Abstract
Somatic hypermutation (SHM) and class-switch recombination (CSR) of Ig genes are dependent upon activation-induced cytidine deaminase (AID)-induced mutations. The scaffolding properties of proliferating cell nuclear antigen (PCNA) and ubiquitylation of its residue K164 have been suggested to play an important role organizing the error-prone repair events that contribute to the AID-induced diversification of the Ig locus. We generated knockout mice for PCNA (Pcna−/−), which were embryonic lethal. Expression of PCNA with the K164R mutation rescued the lethal phenotype, but the mice (Pcna−/−tgK164R) displayed a meiotic defect in early pachynema and were sterile. B cells proliferated normally in Pcna−/−tgK164R mice, but a PCNA-K164R mutation resulted in impaired ex vivo CSR to IgG1 and IgG3, which was associated with reduced mutation frequency at the switch regions and a bias toward blunt junctions. Analysis of the heavy chain V186.2 region after NP-immunization showed in Pcna−/−tgK164R mice a significant reduction in the mutation frequency of A:T residues in WA motifs preferred by polymerase-η (Polη), and a strand-biased increase in the mutation frequency of G residues, preferentially in the context of AID-targeted GYW motifs. The phenotype of Pcna−/−tgK164R mice supports the idea that ubiquitylation of PCNA participates directly in the meiotic process and the diversification of the Ig locus through class-switch recombination (CSR) and somatic hypermutation (SHM).
To mount an effective antibody response, mice and humans create a highly diverse repertoire of antigen binding sites through the rearrangement of the germ line variable (V), diversity (D), and joining (J) Ig locus. Following interaction with antigen, B cells in the germinal centers (GCs) of secondary lymphoid organs express activation-induced cytidine deaminase (AID). AID, together with other enzymes, causes a very high rate (10−5–10−3/base pair/generation) of point mutations in Ig V regions resulting in the affinity maturation and the changes in fine specificity required to produce protective antibodies (1, 2). AID also initiates class-switch recombination (CSR) by mutating the switch regions (SRs) that are located just 5′ of the constant region genes (3, 4). CSR allows antibodies to be distributed throughout the body and to carry out a wide variety of effector functions. AID deaminates deoxycytidines (dC) in single-stranded DNA in the V and SRs to generate deoxyuridine (dU) (1, 2). However, more than half of the mutations in the V and SRs of mice and humans are in A:T bases and are not the result of the direct biochemical action of AID. Rather, these mutations arise during a second phase of SHM and result from the error-prone base excision repair (BER) and mismatch repair (MMR), both of which are recruited to the dU:dG mismatch generated by AID (1, 2, 4).
When critical MMR genes are deleted from mice, most of the mutations in A:T in the V region no longer occur, suggesting that MMR is responsible for the majority of the mutations that arise in A:T bases (1, 2). The remaining mutations in A:T disappear when both MSH2 and UNG (5) are inactivated, indicating that BER is responsible for the remaining mutations in A:T. Deficiency of various MMR proteins leads to different degrees of reduction in CSR (4) and there is virtually no isotype switching in Msh2−/−Ung−/− double knockout mice (5), suggesting that error-prone MMR and BER provide important, but different, and perhaps competing, pathways to create and process the double-stranded DNA breaks (DSB) required for CSR (1, 4). These findings have led to the idea that AID-induced dC-to-dU mutations are repaired in an error-prone fashion by MMR and BER (1). Mutations in A:T bases are reduced in mice and humans deficient in Polη, indicating that Polη is the major translesional error-prone polymerase responsible for the error-prone repair of antibody V regions by MMR and BER (6–8).
A critical and largely unanswered question is how MMR and BER recruit error-prone repair to the V and SRs in centroblast B cells but mediate high-fidelity repair to the rest of the genome in B cells and in general in other cells (9). Studies in yeast and human cells suggest that proliferating cell nuclear antigen (PCNA) that is monoubiquitylated at lysine 164 (10) recruits translesional polymerases, including Polη, to replication forks that are stalled because of DNA lesions (11, 12). PCNA is a DNA-encircling homotrimer that is central to all forms of DNA replication and serves as a sliding platform that recruits MMR and activates polymerases and other factors in a reversible and competitive fashion (13, 14). The monoubiquitylation of PCNA is regulated by a complex mechanism that involves the E2 and E3 ubiquitin ligases, RAD6 and RAD18, and deubiquitylation primarily by USP1 (12, 15, 16). Recent studies in the DT40 chicken B cell line (16, 17) suggest that the recruitment and activation of Polη to the Ig V regions is mediated by PCNA that is monoubiquitylated at residue K164.
As the work described below was being completed it was reported that a mouse expressing PCNA with a lysine-to-arginine mutation at residue 164 that prevented the mono- and polyubiquitylation had a decrease in V region somatic mutations at A:T bases but no significant defect in CSR (18). We have also generated a mouse that expresses PCNA with a lysine-to-arginine mutation at residue 164. The mutant transgene rescues the embryonic lethal phenotype of PCNA-deficient mice, although meiosis progression appears impaired and the mice are sterile. The K164R mutation in PCNA is also responsible for a reduced ability to undergo CSR. Furthermore, an imbalance of A:T vs. C:G mutations at the V region, a reduction in the overall frequency of mutation at the recombined SRs, and an altered pattern of microhomologies at the switch junctions, confirm that ubiquitylation at residue K164 of PCNA plays an important role in the in vivo diversification of the Ig locus.
