Abstract
We have performed a genome-wide analysis of copy number changes in breast and colorectal tumors using approaches that can reliably detect homozygous deletions and amplifications. We found that the number of genes altered by major copy number changes, deletion of all copies or amplification to at least 12 copies per cell, averaged 17 per tumor. We have integrated these data with previous mutation analyses of the Reference Sequence genes in these same tumor types and have identified genes and cellular pathways affected by both copy number changes and point alterations. Pathways enriched for genetic alterations included those controlling cell adhesion, intracellular signaling, DNA topological change, and cell cycle control. These analyses provide an integrated view of copy number and sequencing alterations on a genome-wide scale and identify genes and pathways that could prove useful for cancer diagnosis and therapy.
Keywords: amplification, copy number changes, Digital Karyotyping, high-density SNP arrays, homozygous deletion
It is well accepted that cancer is the result of the sequential mutations of oncogenes and tumor suppressor genes (1). Historically, the discovery of these genes has been accomplished through analyses of individual candidate genes chosen on the basis of functional or biologic data. Recent advances in genomic technologies have permitted simultaneous evaluation of many genes, thereby offering more comprehensive and unbiased information (2, 3). For example, the sequence of large families of genes, and even the human genes in the Reference Sequence (RefSeq) database, have been determined in subsets of human cancers (4, 5). However, the alterations detected by sequencing represent only one category of genetic change that occurs in human cancer. Other alterations include gains (amplifications) and losses (deletions) of discrete chromosomal sequences that occur during tumor progression. Dramatic amplifications of oncogenes such as ERBB2 (6) or MYC (7) and deletions of tumor suppressor genes such as CDKN2A (8), PTEN (9, 10), and SMAD4 (11) have demonstrated the importance of these mechanisms of genetic alteration in particular tumor types. A comprehensive picture of genetic alterations in human cancer should therefore include sequence based alterations together with copy number gains and losses.
Evaluations of copy number changes in cancers using a variety of array types have been reported (12). Several of the more recent studies used oligonucleotide arrays capable of distinguishing >100,000 genomic loci in colon, breast lung, pancreatic, skin cancers, and certain leukemias (13–20). However, identification of focal, high copy number amplifications, or homozygous deletions (HDs) have infrequently been reported, because many prior copy number analyses employing arrays have used genomic DNA purified from primary tumors. Primary tumors contain varying proportions of nonneoplastic cells thereby obscuring focal amplifications, defined by the increased copy number of a small region of the genome, from simple gains of whole chromosome arms. Furthermore, HDs can be difficult to discern in primary tumors because of confounding hybridization signals from nonneoplastic cells within the tumor (17).
Many of the problems encountered with primary tumor samples can be overcome by use of early passage cancer cell lines or xenografts that are devoid of human nonneoplastic cells. Previous studies have shown that the process of generating such in vitro or in vivo cultures is not associated with the development of additional genetic alterations (21). It is now widely recognized that HDs found in cell lines and xenografts represent true genetic alterations that are present in clonal fashion in primary tumors but are difficult to document in the latter as a result of the issues noted above (22, 23).
In the current study, we examined xenografts or cell lines derived from breast and colorectal cancers to obtain high-resolution analyses of copy number and nucleotide alterations. Tumors were evaluated with arrays containing at least 317,000 SNP probes and selected samples were also evaluated with Digital Karyotyping (DK). This latter method provided a highly quantitative measure of gene copy number and was used to validate the sensitivity and specificity of the array data. The sequences of the 18,191 genes from the RefSeq database determined for breast and colorectal cancers were integrated with these results, providing a genome-wide analysis of sequence and copy number alterations.
Results
Optimization of Copy Number Analysis with DK.
DK was used as a standard to develop criteria for assessing amplifications and HDs with Illumina high-density SNP arrays [supporting information (SI)Fig. S1 and Table S1]. Analysis of DK libraries from 18 colorectal tumor samples identified a total of 21 amplification events, and four regions within the autosomal chromosomes where the tag density reached zero, representing HDs (Table S2). As expected, we identified low-amplitude gains and losses of chromosome arms or other large genomic regions. We did not pursue these low-amplitude copy number changes as it is difficult to reliably identify candidate cancer genes from such large regions. To ensure that the copy number changes identified by DK were bona fide amplifications or HDs, we independently examined 12 alterations by quantitative PCR and confirmed the presence of the genomic alterations in every case examined.
We then directly compared DK data to those obtained through genomic hybridization of the same DNA samples to Illumina high-density oligonucleotide array (25, 26). Using fluorescence intensity measurements, we developed an approach to detect HDs and amplifications resulting in ≥12 copies per nucleus (≥6-fold amplification compared with the diploid genome) (see SI Methods).
