Abstract
Dyggve–Melchior–Clausen syndrome and Smith-McCort dysplasia are recessive spondyloepimetaphyseal dysplasias caused by loss-of-function mutations in dymeclin (Dym), a gene with previously unknown function. Here we report that Dym-deficient mice display defects in endochondral bone formation similar to that of Dyggve–Melchior–Clausen syndrome and Smith-McCort dysplasia, demonstrating functional conservation between the two species. Dym-mutant cells display multiple defects in vesicle traffic, as evidenced by enhanced dispersal of Golgi markers in interphase cells, delayed Golgi reassembly after brefeldin A treatment, delayed retrograde traffic of an endoplasmic reticulum-targeted Shiga toxin B subunit, and altered furin trafficking; and the Dym protein associates with multiple cellular proteins involved in vesicular traffic. These results establish dymeclin as a novel protein involved in Golgi organization and intracellular vesicle traffic and clarify the molecular basis for chondrodysplasia in mice and men.
Keywords: bone formation, gene trap mutation, Golgi, proteomics, growth plate
Most mammalian growth occurs ex utero and requires extensive endochondral bone formation, a spatially organized process orchestrated by proliferating growth plate chondrocytes that subsequently differentiate and undergo apoptosis and ossification (1, 2). Inherited defects in bone formation, or osteochondrodysplasias, are responsible for more than 200 clinically distinct diseases that occur with a cumulative incidence exceeding 1 in 10,000 live births (3, 4) caused by mutations in processes critical for osseous bone formation and maintenance. These include growth factor signaling (5), extracellular matrix synthesis (6–13) and remodeling (14–16), protein sorting (17–20), and vesicle traffic (21, 22).
Dyggve–Melchior–Clausen syndrome (DMC) and Smith-McCort dysplasia (SMC) are recessive spondyloepimetaphyseal dysplasias caused by loss-of-function mutations in dymeclin (Dym) (23–25). However, the functions of the DYM gene and the molecular mechanisms responsible for DMC and SMC have not been determined. Aside from potential membrane-spanning sequences, N-myristoylation, and protein trafficking (dileucine) motifs (26), the 74-kDa DYM protein lacks features that would guide predictions as to its functions (23, 25). In addition to being structurally unique, DYM protein sequences are shared only by interspecies orthologues, and no related sequences are present in the Saccharomyces cerevisiae genome.
The present study used genetic and proteomic strategies developed to analyze genes, like Dym, that have been identified by genome sequencing and whose functions are unknown (27–29). In the present study, dymeclin functions were analyzed starting with a mutation induced by gene trapping in murine ES cells (30) and subsequently introduced into the mouse germline. Dym-deficient mice developed chondrodysplasia similar to that of DMC and SMC, providing a murine model of the human diseases and demonstrating functional conservation between the mouse and human proteins. Trafficking defects in Dym-deficient cells, combined with proteomic analysis of Dym-interacting proteins, establish dymeclin as a novel protein involved in Golgi organization and vesicle transport.
Results
Characterization of Dym-Mutant Mice.
A mutation in the Dym gene [supporting information (SI) Fig. S1A] was identified in a screen of ES cell clones mutagenized by the GTR1.3 gene trap vector (30) and was introduced into the mouse germline. The gene trap vector disrupted Dym expression (Fig. S1B) owing to splicing of the first 16 exons to a 3′ exon consisting of the 3′ end of puromycin resistance gene and the IRES-LacZ reporter. The resulting Dym-vector fusion transcripts lack exon 17 and encode a fusion protein that replaces the last 49 aa of Dym with 137 irrelevant residues. Thus, the gene trap allele closely resembles frame-shift mutations in exon 17 observed in some patients with DMC (25, 31).
Mice heterozygous for the Dym mutation were indistinguishable from wild-type mice. Homozygous mutant pups were obtained at Mendelian frequencies (data not shown), but the animals were slightly smaller at birth than wild-type or heterozygous littermates. Differences in body weight became more pronounced with age, and after 12 months the mutants were 75% of the size of wild-type animals (Fig. S2 A and B). All mutant mice exhibited skeletal abnormalities of variable severity characterized by shortened trunk, spine curvature, bite overclosure, shortening of tubular bones, smaller skull, and reduced rib cage volume (Figs. S2C and S3). The lengths and widths of selected bones (scull, vertebrae, femur, tibia, and humerus) were reduced in mutant animals to 88%–93% of wild type (Table S1). Other organs and tissues seemed to have normal histology. As with human patients with SMC, the mice did not display obvious signs of neurologic impairment; however, our analysis did not include specific behavioral tests. In addition, by 6 months of age, 35% (n = 53) of the mutant mice developed unilateral or bilateral hydronephrosis caused by a collagenous obstruction where the urethra joins the kidney (Fig. S4), a phenotype not associated with the human diseases.