Results
Rescue of Lethal Knockout Phenotype in Mice Expressing PCNAK164R Transgene (tgK164R).
To investigate the role of ubiquitylation of PCNA in the diversification of the Ig locus, we generated a transgenic mouse that expresses PCNA with a lysine-to-arginine mutation at residue 164 (K164R) in the absence of any endogenous wild-type (WT) PCNA (Pcna−/−tgK164R). We first generated a null mutation of the Pcna gene by deleting exons 2, 3, and 4 through homologous recombination of one allele in embryonic stem (ES) cells [supporting information (SI) Fig. S1 A and C]. These heterozygous ES cells were used to generate Pcna+/− mice which, when intercrossed, resulted in an early embryonic cell lethal phenotype in Pcna−/− progeny consistent with the critical importance of PCNA in orchestrating DNA replication (13).
Mice that were transgenic for PCNA with the K164R mutation were made by carrying out site-directed mutagenesis of exon 4 in a genomic fragment of the mouse Pcna gene that contained ≈3 kb of the 5′ promoter region, all of the exon and intron sequences, and 875 bp of the 3′ untranslated region (Fig. S1 B and C). This mutated transgene was introduced by pronuclear micro-injection into FVB embryos to generate Pcna+/+tgK164R transgenic mice. The Pcna+/+tgK164R mice were then bred to the heterozygous Pcna+/− mice to obtain transgenic mice that were homozygous for the null mutation at the endogenous Pcna locus and expressed the mutant transgene (Pcna−/−tgK164R). The mutant PCNA (tgK164R) rescued the embryonic lethal phenotype of the PCNA-deficient mice, demonstrating that PCNA that cannot be ubiquitylated at residue K164 is able to mediate DNA replication and repair and allows the apparently normal growth and development of the mouse.
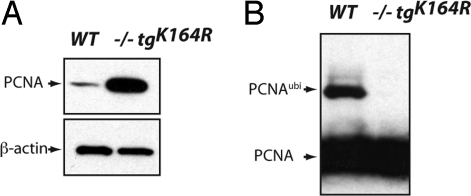
Real-time PCR showed an approximately two- to eightfold increase of steady-state PCNA mRNA in Pcna−/−tgK164R splenic B cells compared to Pcna+/+ cells (Fig. S2). Western blot analysis revealed higher amounts of unmodified PCNA protein in whole cell lysates from mutant B cells (Fig. 1A). Furthermore, modified PCNA at position K164, which has been described to be the only site to accommodate ubiquitylation (19), could not be detected in Pcna−/−tgK164R mice (Fig. 1B).
Fig. 1.
PCNA protein expression in wild-type and Pcna−/−tgK164R mice. Western blot analysis of splenic B cell extracts using anti-PCNA and anti-β-actin antibodies. When compared to WT, a greater amount of unmodified PCNA protein is detected in Pcna−/−tgK164R mice (A). Post-translational modification of lysine 164 is not detected in mutant PCNA protein (B).
Meiotic Defect in Transgenic Pcna−/−tgK164R Mice.
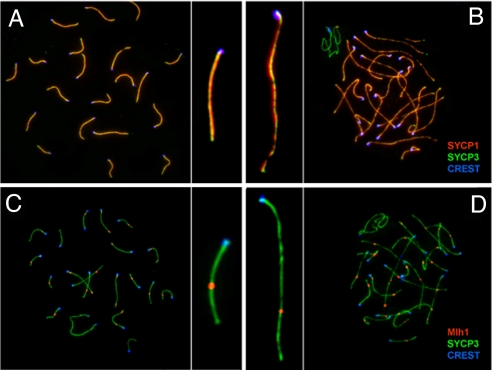
The Pcna−/−tgK164R mice develop normally, but they are sterile. In males, testes size is reduced to <40% of wild-type testes (Fig. S3A). Histopathological analysis revealed that early meiotic progression in Pcna−/−tgK164R males appears normal as indicated by the presence of spermatogonia and early spermatocytes (Fig. S3B). However, meiotic progression is disrupted at early pachynema resulting in the loss of spermatocytes at this stage. Meiotic chromosomes in Pcna−/−tgK164R mice can undergo complete synapsis and form a functional synaptonemal complex as indicated by the synaptonemal complex protein 1 (SYCP1) immunofluorescence, a marker for transversal filament formation (Fig. 2A and 2B) (20). Pcna−/−tgK164R mice also display the localization of Rad51 on meiotic univalent chromosomes at the initial stages of the synaptonemal complex formation during zygonema-to-pachynema transition (data not shown), suggesting that double-strand breaks are being processed and meiotic recombination is initiated. In addition, Mlh1, a key component of meiotic nodules, localized normally in Pcna−/−tgK164R mice during mid pachynema, suggesting crossover formation (Fig. 2 C and D). However, most meiotic chromosomes in mutant mice displayed elongated chromosome axes, suggesting either that meiosis is arrested at an early stage in pachynema in most nuclei or that structural defects in the synaptonemal complex occur (Fig. 2 B and D).
Fig. 2.