Using this approach, all 14 amplification events and 3 HD events identified by DK in three representative tumor samples were detected by Illumina arrays (Table S1 and Fig. S2). In all cases, the genomic boundaries identified by both approaches were similar and within the resolution expected for both methods. No additional copy number changes of the sizes expected to be detected by DK were identified by the array approach in these samples. We did identify 25 additional small HDs (<250 kb in length) that would not have been possible to detect with DK given the number of tags analyzed (24). To independently validate such smaller HDs, we used PCR and Sanger sequencing to examine genes located within small HDs and found that, in each case, multiple exons of each gene could not be amplified or sequenced. These results suggested that our approach for analysis of Illumina array data provided a sensitive and specific method for identification of amplifications and HDs, including relatively small alterations of either type.
Detection of Amplifications and HDs.
A total of 45 breast and 36 colorectal tumors were analyzed by Illumina arrays containing either ≈317,000 or ≈550,000 SNPs (Fig. S1). To determine the fraction of alterations that were likely to be somatic (i.e., tumor-derived), we analyzed these regions in 23 matched normal samples. In the normal samples, no amplifications and only four distinct HDs were detected. We removed these alterations from further analysis, as well as those corresponding to known copy number variation in normal human cells (27, 28). Finally, we removed any copy number changes where the boundaries were identical in two or more samples, because these were likely to represent germ-line variants. Based on this conservative strategy, we estimated that >95% of the 614 amplifications and 463 HDs (Table S3) represented true somatic alterations.
Breast cancers contributed to a majority of the alterations identified, comprising 68% and 80% of the total HDs and amplifications, respectively. Individual colorectal and breast tumors had on average 7 and 18 copy number alterations, respectively. Each colorectal cancer had an average of four HDs and three amplifications. Breast cancers had on average 7 HDs and 11 amplifications. Several of the tumor samples contained copy number alterations that were separated by short nonamplified or deleted sequences, presumably reflecting the complex structure of these alterations (29, 30). The copy number alterations observed encompassed on average 1.7 and 2.4 Mb of colorectal and breast tumor haploid genomic sequence, respectively. The average numbers of protein-coding genes that were affected by amplification or HD were 24 and 9 per breast and colorectal cancer, respectively.
Genes Altered in More than One Tumor.
One of the main challenges in the analysis of somatic alterations in cancers involves the distinction between those changes that are selected for during tumorigenesis (driver alterations) from those that provide no selective advantage (passenger alterations). Even in regions that have multiple copy number alterations, this distinction can be particularly difficult because regions of amplification or HD can contain multiple genes, only a subset of which are presumably the underlying targets. We reasoned that the integration of copy number analyses with sequence data would help reveal driver genes that were more likely to contain genetic alterations. To accomplish this integration, we developed a statistical approach for determining whether the observed genetic alterations of any type in any gene were likely to reflect an underlying mutation frequency that was significantly higher than the passenger rate. To analyze the probability that a given gene would be involved in a copy number alteration, we made the conservative assumption that the frequency of all amplifications and HDs observed in each tumor type represented the passenger mutation frequency (i.e., we assumed that all copy number changes were passengers). The number of actual copy number alterations affecting each gene in all tumors was then compared with the simulated number of expected passenger alterations taking into account gene size, the distribution of SNP locations, and the frequency of passenger amplifications and HDs in breast and colorectal cancers.
We integrated these copy number analyses with the sequence data of the Sjöblom et al. and Wood et al. studies (5, 31). In these studies, the protein coding sequences of 20,857 transcripts from the 18,191 genes in the RefSeq database were determined in breast and colorectal cancer samples, allowing detection of somatic sequence alterations. In the current study, the same 22 breast and colorectal tumor samples were analyzed in parallel by Illumina arrays, together with additional samples of each tumor type (Fig. S1). To integrate these different mutational data for each tumor type, we combined the probability that a gene was a driver gene based on the type and frequency of point mutations observed with the probability that the gene was a driver based on the number of observed amplifications and HDs (Fig. S1).