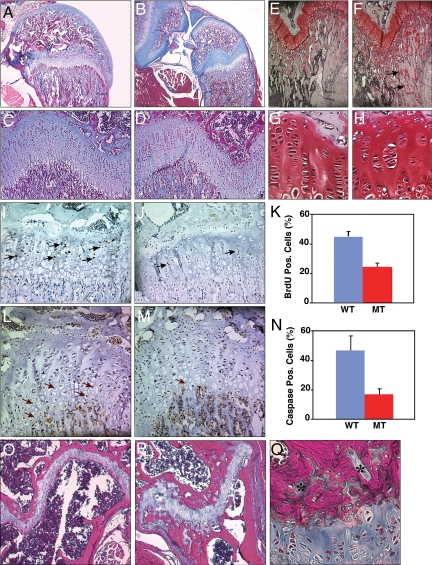
Most long bone growth occurs by postnatal endochondral ossification at the epiphyseal growth plates (1, 2). Secondary ossification centers of homozygous mutant animals at 2 weeks of age were less developed than in wild-type littermates, as illustrated by larger, blue-staining cartilaginous regions (Fig. 1B). At 1 month of age the overall organization of the growth plate in mutant mice was essentially normal, although the hypertrophic zone (i.e., the region just below the stacked chondrocytes; Fig. 1 C and D) in mutant mice was slightly narrower, and the trabecular zone (lower areas of Fig. 1 C and D) seemed to have more collagenous (blue staining) and safranin O-positive unossified extracellular matrix (Fig. 1 C–H). Chondrocytes were sometimes organized into clusters rather than as stacks of cells (Fig. 1 G and H), and the growth plates appeared somewhat hypocellular. Chondrocyte proliferation (Fig. 1 I–K) and the number of caspase 3-positive chondrocytes in the hypertrophic zone were also reduced, suggesting delays in programmed cell proliferation, hypertrophy, and death (Fig. 1 L–N). There was no obvious difference in aggrecan (large aggregating proteo glycan, a major structural component of cartilage) staining (Fig. S5).
Fig. 1.
Growth plate abnormalities in Dym-mutant mice. Sections of proximal tibial and distal femoral epiphysis were examined in wild-type (A, C, E, G, I, L, O) and mutant (B, D, F, H, J, M, P, Q) mice at 2 weeks (A and B), 1 month (C–N), and 6 months of age (O–Q). Sections were stained with either Gomori's trichrome blue (A–D, O–Q) or safranin O (E–H), or were analyzed for BrdU incorporation (I and J) or for caspase 3 expression (L and M). Several islands of chondrocytes within ossified regions (*), and BrdU- and caspase-positive cells (arrows) are indicated. The graphs display the percentage of cells in the proliferative or hypertrophic zones that stained for BrdU incorporation (K) or caspase 3 expression (N), respectively.
By 6 months, the growth plates of mutant mice displayed disorganized patterns of ossification (i.e., the magenta-stained regions of Fig. 1 O and P). Although chondrocytes are normally organized in a compact band separated from trabecular bone by a narrow ossified region (Fig. 1O), the chondrocytes were disorganized and were surrounded by thicker ossified regions (Fig. 1P). The ossified regions also contained numerous small islands of chondrocytes (Fig. 1 P and Q).
Finally, primary mouse embryonic fibroblasts (MEFs) and chondrocytes from mutant animals displayed ultrastructural features resembling those reported for cells from patients with DMC (25, 32). Mutant cells contained large vacuoles, multiple smaller vesicles, and electron-dense bodies (Fig. S6); had less developed Golgi; and presumptive cisternae of the rough endoplasmic reticulum (ER) appeared dilated. In short, histologic phenotypes associated with the murine Dym mutation were remarkably similar to those in DMC patients, suggesting that the functions of the protein are conserved between the two species.
Dymeclin-Interacting Proteins.
Because Dym was a novel and entirely uncharacterized protein, Dym-associated proteins were characterized to guide experiments on biologic function. For this, a tandem affinity (TAP) tagged dymeclin fusion protein (Dym-C-TAP) was expressed in cells, and proteins that copurified through tandem affinity chromatography (Fig. S7) were identified by mass spectrometry (33, 34) (Table S2). Proteins recovered with Dym-C-TAP in two or more experiments included a number of Golgi- and ER-resident proteins, vesicular transport proteins, and nuclear transport proteins (e.g., several subunits of coatomer protein complex, clathrin-adaptor protein [AP] 1 complex subunits, Golgi vesicle docking protein p115, a SNARE protein YKT6, Golgi tethering protein giantin, a subunit of retromer complex VPS35, vesicle fusion factor, N-ethylmaleimide-sensitive factor, clathrin assembly protein PICALM and surfeit 4 [an orthologue of yeast Erv29p, a membrane receptor required for COPII-dependent export of certain proteins from the ER], dynein subunits, and dynamin-like protein 1 [DNM1L]). However, abundant proteins (e.g., cytoskeletal proteins, ribosomal proteins, and translation factors) and proteins involved in removing misfolded and/or overexpressed proteins (e.g., chaperones and proteosome components) frequently associate with TAP-tagged baits and may not reflect physiologic interactions with Dym-C-TAP.