Localization of SYCP1 and MLH1 on meiotic chromosomes during zygonema/pachynema of prophase I in wild-type and Pcna−/−tgK164R mice. Left panel (A and C) wild type, right panel (B and D) Pcna−/−tgK164R. (A and B) Colocalization of SYCP1 (red) and SYCP3 (green) indicates complete synapsis (yellow) of meiotic chromosomes in Pcna−/−tgK164R mice during pachynema. Note the elongated chromosome axis of the mutant chromosomes (B) compared to wild-type chromosomes (A). (C and D) Formation of late recombination nodules during pachynema in wild-type (C) and mutant mice (D) is indicated by MLH1 foci (red). Centromers are detected with anti-CREST antibodies (blue).
Impaired ex Vivo Class-Switch Recombination in Transgenic Pcna−/−tgK164R Mice.
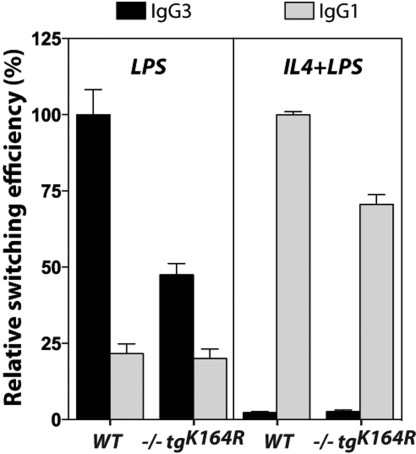
To examine whether the K164R mutation of PCNA has an effect on CSR, we purified splenic B cells from Pcna−/−tgK164R mice and their Pcna+/+ littermates and stimulated them ex vivo with LPS to induce switching from IgM to IgG3, and with LPS plus IL-4 to induce switching to IgG1 (21). The stimulated B cells from individual mice were cultured for 4 days and analyzed by flow cytometry for surface expression of IgG3 and IgG1. Six different experiments were done with two groups of mice: one that consisted of 3-month-old mice that had undergone NP-immunization for the SHM experiments described below, and a second group of unimmunized 8-month-old mice. The groups from the two sets of mice showed similar efficiencies of switching to IgG1 or to IgG3 within each genotype (Fig. S4), so we combined the results from NP-immunized and unimmunized mice (Fig. 3). A representative FACS profile from a PCNA mutant and a wild-type mouse is presented in Fig. S5. The combined results (Fig. 3) show that in Pcna−/−tgK164R mice there is a ≈50% reduction in switching to IgG3 and a ≈25% reduction in switching to IgG1 when compared to their Pcna+/+ littermate controls (P < 0.0001).
Fig. 3.
Reduced ex vivo class-switch recombination in PCNA−/−tgK164R mice. Relative switching to IgG3 and to IgG1 in a total of eight wild-type and eight transgenic mice from six different experiments. The efficiency of switching in the wild-type group within each experiment was defined as 100% and two replicates were assayed for each stimulation. The data shown represent relative mean efficiency of switching ± SEM. P-values were calculated using two-tailed unpaired Student's t tests.
To test if the differences in isotype switching could be the result of a defect in the replication and proliferation of the stimulated B cells expressing the mutated PCNA, the doubling time of the stimulated cells was analyzed by staining with 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) and IgG1 expression was measured at each division (Fig. S6). Throughout the 4 days of stimulation with LPS plus IL-4, the percentage of Pcna−/−tgK164R B cells in each generation was not significantly different from Pcna+/+ controls (Fig. S6A). Consistent with the lack of a defect in replication, the body weight, size of the spleen, and the number of Peyer's patches in Pcna−/−tgK164R mice were comparable to those in Pcna+/+ mice (data not shown). Moreover, the decrease in switching to IgG1 accumulated after the second division, indicating that there was a decrease in switching at each generation (Fig. S6B). In addition, the frequency of B220+PNAhigh GC B cells in the spleen and Peyer's patches was not significantly different between Pcna+/+ and Pcna−/−tgK164R littermates (Fig. S7), suggesting that B cell activation and differentiation are normal in the transgenic Pcna−/−tgK164R mice. Together, these findings confirmed that the inability to ubiquitylate PCNA at residue 164 was not merely delaying the process of CSR, but rather was having a more direct effect on the switching process.
Altered Junctions and Fewer Mutations at the Sμ–Sγ3 Junctions in Pcna−/−tgK164R Mice.
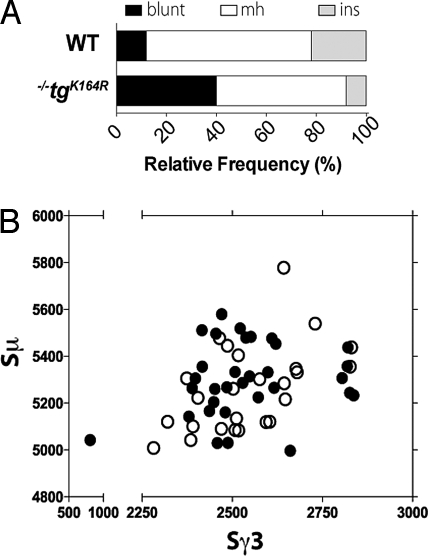
We next asked whether the switching deficiency observed in Pcna−/−tgK164R mice was accompanied by any alteration in the switch junctions. The Sμ–Sγ3 junctional DNA segments from splenic B cells stimulated for 4 days with LPS were compared to Sμ and Sγ3 germline sequences (32 unique junctions from Pcna+/+ and 25 from Pcna−/−tgK164R mice are shown in Fig. S8). There were relatively more blunt Sμ–Sγ3 junctions (P = 0.0285) and fewer microhomologies and insertions in Pcna−/−tgK164R mice when compared to Pcna+/+ littermates (Fig. 4A). However, the remaining Sμ–Sγ3 junctions from Pcna−/−tgK164R mice showed an average length of microhomology (2.4 ± 0.44) similar to the Pcna+/+ control mice (2.1 ± 0.36) (P = 0.6639). There was no significant difference between Pcna+/+ and mutant mice in the utilization of GAGCT or GGGT switching consensus sequences or of WRC/GYW AID hotspots at the breakpoints (Fig. S9A). Furthermore, there was no difference in the parts of the donor μ or recipient γ3 switch regions that participated in the recombination to IgG3 (Fig. 4B).