Table 1 lists the loci that were amplified in at least one tumor and had the highest probability of containing driver genes as determined by the combined mutation analysis (a complete list of amplifications is provided in Table S3 and amplified genes in Table S4). For genes to be considered potential targets of the amplification, the entire coding region of the gene was required to be contained within a focal amplicon. A few candidate genes in this list [e.g., CCNE1 (cyclin E) and ERBB2] were amplified in multiple tumors but were not found to be mutated by sequencing. The majority of candidate genes, however, harbored point mutations in some tumors and amplifications in others. The most striking aspect of this list of candidate genes is that only some of them had been implicated in cancer in the past: of the 19 genes indicated in Table 1, only 8 had been implicated in tumorigenesis. The known cancer genes included MYC, ERBB2 (HER2/NEU), CCNE1, CCND1, EGFR, FGFR2, and IRS2, each of which had been shown to be amplified. In addition, MRE11, which was amplified in breast cancers, has been shown to be mutated in a subset of colorectal cancers and is thought to play an essential role in maintaining chromosomal stability (32). Some genes were shown to be altered in both breast and colorectal cancers, with at least one of the tumors containing amplifications. Interestingly, among these genes, ERBB2 was found to be amplified in both breast and colorectal cancers, and FGFR2 was found to be mutated in breast cancers and amplified in colorectal cancers.
Table 1.
Top candidate cancer genes in breast and colorectal cancer amplifications
| Tumor type | Minimal affected region |
Candidate cancer gene | Total number of amplifications | Total number of point mutations | Passenger probability | ||
|---|---|---|---|---|---|---|---|
| Chr | Left boundary | Right boundary | |||||
| Breast | 19 | 34,933,380 | 35,097,525 | CCNE1 | 4 (5) | 0 | <0.01 |
| 17 | 34,634,168 | 35,387,448 | ERBB2 | 4 (8) | 0 | <0.01 | |
| 11 | 68,626,681 | 69,411,832 | CCND1 | 3 | 0 | <0.01 | |
| 11 | 93,740,661 | 93,972,379 | MRE11A | 1 | 2 | 0.01 | |
| 7 | 25,728,296 | 27,195,245 | HOXA3 | 1 | 2 | 0.01 | |
| 6 | 40,917,990 | 43,889,896 | TREM1 | 1 | 1 | 0.01 | |
| 1 | 149,032,752 | 149,156,966 | FLG2 | 1 (2) | 1 | 0.02 | |
| Colon | 8 | 128,750,181 | 128,848,183 | MYC | 2 | 0 | <0.01 |
| 17 | 19,136,024 | 19,211,040 | EPPB9 | 2 | 0 | <0.01 | |
| 7 | 54,862,624 | 55,406,733 | EGFR | 2 | 0 | <0.01 | |
| 13 | 109,108,212 | 109,557,712 | IRS2 | 1 | 1 | <0.01 | |
| 19 | 57,427,110 | 57,619,851 | ZNF480 | 1 | 1 | <0.01 | |
| 19 | 49,127,007 | 49,207,192 | ZNF155 | 1 | 1 | 0.01 | |
| 15 | 88,561,995 | 89,253,599 | NEUGRIN | 1 | 1 | 0.01 | |
| Combined breast and colon | 19 | 34,933,380 | 35,097,525 | CCNE1 | 5 (6) | 0 | <0.01 |
| 17 | 34,634,168 | 35,387,448 | ERBB2 | 5 (9) | 0 | <0.01 | |
| 6 | 41,419,345 | 42,485,546 | FOXP4 | 2 | 1 | <0.01 | |
| 8 | 37,767,164 | 40,003,731 | GPR124 | 2 (5) | 1 | <0.01 | |
| 20 | 61,788,664 | 61,840,441 | ARFRP1 | 1 (4) | 1 | 0.01 | |
| 10 | 123,231,784 | 123,471,190 | FGFR2 | 1 | 1 | 0.02 | |
| 20 | 29,297,270 | 29,721,415 | HM13 | 1 (4) | 1 | 0.03 | |
Top seven candidate cancer genes for each tumor type are indicated. The combined group corresponds to genes that are altered in both tumor types, with at least one observed amplification. Minimal affected region is defined as the smallest overlapping interval affecting candidate cancer gene. Total number of amplifications indicates the number of focal amplifications or the number of focal and complex amplifications (in parentheses) in the indicated tumor type(s). Candidate cancer gene refers to either known cancer genes or the gene with the highest driver probability within the minimal affected region. Candidate genes were required to be entirely contained within the minimal affected region. The passenger probability provides a combined probability that the number of amplifications and point mutations observed were passengers alterations; these analyses used the intermediate passenger rate for point alterations (see SI Methods for additional information). All amplified genes and their passenger probabilities are indicated in Table S4 and the samples analyzed for such alterations are indicated in Table S6.