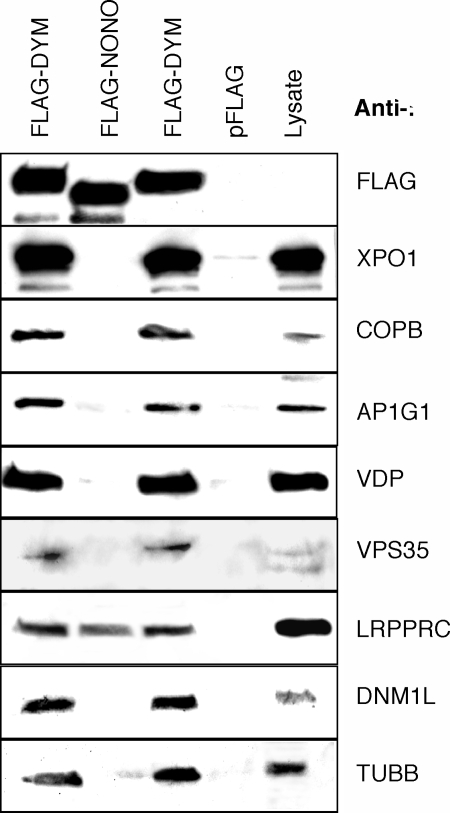
To confirm that proteins identified by mass spectrometry interact with Dym, selected proteins were tested for the ability to coimmunoprecipitate with FLAG-tagged Dym (Fig. 2). Dym coprecipitated with exportin 1, coatomer protein subunit β (COPB), adaptor protein 1 subunit 1 (AP1G1), vesicle docking protein p115 (VDP), vacuolar sorting protein (VPS35), leucine-rich PPR-motif containing protein (LRPPRC), DNM1L, and β tubulin (TUBB). None of these proteins coprecipitated from cells expressing the FLAG sequences alone, and only LRPPRC coprecipitated with FLAG-NONO (non-POU domain containing, octamer-binding protein). The NONO–LRPPRC interaction presumably reflects the ability of both proteins to bind RNA. As controls, the Western blots were also probed for proteins (HRNPA1, MAPK1, SMARCA4, and AURKA) that did not copurify with Dym-C-TAP. Altough all of these proteins were readily detected in cell lysates by Western blotting, none appeared to coprecipitate with FLAG-DYM (data not shown). Moreover, DYM-C-TAP did not cosediment with ribosomes or polyribosomes fractionated by sucrose gradient centrifugation (data not shown).
Fig. 2.
Western blot analysis of Dym-interacting proteins. Proteins immunoprecipitated with an anti-FLAG antibody from 293T cells expressing FLAG-Dym, the FLAG sequence alone, or FLAG-NONO were analyzed by Western blotting using antibodies against the FLAG sequence (FLAG), exportin 1 (XPO1), COPB, AP1G1, VDP, VPS35, LRPPRC, DNM1L, and TUBB. A 293T cell extract (Lysate) was also analyzed to monitor levels of protein expression corresponding to 10% of the input for immunoprecipitation.
Subcellular Localization of Dymeclin and Dym-Interacting Proteins.
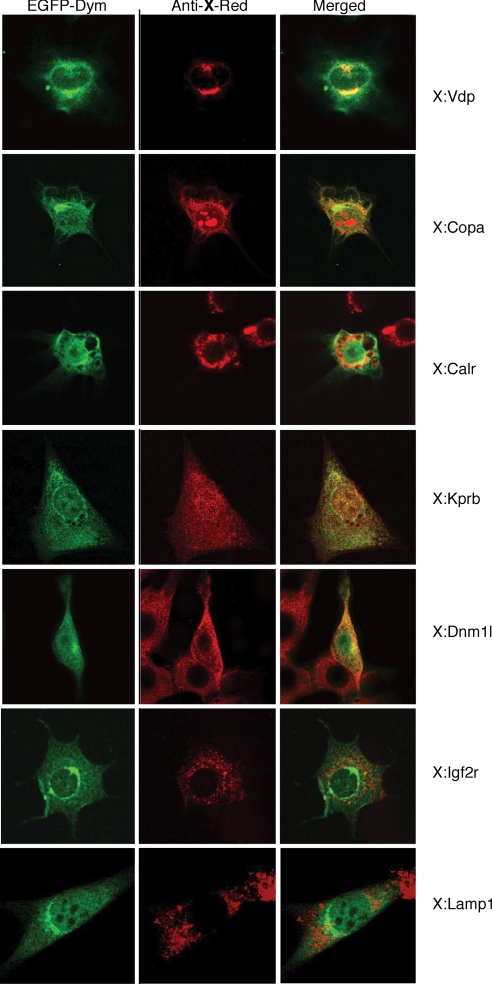
The number and variety of Dym-interacting proteins suggested that the dymeclin protein is located in multiple cellular compartments. Because Dym antibodies were not available, we examined the subcellular localization of a dymeclin-enhanced green fluorescent protein fusion protein (EGFP-Dym) by confocal microscopy. Although EGFP-Dym was distributed throughout the cell, including the nucleus and cytoplasm, the protein was most concentrated in the perinuclear Golgi region (Fig. 3, Left). This region overlapped with areas stained by anti-p115(Vdp), a Golgi and ER-to-Golgi intermediate compartment marker, and partially overlapped with the staining for calreticulin, an ER marker (Fig. 3, Right). Dymeclin also displayed some colocalization with a COPI coat subunit protein (Copa) and with karyopherin (importin) β and dynamin-like protein 1 in punctate structures throughout the cell (Fig. 4). By contrast, Dym fluorescence did not colocalize with the late endosome marker Igf2r or with the lysosome marker Lamp1. Similarly, in cellular fractionation experiments, a Dym-C-TAP fusion protein was distributed in multiple compartments, in contrast to specific reference proteins (Fig. S8). Finally, EGFP-Dym also colocalized with α-tubulin (Fig. S9A), predominantly in the perinuclear Golgi region, and was rendered resistant to extraction by 0.5% Triton X-100 by agents that stabilize microtubules (Fig. S9B). This suggests that a significant amount of the perinuclear dymeclin, like other Golgi proteins, is microtubule associated.
