Abstract
Plasmodium vivax causes over 100 million clinical infections each year. Primarily because of the lack of a suitable culture system, our understanding of the biology of this parasite lags significantly behind that of the more deadly species P. falciparum. Here, we present the complete transcriptional profile throughout the 48-h intraerythrocytic cycle of three distinct P. vivax isolates. This approach identifies strain specific patterns of expression for subsets of genes predicted to encode proteins associated with virulence and host pathogen interactions. Comparison to P. falciparum revealed significant differences in the expression of genes involved in crucial cellular functions that underpin the biological differences between the two parasite species. These data provide insights into the biology of P. vivax and constitute an important resource for the development of therapeutic approaches.
Keywords: comparative genomics, Plasmodium falciparum
It is now increasingly recognized that P. vivax infections contribute significantly to the burden of malaria (1, 2). In all endemic areas except for Africa, P. vivax is often the dominant species, and at least 100 million cases are reported annually (2, 3). Although vivax malaria is clinically less likely than P. falciparum to develop into a life threatening disease, it exerts a substantial toll on the individual's health and economic well being. The chronic, long-lasting nature of the infection contributes substantially to morbidity. Chronicity is because of hypnozoites, dormant liver stages from which fresh blood infection or relapses originate up to 2 years after the infectious bite (4). The presence of hypnozoites make infections by P. vivax difficult to cure radically and pose a serious obstacle to the control and eventual eradication of this parasite.
The description of the P. falciparum genome (5) and staged erythrocytic transcriptome (6, 7) has provided an invaluable resource for the study of this important species. It would be of fundamental and practical interest to do the same for P. vivax because there are important biological and clinical differences between this species and P. falciparum, whose basis is currently unknown (8). For example, the presence of circulating mature erythrocytic stages of P. vivax would suggest that multigene families and processes implicated in antigenic variation and immune evasion are quite different to P. falciparum, whose mature asexual red cell stages generally sequester. Unlike P. falciparum, P. vivax has a selective preference for infecting reticulocytes (9), strongly suggesting an alternate red cell attachment invasion mechanism. In contrast to the rigid, sticky and knobby P. falciparum infected red cell, P. vivax remodels the host-cell membranes to produce a highly deformable erythrocyte characterized by numerous caveola-vesicle complexes (10–12). Finally, the kinetics of gametocyte production in P. vivax is also different than P. falciparum, with P. vivax gametocytes appearing much earlier and being relatively short lived (8). Aside from these notable interspecies differences, there are a number of important phenotypic differences within P. vivax relating to relapse periodicity and chloroquine sensitivity, which the mechanisms behind these differences are still unknown.
Investigations into the biology of P. vivax have been restricted by the lack of a continuous cultivation system. With advances of sequencing technology, the P. vivax genome is now available (13), allowing the construction of a representative microarray. This significant advance, coupled with the ability to mature ex vivo isolates, has opened the way to obtain a high-quality transcriptome of the blood stages. This study aimed to provide a stage-specific transcriptome of the intraerythrocytic developmental cycle (IDC) of P. vivax which can be compared with P. falciparum (6). Although the IDC represents only a relatively small portion of the Plasmodium life cycle, close to two thirds of Plasmodium genes are expressed and transcriptionally regulated during this 48-h development (6, 7). Thus, characterizing the P. vivax IDC transcriptome will provide broad insights into the P. vivax biology and gene functionalities of this parasite. In addition, three separate clinical isolates of P. vivax were used to provide some indication into the magnitude of intraspecies IDC transcriptome variation [see supporting information (SI) Dataset S1, Dataset S2, and Dataset S3].
Results and Discussion
Transcriptional Regulation of P. vivax Genes During the Erythrocytic Stage.
To study the IDC transcriptome of P. vivax, we collected three clinical isolates from acute vivax malaria patients before treatment on the Northwestern border of Thailand (Shoklo Malaria Research Unit, Mahidol University, Mae Sot, Thailand). These synchronous, monoclonal, erythrocytic isolates were cultured ex vivo from the early ring stage to the schizont stage (see Table S1) (12, 14). Transcriptional analysis using a genome-wide long oligonucleotide microarray designed by the recently established algorithm OligoRankPick (15) showed that during the P. vivax IDC, each gene is activated at a particular developmental stage analogous to P. falciparum (Fig. 1) (6). To evaluate the reproducibility and fidelity of the microarray results, one of the IDC transcriptome was replicated in a dye swap experiment, and expression profiles for 5 genes were verified by quantitative RT-PCR (Fig. S1). Similar to P. falciparum, the IDC transcriptome of P. vivax shows that functionally related genes are transcriptionally coregulated and exhibit conservation of the timing of their induction (Fig. 1A).