Table 2 similarly lists the loci that were homozygously deleted in at least one tumor and had the highest probability of containing drivers as determined by the combined mutation analysis (a complete list of HDs is provided in Table S3 and homozygously deleted genes are listed in Table S5). For each of these genes, at least a portion of the coding region was affected by the HD. A number of genes known to be inactivated in colorectal or breast tumorigenesis, such as CDKN2A, PTEN, and TP53 are found in this list. We also identified genes such as CHD5, MAP2K4, SMAD2, and SMAD3 that have been shown to be deleted in other tumor types but not in colorectal or breast cancers. Finally, we discovered a number of genes not known to be affected by HD in any tumor type. For example, HDs and point mutations were found in OMA1 and ZNF521 in colorectal cancers and in MANEA, PCDH8, SATL1, and ZNF674 in breast cancers. During the course of this study, we identified through independent experimentation that PCDH8 is mutated and homozygously deleted in breast cancer (33).
Table 2.
Top candidate cancer genes in breast and colorectal cancer homozygous deletions
| Tumor type | Minimal affected region |
Candidate cancer gene | Total number of homozygous deletions | Total number of point mutations | Passenger probability | ||
|---|---|---|---|---|---|---|---|
| Chr | Left boundary | Right boundary | |||||
| Breast | 9 | 21,963,422 | 21,974,661 | CDKN2A | 6 | 0 | <0.01 |
| X | 41,487,365 | 46,563,031 | ZNF674 | 1 | 2 | 0.01 | |
| X | 82,896,749 | 84,431,661 | SATL1 | 1 | 1 | 0.05 | |
| 6 | 95,085,197 | 96,939,216 | MANEA | 1 | 1 | 0.05 | |
| 13 | 52,176,068 | 60,428,304 | PCDH8 | 1 | 1 | 0.05 | |
| 22 | 17,252,341 | 17,382,662 | PRODH | 1 | 1 | 0.07 | |
| X | 31,030,131 | 31,430,208 | DMD | 1 | 2 | 0.13 | |
| Colon | 10 | 89,635,453 | 89,993,748 | PTEN | 2 | 3 | <0.01 |
| 17 | 7,518,132 | 7,519,370 | TP53 | 1 | 18 | <0.01 | |
| 17 | 10,910,683 | 12,755,698 | MAP2K4 | 3 | 0 | <0.01 | |
| 18 | 43,490,500 | 44,141,990 | SMAD2 | 1 | 3 | <0.01 | |
| 15 | 65,239,008 | 65,323,050 | SMAD3 | 1 | 2 | 0.01 | |
| 18 | 20,178,701 | 21,064,617 | ZNF521 | 1 | 2 | 0.01 | |
| 1 | 57,175,347 | 57,297,452 | OMA1 | 1 | 1 | 0.01 | |
| Combined breast and colon | 10 | 89,697,245 | 89,721,850 | PTEN | 4 | 3 | <0.01 |
| 17 | 7,518,132 | 7,519,370 | TP53 | 1 | 36 | <0.01 | |
| 18 | 56,875,085 | 58,225,845 | CDH20 | 2 | 2 | <0.01 | |
| 1 | 55,485,777 | 57,152,356 | PRKAA2 | 1 | 2 | 0.04 | |
| 18 | 32,281,094 | 32,360,816 | FHOD3 | 1 | 3 | 0.05 | |
| 1 | 6,051,437 | 6,464,455 | CHD5 | 1 | 2 | 0.05 | |
| 6 | 159,338,933 | 159,632,547 | FNDC1 | 1 | 3 | 0.08 | |
Top seven candidate cancer genes for each tumor type are indicated. The combined group corresponds to genes that are altered in both tumor types, with at least one observed homozyous deletion. Minimal affected region is defined as the smallest overlapping interval affecting candidate cancer gene. Candidate cancer gene refers to either known cancer genes or the gene with the highest driver probability within the minimal affected region. Part of the coding sequence of the indicated candidate cancer gene was required to be contained within the minimal deleted region. The passenger probability provides a combined probability that the number of homozygous deletions and point mutations observed were passengers alterations; these analyses used the intermediate passenger rate for point alterations (see SI Methods for additional information). All amplified genes and their passenger probabilities are indicated in Table S4 and the samples analyzed for such alterations are indicated in Table S6.
Pathways Enriched for Copy Number and Point Alterations.