Fig. 3.
Localization of EGFP-Dym. Cells expressing an EGFP-Dym were stained with antibodies to the indicated proteins together with an Alexa594-conjugated secondary antibody and visualized by confocal microscopy: Vdp (p115, vesicular docking protein), Calr (calreticulin), Copa (coatomer subunit protein α), Kprb (karyopherin/importin β), Dnm1 (dynamin like protein 1), Igf2r (insulin-like growth factor 2 receptor), and Lamp1 (lysosomal associated membrane protein 1) marker. EGFP (green) and Alexa594 (red) fluorescence are shown in Left and Middle, respectively, and regions of overlapping fluorescence (yellow) are shown in the merged images on the Right.
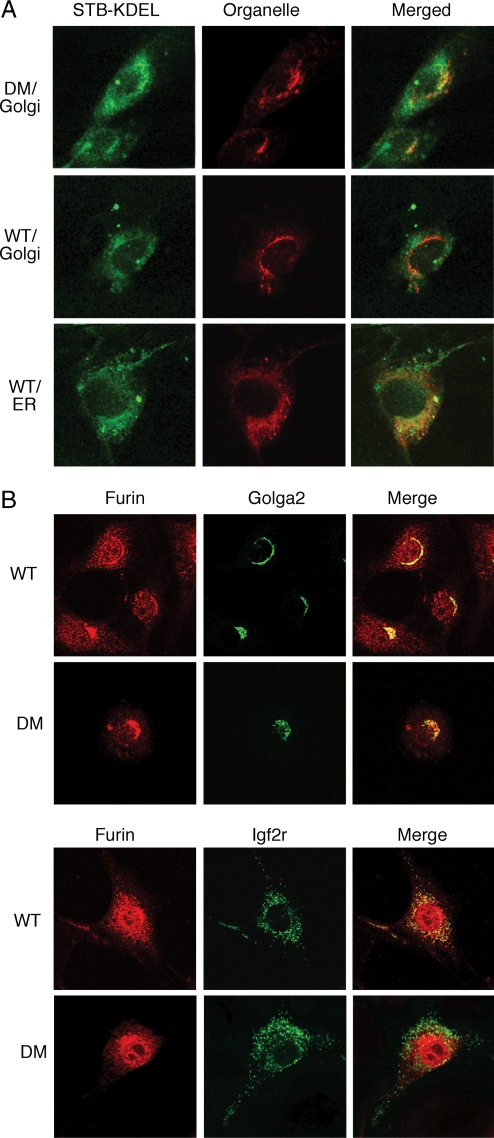
Fig. 4.
Impaired vesicle transport in Dym-mutant fibroblasts. (A) Altered trafficking of STB-KDEL in Dym-mutant cells. Wild-type (WT) and Dym-mutant (DM) embryonic fibroblasts were exposed to fluorescent STB-KDEL, were incubated for 4 h, and were fixed and stained with antibodies for Golgi (Golga2) or ER (calreticulin) marker proteins. Mutant cells displayed delayed retrograde transport, with some STB-KDEL remaining in the Golgi. (B) Altered furin localization in Dym-mutant cells. Wild-type (WT) and Dym-mutant (DM) embryonic fibroblasts were stained with antibodies against furin and Golga2 (Upper) or furin and Igf2r (Lower). Dym-mutant cells displayed pronounced loss of furin staining in late endosomes, as compared with wild-type cells.
Vesicular Transport Defects in Dym-Mutant Cells.
These results indicate that Dym is highly associated with cellular proteins and compartments involved in the transport of vesicles and proteins into and out of the Golgi. Dym involvement in Golgi function was therefore assessed by analyzing Dym-mutant cells for defects in Golgi reassembly after brefeldin A (BfA) treatment, retrograde traffic of an ER-targeted Shiga toxin B subunit, and furin trafficking.