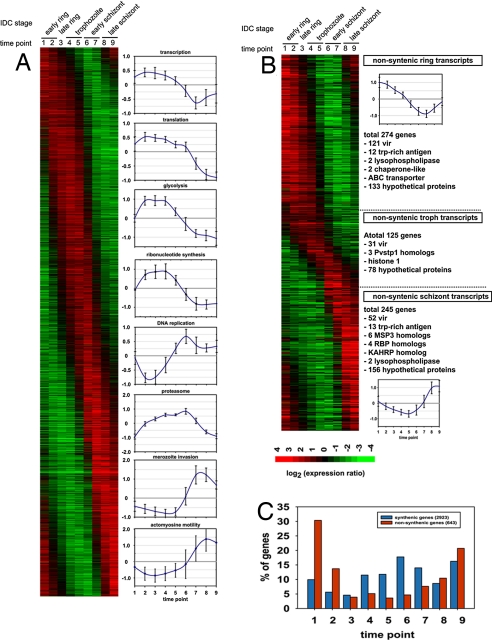
Fig. 1.
Transcriptome of the P. vivax IDC. (A and B) A total 3,566 genes exhibit >2-fold change in mRNA abundance across the P. vivax IDC. The assembled phaseograms represents the transcriptome of a single P. vivax isolate (smru1), and it includes 2,923 genes with a P. falciparum syntenic ortholog (A) and 643 genes without (B). Both IDC transcriptome phaseograms were assembled from the microarray expression profiles in which the values were log2 transformed, and each profile was mean centered. The expression profiles are ordered by the phase of the major periodic component calculated by the Fast Fourier Transformation as described (6). (A) The plots indicate the average expression profiles and standard deviations of expression values for genes that belong to functional groups with synchronized expression during the P. falciparum IDC (6). (B) The nonsyntenic genes are classified into ring, trophozoite, or schizont transcripts based on the time of their peak expression. Representative examples of P. vivax stage specific genes or gene families are listed next to each section of the phaseogram that correspond to individual developmental stages (ring, trophozoite, schizont). (C) The histogram depicts a relative distribution of genes with peak expression in each time point for the syntenic and nonsyntenic gene groups.
Comparison of the P. vivax and P. falciparum IDC Transcriptomes.
Expression of the majority of P. vivax nonsyntenic genes at the schizont-ring transition.
Comparison of the P. vivax and the published P. falciparum IDC transcriptome shows a striking difference in the distribution of IDC-specific gene transcription between P. vivax-P. falciparum syntenic and nonsyntenic genes. Whereas the peaks of expression of syntenic genes are distributed evenly throughout the IDC, with slight bias toward the trophozoite-schizont transition, the global distribution of mRNA abundance of the nonsyntenic genes exhibits a strong preference toward the extremes of the IDC; the schizont-ring stage transition (Fig. 1 B and C). This indicates that the development of the invasive merozoites and the establishment of the parasite in a new host cell represent the key stage at which malaria parasite species-specific differences occur. The vast majority of the P. vivax nonsyntenic genes expressed at the schizont-ring transition (Fig. 1B) can be divided into three broad classes: immune evasion, host-cell invasion, and functionally uncharacterized genes. The vir gene family is the largest gene family in P. vivax, members of which have been implicated in immune evasion (16). Of the 346 vir genes predicted in the P. vivax genome, at least 204 are transcribed during the IDC. Although distinct groups of vir genes are expressed at different IDC stages (Fig. 1B), no correlation was observed between the time of expression and the postulated phylogenic groups (16) (Fig. S2). Members of the pvtrag gene family that have also been linked with P. vivax immune evasion (17) show two distinct phases of transcription (Fig. 1B). These data suggest that during its IDC, P. vivax undergoes two “waves” of antigenic presentation. The first wave is initiated immediately after invasion by expression of a large proportion of the vir and pvtrag genes family (121 vir and 12 pvtrag), and the second wave is timed to schizogony during which another large, but nonoverlapping, group of both gene families (52 vir 13 pvtrag) are expressed. This second wave is potentially reflecting additional needs for antigenic presentation of the nonsequestering P. vivax parasite. This presentation of variant antigens is clearly different from P. falciparum, in which transcription of the majority of the antigenic gene families is silenced in the late stages (18–20). Expression during the schizont-ring transition was also detected for gene families that had undergone lineage-specific evolution. These include genes linked with host-cell invasion like merozoite surface protein 3 (msp3), msp7, and reticulocyte binding proteins (rbp) and two functionally uncharacterized gene families Pv-fam-e (RAD) and Pf-fam-h (PHIST) (Fig. S3). Based on these observations, it is likely that the new members of these gene families, as well as 133 ring- and 156 schizont-specific, nonsyntenic, hypothetical genes expressed during the IDC (Fig. 1B), represent important factors associated with the P. vivax selectivity for young erythrocytes (21) and/or immune evasion.