We examined whether groups of genes belonging to certain cellular pathways were preferentially affected by genetic alterations. For this purpose, we developed a statistical approach that provided a probability that a pathway contained driver alterations, taking into account both the copy number changes and point mutations. Because the net effect of a pathway can be the same whether certain components are amplified or others deleted, all copy number alterations within a gene group were considered together. The analysis was performed by using well annotated GeneGo MetaCore databases (34). For each gene group, we considered whether the component genes were more likely to be affected by point mutations, amplifications, or HDs, as compared with all genes analyzed using a modified version of gene set enrichment analysis (GSEA) (35) rather than the total number of mutations within individual groups. This approach limits the effects of single highly mutated genes and requires the involvement of multiple genes to score a pathway as significantly affected.
These analyses identified gene groups that were enriched for genetic alterations in these tumor types (Table 3). In particular, the EGFR and ERBB gene pathways were enriched for alterations in both tumor types, involving various components of the PI3 kinase pathway (Fig. 1). One-third of genes in these combined pathways were mutated by sequence alterations, amplifications, or HDs. Enrichment of alterations in other canonical gene groups including Notch and G1-S cell cycle transition pathways were also detected. The latter group included HDs of CDKN2A and CDKN2B genes and amplifications of cyclin D1, cyclin D3, and cyclin E3 genes in breast cancers. For all these gene groups, new genes were identified that had not been implicated by genetic alterations in these cellular processes. Finally, a variety of gene groups not known to be enriched for copy number changes in tumorigenesis were identified. These included genes implicated in cell–cell interaction and adhesion, including cadherins and metalloproteases. As an example, in colorectal cancers, a total of 33 cadherin and protocadherin genes were detected as being affected by copy number or sequence changes. In breast cancers, there was also enrichment in genes implicated in DNA topological control, including alterations in a number of topoisomerases (TOP1, TOP2A, TOP2B, and TOP3A) and helicases. Pathways showing significant enrichment for genetic alterations are listed in Table S7.
Table 3.
Candidate cancer pathways altered in breast and colorectal cancers
| Pathways | Category | Total number of genes in pathway | Number of altered genes | Number of point mutations | Number of amplifications | Number of homozygous deletions | Q value |
|---|---|---|---|---|---|---|---|
| Breast | |||||||
| Cell cycle_ATM/ATR regulation of G1/S | MA | 37 | 10 | 26 | 8 | 0 | 1.13E-04 |
| DNA topological change | GO | 10 | 7 | 5 | 2 | 0 | 4.50E-04 |
| Development_EGFR signaling via PIP3 | MA | 23 | 5 | 8 | 4 | 2 | 1.38E-03 |
| Cell–cell adhesion | GO | 67 | 16 | 20 | 1 | 5 | 1.67E-03 |
| Signal transduction_AKT signaling | MA | 50 | 12 | 31 | 6 | 2 | 3.00E-03 |
| T cell proliferation | GO | 10 | 4 | 2 | 3 | 0 | 3.57E-03 |
| Cell cycle_G1-S Growth factor regulation | GG | 153 | 27 | 19 | 21 | 13 | 3.59E-03 |
| Signal transduction_ERBB-family | GG | 74 | 13 | 15 | 9 | 2 | 9.79E-03 |
| Development_Leptin signaling via PI3K-dependent pathway | MA | 48 | 11 | 12 | 5 | 0 | 1.40E-02 |
| Cell adhesion_Role of CDK5 | MA | 12 | 5 | 3 | 4 | 2 | 1.42E-02 |
| Regulation of angiogenesis | GO | 11 | 3 | 2 | 4 | 1 | 1.42E-02 |