BfA causes the Golgi in interphase cells to redistribute to the ER by COPI-vesicle-independent retrograde traffic (35). Golgi membranes reassemble after removal of the drug by anterograde (ER-to-Golgi) traffic. This can be visualized by staining cells for Golga2 (GM130), a cis-Golgi marker protein. Although no differences in Golgi disassembly were observed when comparing wild-type and mutant cells, a higher proportion of untreated Dym-mutant cells appeared to have less-compact, fragmented Golgi (Fig. S10). In addition, relocation of Golga2 between the ER and Golgi was delayed in mutant cells recovering from BfA treatment. Thus, in wild-type cells, most Golga2 fluorescence returned to the normal distribution within 30 min after BfA washout, whereas localization of Golga2 in mutant cells was not restored even after 60 min (Fig. S10).
KDEL-tagged Shiga toxin subunit B (STB-KDEL) is internalized by endocytosis and traffics to the Golgi and then to the ER by COPI-dependent retrograde traffic that involves recognition of the KDEL ER-retrieval motif (36). Mouse fibroblasts were exposed to fluorescent STB-KDEL for various times, fixed, and stained for Golgi (Golga2) or ER marker (calreticulin) proteins. By 4 h much of the STB-KDEL in wild-type cells was located in the ER, as expected (Fig. 4A). By contrast, some of STB-KDEL in mutant cells remained in the Golgi, suggesting that Dym deficiency impairs retrograde traffic between the Golgi and ER (Fig. 4A).
Interactions between dymeclin and proteins of the clathrin-adaptor protein complex 1 (AP1) and the retromer complex subunits, which participate in the trafficking of proteins between the trans-Golgi network (TGN) and endosomal compartments (37), led us to examine wild-type and Dym-mutant cells for differences in the furin localization. Although furin is concentrated in TGN, the protein is transported to the plasma membrane and recycles via endosomes back to the TGN. In wild-type MEFs, furin was distributed throughout the cell, including the TGN and late endosomes, as evidenced by colocalization with Golga2 and Igf2r (Fig. 4B). However, in Dym-deficient MEFs, furin was primarily localized to the Golgi, and levels of the protein in late endosomes were considerably diminished (Fig. 4B).
Discussion
The present study describes the first genetic and proteomic analysis of mammalian dymeclin, a gene of unknown function previously identified as the cause of two recessive genetic diseases in man, DMC and SMC (23, 38). The dymeclin mutation was identified from a screen of ES cells with mutations induced by the GTR1.3 poly(A) trap retrovirus (30). The mutant allele with the provirus is inserted into the last intron of the Dym gene, structurally resembles the mutation observed in human cases of DMC, and encodes a protein in which the carboxy-terminal 49 aa have been replaced by residues of an irrelevant sequence. Dym-deficient mice developed osteochondrodysplasia similar to that of DMC and SMC (25, 31). Thus, the mouse and human proteins are functionally conserved.
The Golgi complex functions as the principle organelle engaged in the processing, sorting, and transport of secreted proteins in mammalian cells. The complex is maintained by vesicle traffic from the ER and by processes to recover proteins and membranes that have been carried to downstream compartments. The Dym protein was expressed at highest levels in the Golgi, associated with cellular proteins involved in vesicular traffic to and from the Golgi complex, and colocalized in vesicles containing the proteins with which it was found to associate. These could include all vesicles with coat proteins but not late endosomes or lysosomes. Moreover, Dym-mutant fibroblasts displayed multiple defects in vesicular traffic, including enhanced dispersal of Golgi markers in interphase cells, delayed Golgi reassembly after BfA treatment, delayed retrograde (Golgi-to-ER) traffic of an ER-targeted Shiga toxin B subunit, and furin mislocalization involving loss of the protein from late endosomes. These defects are consistent with the activities of Dym-associated proteins, notably p115/VDP (anterograde or ER-to-Golgi traffic), COPB (retrograde or Golgi-to-ER and intra-Golgi transport), VPS35 and AP1G1 (endosome–Golgi protein sorting and traffic), DNM1L (membrane fission), and β-tubulin (Golgi assembly and organization and vesicle transport). These results establish dymeclin as a novel protein that participates in the traffic of vesicles and proteins into and out of the Golgi and provide a molecular basis for osteochondrodysplasias in mice and men.
The Dym protein sequence is predicted to contain multiple membrane-spanning domains and dileucine protein trafficking motifs but otherwise lacks features that would shed light on its function (23–25). In principle, Dym could influence vesicle traffic at a number of levels, including the budding, movement, sorting, and fusion of vesicle membranes, possibly by acting as a membrane-spanning adaptor. Dym could also play a direct role in Golgi organization, because Golgi membranes are more dispersed in Dym-mutant cells, and Dym-related sequences are not present in S. cerevisiae, whose Golgi do not acquire the stacked organization observed in interphase mammalian cells (39). However, similar changes in Golgi organization frequently accompany disruptions of vesicular traffic, as illustrated by attenuating any of the Dym-associated proteins p115/VDP, COPB, AP1, VPS35, and TUBB.
Skeletal dysplasias comprise a large and diverse group of genetic disorders distinguished by clinical/radiological features (3) and underlying genetic defects (40). The present study establishes the importance of intracellular vesicle traffic in mammalian postnatal bone formation, a highly organized process involving chondrocyte proliferation, differentiation, apoptosis, and calcification (1, 2). In principle, transport defects could affect endochondral bone formation at a number of levels, including the synthesis, processing, secretion, or uptake of growth factors, extracellular matrix, and matrix-remodeling proteases (5–16). Such factors also spatially regulate chondrocyte proliferation and apoptosis within the growth plate, consistent with reductions in BrdU and caspase 3-positive cells and growth plate histology in Dym-mutant mice.