Fig. 4.
Bias toward blunt Sμ–Sγ3 switch junctions and normal distribution of breakpoints in PCNA−/−tgK164R mice. (A) A significant increase in the relative number of blunt junctions in PCNA−/−tgK164R mice. The graph depicts the relative frequency of junctions with blunt joins, small (1–9 bp) microhomologies (mh) or insertions (ins) at the junction. (B) Scatter analysis of the Sμ–Sγ3 breakpoints from in vitro LPS-stimulated splenic B cells. The x-axis indicates the position of the Sγ3 breakpoint and the y-axis of the Sμ breakpoint. Open circles denote breakpoints from PCNA−/−tgK164R mice, filled circles from their littermate controls.
There was also a significant decrease in the overall frequency of mutation at the donor Sμ region (1 × 10−3 mutations/base in the mutant mice vs. 6.8 × 10−3 in the WT; P < 0.001) and also at the acceptor Sγ3 region (0.8 × 10−3 mutations/base in the mutant mice vs. 2.7 × 10−3 in the WT mice; P = 0.0395) (Table 1). Furthermore, the few mutations detected at the Sμ and Sγ3 regions in the Pcna−/−tgK164R mice were almost exclusively at C:G pairs (Fig. S9 B and C).
Table 1.
Mutation frequency analysis of Sμ–Sγ3 junctions
| Sμ region |
Sγ3 region |
|||
|---|---|---|---|---|
| WT (N = 4) | TG (N = 4) | WT (N = 4) | TG (N = 4) | |
| Sequences analyzed | 32 | 25 | 32 | 25 |
| Mutated sequences | 18 (56.2%) | 5 (20%) | 9 (28.2%) | 4 (16%) |
| Unique sites* | 10,394 | 6,909 | 5,949 | 4,945 |
| Mutated sites† | 70 | 7 | 16 | 4 |
| Overall frequency | 6.8 × 10−3 | 1.0 × 10−3 | 2.7 × 10−3 | 0.8 × 10−3 |
*Theoretical maximum number of unique mutations: total number of nucleotides sequenced from unique recombined junctions.
†Observed number of unique mutations. Because only recombined junctions were scored, every mutation was considered independent.
Altered Pattern of V Region Somatic Hypermutation in Transgenic Pcna−/−tgK164R Mice.
To investigate whether ubiquitylation of PCNA at residue K164 is involved in somatic hypermutation of the V regions of the Ig genes, four Pcna−/−tgK164R and three Pcna+/+ mice were immunized with NP30-CGG and boosted 4 weeks later. One week after boost, splenic B cells were analyzed for the pattern of somatic mutations in the rearranged V186.2 gene, which is a member of the J558 gene family that dominates the response to NP-immunization (22). cDNA was generated from the spleens of the immunized mice using a high-fidelity reverse transcriptase and the rearranged V186.2 genes, including the 5′ region of the Cγ1, were amplified using nested PCR, subcloned and sequenced. The vast majority of the sequences could be assigned to germline V186.2 gene, and the 15 sequences (of a total of 173) that showed higher homology to other members of the family (i.e., V24.8, V23, and V3 genes) were excluded from the mutation analysis.
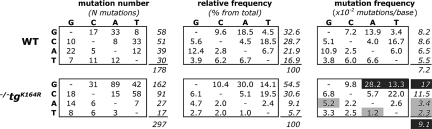
Unique mutations in the nontranscribed strand of the V186.2 region (273 bp) were compiled from a total of 77 and 81 sequences from WT and Pcna−/−tgK164R mice, respectively. The experimental error rate because of reverse transcription and PCR was estimated by analysis of the first 39 bp from the Cγ1 segment adjacent to the V186.2 region to be 0.1 × 10−2 mutations/base (9 mutations in 6,162 bp). The overall frequency of unique mutations at the V186.2 gene in the Pcna−/−tgK164R mice was 9.1 × 10−2 mutations/base, which was significantly higher than the 7.2 × 10−2 mutations/base detected in the Pcna+/+ mice (P = 0.0152) (Table 2, Fig. 5 and Table S1). The overall frequencies of transitions and transversions, however, were similar in both mutant and WT mice (Table 2 and Table S1).
Table 2.