| Development_ERBB-family signaling | MA | 44 | 10 | 13 | 8 | 0 | 1.47E-02 |
| Regulation of cell migration | GO | 30 | 9 | 8 | 4 | 1 | 1.47E-02 |
| Cell cycle_Brca1 as a transcription regulator | MA | 26 | 7 | 22 | 4 | 1 | 1.58E-02 |
| Cell differentiation | GO | 391 | 59 | 69 | 22 | 9 | 1.58E-02 |
| Cell adhesion_Endothelial cell contacts | MA | 34 | 10 | 10 | 2 | 0 | 1.65E-02 |
| Cell projection organization and biogenesis | GO | 21 | 7 | 6 | 3 | 0 | 1.65E-02 |
| Signal transduction_NOTCH signaling | GG | 197 | 27 | 46 | 11 | 2 | 1.79E-02 |
| Development_Hemopoiesis, Erythropoietin pathway | GG | 146 | 25 | 28 | 14 | 0 | 1.99E-02 |
| Signal transduction_Leptin signaling | GG | 101 | 20 | 16 | 11 | 0 | 2.10E-02 |
| Colon | |||||||
| Development_EGFR signaling via PIP3 | MA | 23 | 9 | 12 | 5 | 6 | 8.79E-05 |
| Negative regulation of cell cycle | GO | 105 | 17 | 64 | 3 | 5 | 5.66E-04 |
| Development_EGFR signaling via small GTPases | MA | 34 | 9 | 25 | 6 | 4 | 1.20E-03 |
| Cell cycle_G1-S Growth factor regulation | GG | 153 | 20 | 52 | 10 | 5 | 1.65E-03 |
| p38-MAPK cascade activation via IGF1R and EGFR | MA | 17 | 6 | 9 | 0 | 4 | 3.69E-03 |
| Cell adhesion_Cadherins | GG | 210 | 29 | 65 | 3 | 6 | 7.98E-03 |
| Defense response to bacterium | GO | 70 | 14 | 9 | 7 | 0 | 9.24E-03 |
| Development_CNTF receptor signaling | MA | 33 | 7 | 9 | 0 | 2 | 1.12E-02 |
| Cytoskeleton remodeling_Role of PDGFs in cell migration | MA | 20 | 5 | 8 | 1 | 1 | 1.98E-02 |
| T cell proliferation | GO | 10 | 2 | 0 | 1 | 2 | 2.00E-02 |
| Regulation of angiogenesis | GO | 11 | 4 | 2 | 3 | 0 | 2.03E-02 |
| Signal transduction_AKT signaling | MA | 50 | 9 | 28 | 4 | 4 | 2.09E-02 |
| DNA fragmentation during apoptosis | GO | 15 | 3 | 1 | 3 | 0 | 2.18E-02 |
| Proteolysis | GO | 400 | 40 | 55 | 3 | 3 | 2.32E-02 |
| Signal transduction_NOTCH signaling | GG | 197 | 26 | 50 | 8 | 10 | 3.02E-02 |
| Signal transduction_ERBB-family | GG | 74 | 13 | 30 | 9 | 6 | 3.02E-02 |
| Cell adhesion_Cell-matrix interactions | GG | 158 | 19 | 31 | 4 | 0 | 3.02E-02 |
| Cytoskeleton remodeling_Role of Activin A | MA | 18 | 5 | 10 | 0 | 5 | 3.85E-02 |
| Transcription_Receptor-mediated HIF regulation | MA | 28 | 6 | 11 | 0 | 3 | 3.89E-02 |
| Signal transduction_Androgen receptor | GG | 91 | 15 | 20 | 3 | 2 | 4.60E-02 |
Pathways correspond to GeneGO MetaCore pathway maps (MA), Gene Ontology groups (GO), or GeneGo groups (GG). The top 20 entries having at least two copy number alterations are indicated for each tumor type. Only pathways containing between 10 and 500 genes are included in this table. ″Alterations in other tumor type″ corresponds to the number of copy number changes and point mutations observed in colorectal cancer when considering the pathways enriched for alterations in breast cancer and vice-versa for the pathways enriched for alterations in colorectal cancer. ″Q value″ corresponds to the probability of obtaining the observed enrichment of alterations in each pathway through passenger alterations alone as described in SI Methods.
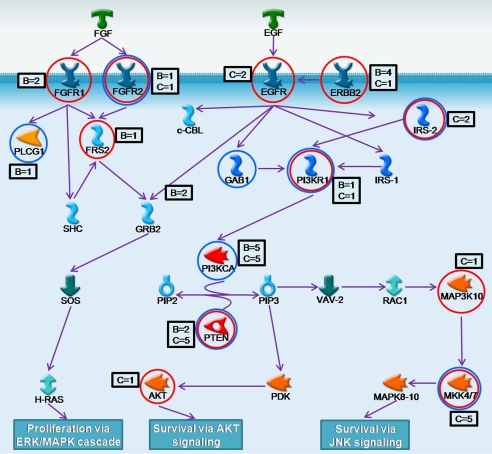
Fig. 1.
Alterations in the combined FGF, EGFR, ERBB2, and PI3K pathways. Genes affected by copy number alterations are circled in red, whereas those altered by point mutations are circled in blue. The number of breast (B) and colorectal (C) tumors containing alterations are indicated in boxes adjacent to each gene.
Discussion
The integrated mutational analysis described herein provides a global picture of the genetic alterations of breast and colorectal cancers. The combination of sequencing and copy number analysis permits the identification of genes and pathways that may not be easily detected by either analysis alone.