Although quite similar, the mouse Dym mutation did not produce an exact phenocopy of DMC. First, Dym-mutant mice displayed no obvious behavioral impairment, whereas mental retardation is a common feature of DMC. Additional studies will be required to determine whether mouse Dym affects cognitive functions in less obvious ways. Second, ≈35% of Dym-mutant mice developed hydronephrosis, a phenotype not observed in DMC. Although the reasons for these differences are not clear, genetic background may play an important role, considering the incomplete penetrance of both phenotypes.
Previous studies attributed cranio-lenticulo-sutural dysplasia and spondyloepiphyseal dysplasia tarda to mutations affecting SEC23A and SEDL (17, 18, 21), components of COPII-coated vesicles and transport protein particle, respectively, that function in ER-to-Golgi vesicle transport. SEDL, which structurally resembles SNAREs, may function as a receptor for COPII-coated vesicles. However, vesicle transport defects associated with SEC23A and SEDL mutations have not been characterized in mammalian cells and have only been inferred on the basis of the analysis of yeast or zebrafish orthologues (22, 41, 42). By contrast, the present study provides a mammalian model of osteochondrodysplasia associated with characterized defects in mammalian vesicle transport. Dymeclin illustrates a novel component of mammalian vesicle transport that is not conserved in S. cerevisiae. Additional examples are likely to be found among the genes responsible for other osteochondrodysplasias in man.
Materials and Methods
Dym-Mutant Mice.
The Dym mutation was generated in ES cells by tagged sequence mutagenesis (30) and was introduced into the germ line. The position of gene trap vector insertion within the Dym gene was determined by sequencing 3′ RACE and inverse PCR products (30). Animals were genotyped by Southern blot hybridization or by PCR.
Tissue pathology was assessed from 5-μm paraffin-embedded sections stained with hematoxylin and eosin. Apoptotic cells in paraffin sections were stained by using an antiactivated caspase 3 antibody together with biotinylated secondary antibodies and streptavidin-conjugated horseradish peroxidase. Cell proliferation was monitored by BrdU labeling. Animals injected IP with 100 μg BrdU per gram of body weight were killed after 4 h, and paraffin sections were stained for incorporated BrdU as described by the reagent supplier (Zymed). The percentage of activated caspase 3 and BrdU-positive chondrocytes in hyperplastic and proliferative zones was determined from five frames of wild-type and mutant growth plates.
Cell Culture and DNA Transfection.
MEFs isolated from E12 embryos were grown in DMEM supplemented with 10% FBS and antibiotics. Ribcage chondrocytes were isolated from 2-week-old mice. Cartilage was minced with a razor blade and digested with pronase (1 mg/ml in F12/MEM) for 30 min at 37°C and after washing with PBS, with collagenase (1 mg/ml in F12/MEM) at 37°C overnight. Dissociated cells were strained, washed two times with PBS, and plated on collagen-coated plates in F12/MEM containing 10% FBS and antibiotics.
HEK293T human kidney cells and NIH 3T3 mouse fibroblasts were cultured in DMEM with 10% FBS and antibiotics and transfected using Polyfect reagent as recommended by the supplier (Qiagen). GFP-Dym, FLAG-Dym, and Dym-C-TAP constructs were made by cloning the PCR-amplified Dym ORF into EGFP-C2, pFLAG-CMV-6a, and pcDNA1-C-TAP vectors, respectively. FLAG-NONO vector was obtained by cloning amplified NONO coding sequences into the pFLAG-CMV-6a vector.
Immunochemistry.
HEK293T cells transfected with FLAG-Dym, FLAG-NONO, and pFLAG-CMV-6a plasmids were lysed in IPP150 buffer with 0.1% of Nonidet P-40 48 h after transfection. Lysates were cleared by centrifugation and incubated with anti-FLAG affinity gel for 4 h at 4°C. Anti-FLAG beads were then washed five times with IPP150 buffer, resuspended in Laemmli buffer, and analyzed by Western blotting.
For protein localization experiments, NIH 3T3 cells grown in eight-well chamber slides (Nunc Lab-Tek Chamber Slide System; Fisher Scientific) were transfected with a GFP-Dym expression vector. Forty-eight hours after transfection, the cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min, permeabilized for 5 min with 0.5% Triton X-100 in PBS, washed with PBS, and treated with 1% BSA in PBS for 1 h (all at room temperature). The cells were incubated overnight with primary antibodies at 4°C and then rinsed three times with PBST (0.02% Tween in PBS) for 10 min. The appropriate Alexa 594-conjugated secondary antibodies were applied in the dark for 2 h at room temperature. After three washes with PBST of 10 min each, the cells were mounted with ProLong Antifade reagent (Invitrogen). Negligible fluorescence was observed in control experiments omitting primary antibody. Slides were analyzed on a Zeiss LSM 510 META confocal laser scanning upright microscope using Zeiss imaging software. For display, images were converted into TIFF format and assembled using Adobe Photoshop 7.0.