Unique mutation frequency analysis of the V186.2 region
| WT (N = 3) | TG (N = 4) | P | ||
|---|---|---|---|---|
| Sequences analyzed | 77 | 81 | ||
| Mutated sequences | 75 (97%) | 81 (100%) | ||
| Unique sites* | 2,457 | 3,276 | ||
| Mutated sites† | 178 | 297 | ||
| Overall frequency | 7.2 × 10−2 | 9.1 × 10−2 | 0.0152 | |
| G/C mutations | 8.3 × 10−2 | 14.5 × 10−2 | <0.0001 | |
| A/T mutations | 6.0 × 10−2 | 2.9 × 10−2 | <0.0001 | |
| Transversions | 4.8 × 10−2 | 5.9 × 10−2 | 0.1476 | |
| Transitions | 12.1 × 10−2 | 15.3 × 10−2 | 0.0528 | |
| AID hotspot‡ | WRC/ | 15.6 × 10−2 | 21.2 × 10−2 | 0.1641 |
| /GYW | 19.4 × 10−2 | 36.7 × 10−2 | 0.0002 | |
| AID coldspot‡ | SYC/ | 1.8 × 10−2 | 1.4 × 10−2 | 1.0000 |
| /GRS | 1.8 × 10−2 | 4.2 × 10−2 | 0.2262 | |
| Polη hotspot‡ | WA/ | 11.6 × 10−2 | 6.0 × 10−2 | 0.0343 |
| /TW | 6.9 × 10−2 | 4.0 × 10−2 | 0.2527 |
*Theoretical maximum number of unique mutations: number of sites × the number of mice in the category × 3 (because 3 possible substitutions can occur at each site).
†Observed number of unique mutations: within each mouse, identical mutations were counted once.
‡Underlined site of the motif is scored. W = A/T, R = A/G, Y = C/T, and S = G/C.
Fig. 5.
Detailed mutations in the V186.2 region from WT and PCNA−/−tgK164R mice. Left panel, absolute number of unique mutations classified by base pair from (y-axis) → to (x-axis); middle panel, percentages of the total number of unique mutations; right panel, mutation frequency corrected for base composition (number of unique mutations/theoretical maximum number of unique mutations). For each category a contingency table was assigned and a χ2 test was applied (see Table S1). Black boxes denote statistically significant increase of mutation frequency compared to WT; gray boxes denote significant decrease.
In agreement with earlier studies of V regions (1), the Pcna+/+ group showed a similar frequency of unique mutations at C:G and A:T residues (Table 2). However, there were significantly more mutations at C:G than at A:T residues in the Pcna−/−tgK164R group. The reason for this imbalance was that the frequency of unique mutations at A:T was reduced by approximately twofold in the Pcna−/−tgK164R mice (2.9 × 10−2 mutations/base in the mutant mice vs. 6.0 × 10−2 in the WT, P < 0.0001) (Table 2 and Table S1), while the frequency at C:G was increased by approximately twofold in the Pcna−/−tgK164R mice (14.5 × 10−2 mutations/base in the mutant mice vs. 8.3 × 10−2 in the WT, P < 0.0001). This suggests that ubiquitylation of PCNA affects the regulation of the balance between the mechanisms that target A:T and C:G in the V region.
The WA motif, and its complementary TW (W = A or T), has been shown to be a target for Polη on the Ig locus (23, 24), with a preference for A-to-G substitutions on WA motifs of the nontranscribed strand (25). For the WA motif, there was a significant decrease of mutations in the Pcna−/−tgK164R mice compared to Pcna+/+ (P = 0.0343) (Table 2), but no difference was detected for the frequency of mutation at the TW motif (P = 0.2527) that reflects WA targeting on the transcribed strand. Additional evidence for an impairment of Polη activity in the Pcna−/−tgK164R mouse became apparent when the different types of substitutions were examined (Fig. 5 and Table S1). A significant decrease in A-to-G and T-to-A mutations (P < 0.05), which are largely generated by Polη (7), was observed in the Pcna−/−tgK164R mice.
On the other hand, the increase in the frequency of C:G mutations in the Pcna−/−tgK164R mice was attributable to a significant increase in G-to-A and G-to-T substitutions (P ≤ 0.0001), whereas no difference was detected in the overall mutation frequency at C (P = 0.0940) (Fig. 5 and Table S1). This bias toward mutation of G residues was also observed when we examined the frequency of mutation occurring in WRC motifs (Table 2), and in the complementary GYW (R = A or G, Y = C or T) motifs, which have been described as hotspots for AID targeting (26, 27). A significant increase in mutability of the V186.2 nontranscribed strand at GYW motifs (P = 0.0002), which reflects the targeting of WRC on the transcribed strand, was not accompanied by the same increase in WRC mutations (P = 0.1641). A role for AID in the strand-biased targeting of C in hotspots in the transcribed strand was further supported by the absence of difference in the frequency of mutation of SYC/GRS AID coldspots (S = G or T) (P ≥ 0.2262) (Table 2). In summary, the K164R mutation of PCNA reveals a strand bias in C:G mutations and an excess of targeting of GYW motifs on the nontranscribed strand of the V186.2 region. This may be interpreted to mean that Cs within the context of WRC motifs on the transcribed strand are targeted more frequently for mutation when PCNA cannot be ubiquitylated.