The analysis of copy number changes can also provide general insights into the functional effects of point mutations. Single-nucleotide substitutions in genes that are observed to be deleted are more likely to be inactivating, whereas substitutions in genes that are amplified are more likely to be activating. This was confirmed by the observation of HDs and point mutations in TP53, SMAD2, SMAD3, and PTEN, all of which are thought to be tumor suppressors. If copy number changes faithfully reflect the overall effect of target genes, one would expect to infrequently see both amplifications and HDs of the same set of genes in human tumors. Accordingly, we observed an underrepresentation of genes that are homozygously deleted in one tumor and amplified in another [only 2 of the 1,148 altered genes identified were altered by both amplification and HD (P < 0.01, binomial test)].
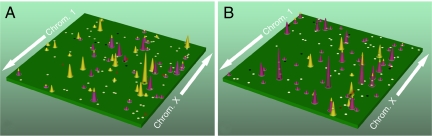
These studies have several implications for future large-scale genomic analyses (36). One is that the complexity of genetic alterations in human cancer increases when considering both point alterations and copy number changes. In addition to a median of 84 and 76 genes altered by point mutation, breast and colorectal cancers have a median of 24 and 9 genes altered by a major copy number change. These observations support a view of the breast and colorectal cancer genomic landscape where a few commonly affected “gene mountains” are scattered among a much larger number of “gene hills” that are infrequently altered by either point mutation or copy number changes. An example of a cancer genome landscape that incorporates copy number changes, illustrated in Fig. 2, shows gene mountains and hills that result from the combined analysis.
Fig. 2.
Genomic landscape of copy number and nucleotide alterations in two typical cancer samples. A indicates breast cancer alterations, whereas B indicates colorectal cancer alterations. The telomere of the short arm of chromosome 1 is represented in the rear left corner of the green plane and ascending chromosomal positions continue in the direction of the arrow. Chromosomal positions that follow the front edge of the plane are continued at the back edge of the plane of the adjacent row and chromosomes are appended end to end. Peaks indicate the 60 highest-ranking candidate cancer genes for each tumor type, with peak heights reflecting the passenger probability scores. The yellow peaks correspond to genes that are altered by copy number changes, whereas those altered only by point mutations are purple. The dots represent genes that were altered by copy number changes (red squares) or point mutations (white circles) in the B9C breast or Mx27 colorectal tumor samples. Altered genes participating in significant gene groups or pathways (Table S6) are indicated as black circles or squares.
Although cancer genome landscapes are complex, they may be better understood by placing all genetic alterations within defined cellular pathways. Our analyses identified several converging gene pathways, including the ERBB2, EGFR, and PI3K pathways, that were affected by copy number changes and point alterations in both breast and colorectal cancers. In addition, many pathways implicated in colorectal tumor progression (Notch, AKT, and MAPK) were enriched for alterations. Interestingly, many gene groups contained genes that were both amplified and others that were deleted, suggesting that different genes within the same group or pathway may be affected through alternate mechanisms. This is consistent with the observation that most signaling pathways contain both positive and negative regulators and alterations in any of these can lead to dysregulated signaling.
The copy number and sequence alterations reported here should be placed in the context of other analyses to reveal the full compendium of molecular changes in a tumor cell. One limitation of our approach is that the copy number analyses we performed may have missed very small regions (<20 kb) that were amplified or deleted. Use of arrays with higher numbers of SNPs or larger DK libraries generated by using next-generation sequencing approaches will help improve the sensitivity of these analyses. Additionally, the incorporation of approaches that detect structural changes (e.g., translocations) and epigenetic alterations will likely prove to be useful. Finally, the statistical techniques we developed highlight the best candidates for future functional studies, but it remains possible that specific loci are more likely to be altered by copy number changes than others because they are located near fragile sites or other hotspots for recombination (37). Therefore, these genetic analyses can only identify candidate genes that may play a role in cancer and do not definitively implicate any gene in the neoplastic process.
Several of the pathways identified affected a relatively high fraction of tumors and may be useful for cancer diagnosis or therapy. Alterations in signaling pathways of FGFR, EGFR, ERBB2, and PI3K were detected in nearly two-thirds of breast and colorectal tumors that were comprehensively examined in this study. These data suggest that the ERBB2 inhibitors may be useful not only in breast cancers but also in selected colorectal cancer patients in combination with existing therapeutic agents. Additionally, a significant fraction of the breast tumors analyzed had genetic alterations in genes regulating DNA topology. Although TOP2A is coamplified with ERBB2 and therefore does not represent the likely driver of this amplicon, alterations of TOP2A may still be of clinical utility. Because high doses of anthracyclines may improve clinical outcomes in breast cancer patients with TOP2A amplifications (38, 39), our observations suggest that the additional alterations that we identified could be used to select patients that may respond to topoisomerase-targeted therapies. In a similar fashion, tumor cells deficient in certain cellular processes as a result of HDs could be targeted pharmacologically through synthetic lethality. In a general sense, our discovery that a typical colorectal or breast cancer has four to seven genes homozygously deleted suggests that further development of strategies targeting such HDs (40) could be widely applicable.