Protein Trafficking in Dym-Mutant Cells.
Wild-type and Dym-deficient MEFs were grown on chamber slides and treated with of BfA (5 mg/ml in growth media), and the distribution of Golga2 was determined by immunofluorescence. For BfA washout experiments, the cells were treated with BfA for 30 min, washed with PBS, and incubated in growth media for different times before analysis.
Trafficking of Alexa 488-labeled STB-KDEL with an N-glycosylation site was assessed in wild-type and Dym-deficient MEFs. The cells were incubated for 45 min at 4°C with 50 nM STB-KDEL (36) (kindly provided by L. Johannes, Institute Curie, Paris, France) in growth medium, washed with ice-cold growth medium, and transferred to 37°C, and the distribution of STB-KDEL in relation to Golga2 and calreticulin was determined by fluorescence microscopy. Golga2 (GM130) and Igf2r (CI-MPR) or furin were stained using Alexa 488-congugated antimouse or Alexa 594-congugated antirabbit antibodies, respectively.
Supplementary Material
Acknowledgments.
We thank L. Johannes, Institute Curie, Paris, France for kindly providing STB-KDEL; and Abudi Nashabi for technical assistance. This work was supported by Public Health Service Grants (P01HL68744 to H.E.R. and GM64779, HL68744, ES11993, and CA098131 to A.J.L.), and additional support was provided by the Department of Microbiology and Immunology and by Cancer Center Support grant P30CA68485.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804259105/DCSupplemental.
References
- 1.Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 2.Ortega N, Behonick DJ, Werb Z. Matrix remodeling during endochondral ossification. Trends Cell Biol. 2004;14:86–93. doi: 10.1016/j.tcb.2003.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall CM. International nosology and classification of constitutional disorders of bone. Am J Med Genet. 2002;113:65–77. doi: 10.1002/ajmg.10828. [DOI] [PubMed] [Google Scholar]
- 4.Mortier GR. The diagnosis of skeletal dysplasias: A multidisciplinary approach. Eur J Radiol. 2001;40:161–167. doi: 10.1016/s0720-048x(01)00397-7. [DOI] [PubMed] [Google Scholar]
- 5.Hurvitz JR, et al. Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat Genet. 1999;23:94–98. doi: 10.1038/12699. [DOI] [PubMed] [Google Scholar]
- 6.Murray LW, Bautista J, James PL, Rimoin DL. Type II collagen defects in the chondrodysplasias. I. Spondyloepiphyseal dysplasias. Am J Hum Genet. 1989;45:5–15. [PMC free article] [PubMed] [Google Scholar]
- 7.Muragaki Y, et al. A mutation in the gene encoding the alpha 2 chain of the fibril-associated collagen IX, COL9A2, causes multiple epiphyseal dysplasia (EDM2) Nat Genet. 1996;12:103–105. doi: 10.1038/ng0196-103. [DOI] [PubMed] [Google Scholar]
- 8.Paassilta P, et al. COL9A3: A third locus for multiple epiphyseal dysplasia. Am J Hum Genet. 1999;64:1036–1044. doi: 10.1086/302328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czarny-Ratajczak M, et al. A mutation in COL9A1 causes multiple epiphyseal dysplasia: Further evidence for locus heterogeneity. Am J Hum Genet. 2001;69:969–980. doi: 10.1086/324023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs MD, et al. Pseudoachondroplasia and multiple epiphyseal dysplasia due to mutations in the cartilage oligomeric matrix protein gene. Nat Genet. 1995;10:330–336. doi: 10.1038/ng0795-330. [DOI] [PubMed] [Google Scholar]
- 11.Hecht JT, et al. Mutations in exon 17B of cartilage oligomeric matrix protein (COMP) cause pseudoachondroplasia. Nat Genet. 1995;10:325–329. doi: 10.1038/ng0795-325. [DOI] [PubMed] [Google Scholar]
- 12.Chapman KL, et al. Mutations in the region encoding the von Willebrand factor A domain of matrilin-3 are associated with multiple epiphyseal dysplasia. Nat Genet. 2001;28:393–396. doi: 10.1038/ng573. [DOI] [PubMed] [Google Scholar]
- 13.Gleghorn L, Ramesar R, Beighton P, Wallis G. A mutation in the variable repeat region of the aggrecan gene (AGC1) causes a form of spondyloepiphyseal dysplasia associated with severe, premature osteoarthritis. Am J Hum Genet. 2005;77:484–490. doi: 10.1086/444401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hastbacka J, et al. Atelosteogenesis type II is caused by mutations in the diastrophic dysplasia sulfate-transporter gene (DTDST): Evidence for a phenotypic series involving three chondrodysplasias. Am J Hum Genet. 1996;58:255–262. [PMC free article] [PubMed] [Google Scholar]