Discussion
Lysine 164 in PCNA has been previously shown in yeast and cultured animal cells to be the site of monoubiquitylation that can facilitate and coordinate the recruitment and activation of translesional polymerases to process DNA lesions through error-prone or error-free pathways (10–12). Transgenic expression of PCNA with a lysine-to-arginine mutation at that residue (tgK164R) rescued the lethal phenotype of the Pcna−/− mice, indicating that the post-translational modification of PCNA at residue K164 is not essential for growth and development. The development of GC B cells (B220+PNAhigh cells) also appeared to be normal. However, the development of reproductive germinal cells was severely affected and resulted in sterility both in the Pcna−/−tgK164R mice described in this paper and in a knockin mouse in which Pcna, with the same K164R mutation, has replaced the wild-type gene (18), suggesting that the block in meiosis that we observed in the Pcna−/−tgK164R mice is not because of overexpression of the transgene. Although meiotic chromosomes in mutant males are able to undergo synapsis, form a functional synaptonemal complex, process DSBs during the initiation of meiotic recombination, and initiate crossover formation, we failed to observe any meiotic nuclei at stages beyond pachynema and the chromosome axes appeared elongated, suggesting a buildup of early pachynema nuclei.
On the basis of previous work with yeast (12), human cells (11), and DT40 chicken cells (16, 17, 19), we examined whether the K164R mutation would interfere with the second phase of SHM that is responsible for the mutations in A:T residues, which have been attributed to the recruitment of error-prone repair mediated primarily by Polη (6, 8, 28, 29). We found that in the V186.2 region of Pcna−/−tgK164R mice, there is a significant reduction in the frequency of mutations in A:T residues compared to their Pcna+/+ littermates, especially in the WA motifs that are on the nontranscribed strand and are preferentially targeted by Polη (7, 25). Physical interaction with PCNA is essential in vivo and in vitro for the activity of Polη, stimulating its error-prone activity (30), and MSH2/MSH6 stimulate the activity of Polη in B cell lysates (31). However, it is still controversial whether ubiquitylation of PCNA at residue K164 enhances the binding affinity for Polη (12, 32) or is dispensable for the access of Polη to PCNA (33) but disrupts the binding of other factors that interfere with the recruitment of error-prone polymerases (30). Whatever the mechanism, the results reported here and in the knockin PcnaK164R/K164R mice support the notion that ubiquitylation of PCNA at residue 164 plays an important role in the introduction of Polη-mediated mutations at A:T bases in the Ig V and SRs. The approximately twofold reduction of mutation frequency in A:T bases (or ≈62% reduction in relative frequency) that we show here is comparable to that reported for Polη-deficient mice (6, 7). Langerak et al. reported an even greater reduction in mutations in A:T (18). This difference may reflect the different V regions or types of B cells analyzed or the fact that we corrected for base composition and scored only unique mutations. Nevertheless, the combined findings suggest the existence of a PCNAubi-independent pathway.
The decrease in mutations at A:T in the V region of the Pcna−/−tgK164R mice is associated with an increase in mutations at C:G residues that is largely attributable to G-to-T transversions and G-to-A transitions located in GYW AID hotspots. This suggests that when PCNA cannot be ubiquitylated, there is a strand-biased hypermutation process that favors C in WRC motifs in the transcribed strand. It is possible that AID targeting to the transcribed strand is favored when PCNA cannot be ubiquitylated and is followed by recruitment of error-prone repair to resolve the AID induced dU:dG mismatches or that short-patch BER might be favored when PCNA cannot be ubiquitylated, and this might happen in a strand-biased manner. This is supported by studies suggesting that MMR might act preferentially on the top strand displacing BER to the bottom strand (34, 35). Because no significant change was detected in the mutation frequency of C sites (especially in C-to-G mutations) or in G-to-C mutations, which could be contributed by the error-prone polymerase Rev1 (36, 37), it seems unlikely that Rev1 activity is impaired by the loss of PCNA ubiquitylation or that Rev1 is responsible for the increase of mutations at C:G pairs in the Pcna−/−tgK164R mice. Therefore, other error-prone polymerases may be favored in the absence of ubiquitylation of PCNA and contribute to the strand bias in mutations at C:G residues.
In general, our findings are consistent with those reported in the comparable knockin mouse (18) that does not overexpress PCNA. This suggests that the K164R mutation rather than the overexpression of the mutant PCNA in the transgenic mouse studied here is responsible for the changes in the characteristics of the V region mutations in the Pcna−/−tgK164R mice. While the knockin mouse did not have a significant defect in switching (18), the B cells from Pcna−/−tgK164R mice have a reduced ability to undergo ex vivo class switching to IgG1 and IgG3. This may reflect some technical difference in the way the experiments were done or analyzed, but the knockin mice do show a tendency toward a decrease in switching (18). In addition, we do not think that the increased switching phenotype in the transgenic mice is a result of the overexpression of the mutant PCNA because the frequency of switching was the same in two Pcna+/+tgK164R mice, which overexpressed total PCNA protein, and in their Pcna+/+ littermates (data not shown).
