignment of divergent protein sequences reveals strong secondary structure element conservation. Nucleic Acids Res. 2004;32:4015–4025. doi: 10.1093/nar/gkh728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase-activating enzyme: A Mossbauer spectroscopic study. J Am Chem Soc. 2002;124:912–913. doi: 10.1021/ja017562i. [DOI] [PubMed] [Google Scholar]
- 35.Wong KK, et al. Molecular properties of pyruvate formate-lyase-activating enzyme. Biochemistry. 1993;32:14102–14110. doi: 10.1021/bi00214a005. [DOI] [PubMed] [Google Scholar]
- 36.Kacprzak S, Reviakine R, Kaupp M. Understanding the electron paramagnetic resonance parameters of protein-bound glycyl radicals. J Phys Chem. 2007;111:820–831. doi: 10.1021/jp0674571. [DOI] [PubMed] [Google Scholar]
- 37.Broderick JB, et al. Pyruvate formate-lyase-activating enzyme: Strictly anaerobic isolation yields active enzyme containing a [3Fe-4S]+ cluster. Biochem Biophys Res Commun. 2000;269:451–456. doi: 10.1006/bbrc.2000.2313. [DOI] [PubMed] [Google Scholar]
- 38.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation, and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 40.McRee DE. XtalView/Xfit: A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Chen R, Li L, Weng Z. ZDOCK: An initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 43.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.