- 15.Thiele H, et al. Loss of chondroitin 6-O-sulfotransferase-1 function results in severe human chondrodysplasia with progressive spinal involvement. Proc Natl Acad Sci USA. 2004;101:10155–10160. doi: 10.1073/pnas.0400334101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy AM, et al. MMP13 mutation causes spondyloepimetaphyseal dysplasia, Missouri type (SEMD(MO) J Clin Invest. 2005;115:2832–2842. doi: 10.1172/JCI22900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gedeon AK, et al. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat Genet. 1999;22:400–404. doi: 10.1038/11976. [DOI] [PubMed] [Google Scholar]
- 18.Jang SB, et al. Crystal structure of SEDL and its implications for a genetic disease spondyloepiphyseal dysplasia tarda. J Biol Chem. 2002;277:49863–49869. doi: 10.1074/jbc.M207436200. [DOI] [PubMed] [Google Scholar]
- 19.Raas-Rothschild A, et al. Molecular basis of variant pseudo-hurler polydystrophy (mucolipidosis IIIC) J Clin Invest. 2000;105:673–681. doi: 10.1172/JCI5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudo M, Brem MS, Canfield WM. Mucolipidosis II (I-cell disease) and mucolipidosis IIIA (classical pseudo-hurler polydystrophy) are caused by mutations in the GlcNAc-phosphotransferase alpha/beta -subunits precursor gene. Am J Hum Genet. 2006;78:451–463. doi: 10.1086/500849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyadjiev SA, et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 22.Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet. 2006;38:1198–1203. doi: 10.1038/ng1880. [DOI] [PubMed] [Google Scholar]
- 23.Cohn DH, et al. Mental retardation and abnormal skeletal development (Dyggve–Melchior–Clausen dysplasia) due to mutations in a novel, evolutionarily conserved gene. Am J Hum Genet. 2003;72:419–428. doi: 10.1086/346176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehtesham N, et al. Evidence that Smith-McCort dysplasia and Dyggve–Melchior–Clausen dysplasia are allelic disorders that result from mutations in a gene on chromosome 18q12. Am J Hum Genet. 2002;71:947–951. doi: 10.1086/342669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Ghouzzi V, et al. Mutations in a novel gene Dymeclin (FLJ20071) are responsible for Dyggve–Melchior–Clausen syndrome. Hum Mol Genet. 2003;12:357–364. doi: 10.1093/hmg/ddg029. [DOI] [PubMed] [Google Scholar]
- 26.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 27.Hicks GG, et al. Functional genomics in mice by tagged sequence mutagenesis. Nat Genet. 1997;16:338–344. doi: 10.1038/ng0897-338. [DOI] [PubMed] [Google Scholar]
- 28.Skarnes WC, et al. A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 30.Lin Q, et al. Mutagenesis of diploid mammalian genes by gene entrapment. Nucleic Acids Res. 2006;34:e139. doi: 10.1093/nar/gkl728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paupe V, et al. Recent advances in Dyggve–Melchior–Clausen syndrome. Mol Genet Metab. 2004;83:51–59. doi: 10.1016/j.ymgme.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Engfeldt B, et al. Dyggve–Melchior–Clausen dysplasia. Morphological and biochemical findings in cartilage growth zones. Acta Paediatr Scand. 1983;72:269–274. doi: 10.1111/j.1651-2227.1983.tb09710.x. [DOI] [PubMed] [Google Scholar]
- 33.Link AJ, Fleischer TC, Weaver CM, Gerbasi VR, Jennings JL. Purifying protein complexes for mass spectrometry: Applications to protein translation. Methods. 2005;35:274–290. doi: 10.1016/j.ymeth.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 34.McAfee KJ, Duncan DT, Assink M, Link AJ. Analyzing proteomes and protein function using graphical comparative analysis of tandem mass spectrometry results. Mol Cell Proteomics. 2006;5:1497–1513. doi: 10.1074/mcp.T500027-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: Insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johannes L, Tenza D, Antony C, Goud B. Retrograde transport of KDEL-bearing B-fragment of Shiga toxin. J Biol Chem. 1997;272:19554–19561. doi: 10.1074/jbc.272.31.19554. [DOI] [PubMed] [Google Scholar]
- 37.Robinson MS. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Burns C, Powell BR, Hsia YE, Reinker K. Dyggve–Melchior–Clausen syndrome: Report of seven patients with the Smith-McCort variant and review of the literature. J Pediatr Orthop. 2003;23:88–93. [PubMed] [Google Scholar]
- 39.Barr FA, Egerer J. Golgi positioning: Are we looking at the right MAP? J Cell Biol. 2005;168:993–998. doi: 10.1083/jcb.200501088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Superti-Furga A, Bonafe L, Rimoin DL. Molecular-pathogenetic classification of genetic disorders of the skeleton. Am J Med Genet. 2001;106:282–293. [PubMed] [Google Scholar]
- 41.Gecz J, Shaw MA, Bellon JR, de Barros Lopes M. Human wild-type SEDL protein functionally complements yeast Trs20p but some naturally occurring SEDL mutants do not. Gene. 2003;320:137–144. doi: 10.1016/s0378-1119(03)00819-9. [DOI] [PubMed] [Google Scholar]
- 42.Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.