During CSR, MMR and BER pathways are involved in the generation of DNA double-strand breaks appropriate for end-joining recombination (4). All classical nonhomologous end-joining (NHEJ) factors tested to date have been shown to play a role in CSR (38). An alternative end-joining pathway during CSR, which involves microhomologies and the absence of blunt junctions, has recently been uncovered in the absence of specific NHEJ factors (39). The decrease in switching reported here for the Pcna−/−tgK164R mice was associated with a bias toward the use of Sμ–Sγ3 blunt junctions and a reduced frequency of nonblunt junctions (i.e., microhomologies and insertions). This observation suggests that there is a reduction in the efficiency of microhomology-mediated repair when PCNA cannot be ubiquitylated and resembles the results observed in Exo1−/− (40) and Msh2−/− mice (41). In contrast, an increased frequency of longer microhomologies has been observed in Msh2−/−Mlh1−/− (42) or in Mlh1−/− or Pms2−/− mice (41), which has been interpreted to suggest that MLH1/PMS2 heterodimer prevents long stretches of junctional homology while MSH2 acts subsequently or independently on the end processing of the junctions (42). As opposed to the preference of CSR for consensus motifs in Msh2−/− mice (43), the targeting to the GAGCT or GGGT consensus sequences or the WRC/GYW AID hotspots at the sites of recombination or the location of those sites was not affected in Pcna−/−tgK164R mice. Our data suggest: (i) that PCNA might function in coordination with MMR complexes during the first steps of DSB generation at the SRs, which would be consistent with previous studies suggesting that binding to PCNA is a mechanism of MMR recruitment and (ii) that ubiquitylation of PCNA plays an important, but not absolutely essential, role in the end-joining pathways that operate during CSR. Furthermore, the changes we observed in the recombined switch junctions might reflect a defect in the processing of DSBs, which could also result in a defect in meiotic process(es) and the activation of the pachynema checkpoint.
Because defective human Polη (28) and Polη−/− mice (35) do not show a deficiency in CSR, our results also suggest that additional factors, besides Polη, are recruited and/or regulated by ubiquitylation of PCNA at lysine 164 and are responsible for the defect in mutation of SRs and the impairment of CSR shown in Pcna−/−tgK164R mice. One possibility is that the absence of ubiquitylated PCNA may drive the BER reaction within SRs toward a high-fidelity Polβ-dependent short-patch BER, as opposed to long-patch BER, compromising the efficiency of CSR. In fact, it has been shown that Polβ plays an inhibitory role in CSR by error-free repair of breaks in SRs (44). Although Polβ is not critically involved in SHM (45), there are increased mutations in the SRs of Polβ−/− B cells (44). The fact that Pcna−/−tgK164R mice show a significant reduction in the overall frequency of mutation at the SRs might be explained by an equilibrium/competition between Polη error-prone and Polβ error-free repair at the SRs of the Ig locus.
Materials and Methods
Mice.
Pcna−/−tgK164R and wild-type littermate mice were housed in a pathogen-free facility. All protocols involving animals have been approved by the Animal Care and Use Committee of Albert Einstein College of Medicine (AECOM) in accordance with the US Public Health Service Animal Welfare Policy. A group of four wild-type and four Pcna−/−tgK164R unimmunized 8-month-old mice, and a second group of four wild-type and four Pcna−/−tgK164R NP-immunized 3-month-old mice were used for this work. Genotyping strategy is shown in SI Text.
Analysis of PCNA Expression by Western Blot.
Protein was extracted from splenic resting B cells using Novex Tris-Glycine SDS sample buffer (Invitrogen), and SDS/PAGE was performed under reducing conditions using 4−20% Novex Tris-Glycine gels (Invitrogen). PCNA and β-actin were detected with PC10 (Santa Cruz Biotechnology) and AC-74 (Sigma) mouse IgG2a monoclonal antibodies, respectively. Analysis of PCNA expression by real-time PCR is shown in SI Text.
Analysis of Chromosome Spreads.
Chromosome spreads were prepared as previously described (46) and subjected to indirect immunofluorescence using antibodies against MutL homolog (Mlh1), RecA homolog (Rad51), centromers (CREST), and synaptonemal complex proteins (SYCP1 and SYCP3). Secondary antibodies (Jackson Immunochemicals) were conjugated to fluoroscein, Cy3, or Cy5 and images were captured with an Olympus BX61 upright microscope (coolsnap HQ camera) and processed with IP Lab acquisition software.
Ex Vivo Class-Switching Assay.
Splenic B cells from immunized and nonimmunized PCNA−/−tgK164R and wild-type mice were isolated and depleted of T cells by complement-mediated lysis (21). Splenocytes were stimulated with either 50 μg/ml of LPS (Sigma-Aldrich) or LPS plus 50 ng/ml of rIL-4 (R&D Systems). After 4 days in culture, surface IgM and IgG were stained and analyzed by FACS as previously described (47). B cell proliferation analysis is shown in SI Text.
Switch Junction Analysis.
Genomic DNA was extracted from splenic B cells stimulated in vitro for 4 days with LPS. Junctional Sμ–Sγ3 regions were amplified using high-fidelity PfuTurbo DNA polymerase (Stratagene) in two sequential rounds with specific primers as previously described (48, 49). Detailed description of cloning and sequence analysis of switch junctions is shown in SI Text.
Hypermutation Analysis.
Littermates of 6-week-old PCNA−/−tgK164R and wild-type mice were immunized i.p. with (4-hydroxy-3-nitrophenyl)acetyl (NP)30-CGG (BioSearch Technologies) in alum (Pierce) (40) and boosted 4 weeks after primary immunization. RNA was prepared 7 days after boost from splenic B cells homogenized in TRIzol (Invitrogen) and cDNA was synthesized using oligo(dT) and AccuScript high-fidelity reverse transcriptase (Stratagene). A nested PCR (PCR) was performed with high-fidelity PfuTurbo DNA polymerase (Stratagene) to amplify VH186.2 joined to the Ig IgG1 constant region as previously described (40). Cloning and sequencing of the PCR products were done as described for switch junction analysis. Sequence alignment was done with SeqMan 5.07 software (DNASTAR Inc.), excluding primer areas and using GenBank sequences J00530.1 (nucleotides 224–496) for the V186.2 region, and NC_000078 (nucleotides 1–39) for the Cγ1 region, as consensus sequences. Analysis of mutated sequences was done using the SHMTool webserver (http://scb.aecom.yu.edu/cgi-bin/p1).