Materials and Methods
DNA samples from tumor-derived xenografts and cell lines were obtained and purified, and analyzed using DK and Illumina SNP arrays (24, 41). Bioinformatic analyses were used to determine focal amplifications and HDs. Statistical methods were used to determine the likelihood that genetic alterations occurred at a frequency higher than the passenger rate and to identify gene groups enriched for copy number and sequence alterations. Detailed information for materials and methods is described in SI Methods.
Supplementary Material
Acknowledgments.
We thank R. Ashworth and A. Scott for assistance with Illumina analyses, S. Bentivegna for assistance with DK, D. H. Nguyen for the artwork in Fig. 2, and M. Newton for sharing software for non-id Bernoulli calculations. This work was supported by the Virginia and D. K. Ludwig Fund for Cancer Research; National Institutes of Health Grants CA121113, CA 57345, CA 43460, and CA 109274; National Cancer Institute Division of Cancer Prevention Contract HHSN261200433002C; the Pew Charitable Trusts, and the Avon Foundation.
Footnotes
Conflict of interest statement: Under separate licensing agreements between Beckman Coulter and the Johns Hopkins University and Genzyme Corporation and the Johns Hopkins University, V.E.V., K.W.K., and B.V. are entitled to a share of royalties received by the University on sales of products described in this article. V.E.V., K.W.K., and B.V. and the University own Genzyme Corporation stock, which is subject to certain restrictions under University policy. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808041105/DCSupplemental.
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Bardelli A, Velculescu VE. Mutational analysis of gene families in human cancer. Curr Opin Genet Dev. 2005;15:5–12. doi: 10.1016/j.gde.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Strausberg RL, Levy S, Rogers YH. Emerging DNA sequencing technologies for human genomic medicine. Drug Discov Today. 2008;13:569–577. doi: 10.1016/j.drudis.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 6.Slamon DJ, et al. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 7.Collins S, Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982;298:679–681. doi: 10.1038/298679a0. [DOI] [PubMed] [Google Scholar]
- 8.Kamb A, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 9.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 10.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 11.Hahn SA, et al. Dpc4, a Candidate Tumor Suppressor Gene At Human Chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 12.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37(Suppl):S11–7. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 13.Camps J, et al. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008;68:1284–1295. doi: 10.1158/0008-5472.CAN-07-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haverty PM, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 16.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 17.Harada T, et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27:1951–1960. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 19.Nagayama K, et al. Homozygous deletion scanning of the lung cancer genome at a 100-kb resolution. Genes Chromosomes Cancer. 2007;46:1000–1010. doi: 10.1002/gcc.20485. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 21.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cairns P, et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- 23.Liggett WH, Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16:1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 24.Wang TL, et al. Digital karyotyping. Proc Natl Acad Sci USA. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steemers FJ, et al. Whole-genome genotyping with the single-base extension assay. Nat Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- 26.Peiffer DA, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 28.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 29.Volik S, et al. End-sequence profiling: Sequence-based analysis of aberrant genomes. Proc Natl Acad Sci USA. 2003;100:7696–7701. doi: 10.1073/pnas.1232418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64:2998–3001. doi: 10.1158/0008-5472.can-04-0587. [DOI] [PubMed] [Google Scholar]
- 33.Yu JS, et al. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27:4657–4665. doi: 10.1038/onc.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007;296:50–57. [PubMed] [Google Scholar]
- 37.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192:1–17. doi: 10.1016/s0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 38.Tanner M, et al. Topoisomerase IIalpha gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J Clin Oncol. 2006;24:2428–2436. doi: 10.1200/JCO.2005.02.9264. [DOI] [PubMed] [Google Scholar]
- 39.Coon JS, et al. Amplification and overexpression of topoisomerase IIalpha predict response to anthracycline-based therapy in locally advanced breast cancer. Clin Cancer Res. 2002;8:1061–1067. [PubMed] [Google Scholar]
- 40.Varshavsky A. Targeting the absence: Homozygous DNA deletions as immutable signposts for cancer therapy. Proc Natl Acad Sci USA. 2007;104:14935–14940. doi: 10.1073/pnas.0706546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leary RJ, Cummins J, Wang TL, Velculescu VE. Digital karyotyping. Nat Protoc. 2007;2:1973–1986. doi: 10.1038/nprot.2007.276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.