Supplementary Material
Acknowledgments.
The authors thank M. Sadowsky, B. Birshtein, and J. Stavnezer for helpful discussions; R. Sellers for comparative pathology services; and M. Fan for additional technical support. This work was supported by Postdoctoral Fellowship EX-2006-0732 from the Spanish Ministry of Education and Science (to S.R.); the Medical Scientist Training Program T32GM007288 (to J.U.P. and F.L.K.); and National Institutes of Health Grants CA72649 and CA102705 (to M.D.S.), CA76329 and CA93484 (to W.E., E.A., and U.W.), and AG028872 and P01-G027734 (to T.M. and A.B.). Support also came from the Harry Eagle Chair provided by the National Women's Division (to M.D.S.) and the Seaver Foundation Center for Bioinformatics (to T.M. and A.B.), both at Albert Einstein College of Medicine.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808182105/DCSupplemental.
References
- 1.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 2.Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 4.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 6.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda K, et al. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J Biol Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- 8.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amsterdam) 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 10.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 11.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 12.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: Functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 15.Ulrich HD. Deubiquitinating PCNA: A downside to DNA damage tolerance. Nat Cell Biol. 2006;8:303–305. doi: 10.1038/ncb0406-303. [DOI] [PubMed] [Google Scholar]
- 16.Bachl J, Ertongur I, Jungnickel B. Involvement of Rad18 in somatic hypermutation. Proc Natl Acad Sci USA. 2006;103:12081–12086. doi: 10.1073/pnas.0605146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arakawa H, et al. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson LJ, et al. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr Rev. 2006;27:398–426. doi: 10.1210/er.2005-0017. [DOI] [PubMed] [Google Scholar]
- 21.Schrader CE, Edelmann W, Kucherlapati R, Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J Exp Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss U, Rajewsky K. The repertoire of somatic antibody mutants accumulating in the memory compartment after primary immunization is restricted through affinity maturation and mirrors that expressed in the secondary response. J Exp Med. 1990;172:1681–1689. doi: 10.1084/jem.172.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda T, et al. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 25.Mayorov VI, Rogozin IB, Adkison LR, Gearhart PJ. DNA polymerase eta contributes to strand bias of mutations of A versus T in immunoglobulin genes. J Immunol. 2005;174:7781–7786. doi: 10.4049/jimmunol.174.12.7781. [DOI] [PubMed] [Google Scholar]
- 26.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 27.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophy Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 28.Zeng X, Negrete GA, Kasmer C, Yang WW, Gearhart PJ. Absence of DNA polymerase eta reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions. J Exp Med. 2004;199:917–924. doi: 10.1084/jem.20032022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faili A, et al. DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J Exp Med. 2004;199:265–270. doi: 10.1084/jem.20031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson TM, et al. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J Exp Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of polymerase eta in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolaishvili-Feinberg N, et al. Ubiquitylation of proliferating cell nuclear antigen and recruitment of human DNA polymerase eta. Biochemistry. 2008;47:4141–4150. doi: 10.1021/bi702329h. [DOI] [PubMed] [Google Scholar]
- 34.Unniraman S, Schatz DG. Strand-biased spreading of mutations during somatic hypermutation. Science. 2007;317:1227–1230. doi: 10.1126/science.1145065. [DOI] [PubMed] [Google Scholar]
- 35.Martomo SA, et al. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc Natl Acad Sci USA. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen JG, et al. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross AL, Sale JE. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Jolly CJ, Cook AJ, Manis JP. Fixing DNA breaks during class switch recombination. J Exp Med. 2008;205:509–513. doi: 10.1084/jem.20080356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 40.Bardwell PD, et al. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 41.Schrader CE, Vardo J, Stavnezer J. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J Exp Med. 2002;195:367–373. doi: 10.1084/jem.20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrader CE, Vardo J, Stavnezer J. Mlh1 can function in antibody class switch recombination independently of Msh2. J Exp Med. 2003;197:1377–1383. doi: 10.1084/jem.20022190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehrenstein MR, Neuberger MS. Deficiency in msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: Parallels with somatic hypermutation. EMBO J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X, Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J Exp Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esposito G, et al. Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc Natl Acad Sci USA. 2000;97:1166–1171. doi: 10.1073/pnas.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolas NK, et al. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol. 2005;171:447–458. doi: 10.1083/jcb.200506170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, et al. The mismatch repair protein Msh6 influences the in vivo AID targeting to the Ig locus. Immunity. 2006;24:393–403. doi: 10.1016/j.immuni.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 48.Wu X, et al. A role for the MutL mismatch repair Mlh3 protein in immunoglobulin class switch DNA recombination and somatic hypermutation. J Immunol. 2006;176:5426–5437. doi: 10.4049/jimmunol.176.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehrenstein MR, Rada C, Jones AM, Milstein C, Neuberger MS. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc Natl Acad Sci USA. 2001;98:14553–14558. doi: 10.1073/pnas.241525998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.