Transcriptional shift of several conserved functions in the P. vivax IDC.
Pearson correlation comparisons between the P. vivax and P. falciparum IDC transcriptomes (Fig. S4) revealed that although the majority of P. vivax genes exhibit identical transcriptional regulation, considerable differences exist for a small portion of the P. vivax genome (Fig. 2). Whereas ≈68% of the syntenic genes that include enzymes with basic metabolic function (dhfr-ts) or genes with highly conserved function (msp1) (Fig. 2B) exhibit identical expression profiles (Fig. 2A, “high correlation”), ≈22% show a partial shift in mRNA abundance across the IDC (Fig. 2A, “low-to-no correlation”). For example, msp8, which is associated with the establishment of early rings in P. falciparum (22), is transcribed throughout the entire ring and trophozoite stages reflecting an extended requirement for this protein in P. vivax. Strikingly, up to 11% of the syntenic genes show dramatically altered expression during the P. vivax IDC as compared with P. falciparum (Fig. 2A, “negative correlation”). One of the interesting examples is knob-associated, histidine-rich protein (KAHRP), which is involved in the formation of protein dense protrusions (“knobs”) on the surface of the P. falciparum infected erythrocytes and has been found to be essential for cytoadherence under physiological flow conditions (23). P. vivax, which is devoid of knobs, has two proteins (Pv081835 and Pv003520) that carry a short N-terminal segment (≈60 aa, ≈10% of PfKAHRP length) that is highly homologous to PfKAHRP. (Note: Throughout this article, gene identifiers, indicated as Pvxxxxxx, refer to GenBank identifiers, PVX_xxxxxx.) Moreover, Pv003520 is syntenic to pfkahrp, located just before the syntenic break between chromosome 2 and 4 of P. falciparum and P. vivax, respectively. This suggests that pfkahrp and its P. vivax counterparts share a common ancestor. In contrast to PfKAHRP, which is expressed during the ring stage, both Pv081835 and Pv003520 are expressed in the late schizont stage (Figs. 1B and 2B). Taken together, this represent a case where diversion of the amino acid sequence is coupled with the shift in the transcriptional regulation and likely reflects functional evolution of these proteins between the two Plasmodium species.
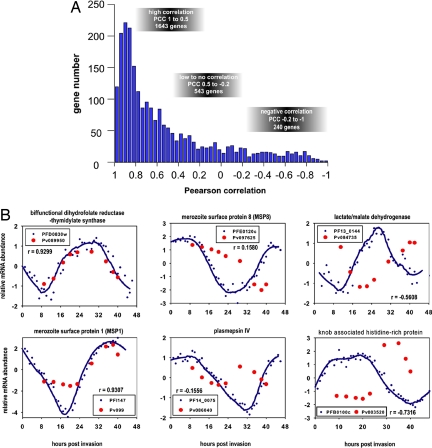
Fig. 2.
Comparative analyses of P. vivax and P. falciparum IDC transcriptomes. Using the best fit Pearson correlations, we correlate gene expression data in TP1–9 in P. vivax to the expression data in TP 9, 13, 17, 20, 23, 29, 35, 40, and 43 in the P. falciparum transcriptome (Fig. S2). (A) Histogram of the overall distribution of Pearson correlation coefficients (PCCs) for 2,426 P. vivax–P. falciparum gene pairs, which are syntenic between both species (2,923) and whose expression is also detected in both IDC transcriptomes. The PCC distribution is calculated based on the IDC transcriptome of the P. vivax smru1 isolate and the P. falciparum HB3 strain. To evaluate the timing of transcription, the mRNA expression ratios were log2 transformed, and each expression profile was mean centered. PCCs were calculated based on the visual inspections of the corresponding expression profiles, and we defined arbitrary PCCs thresholds that divide the genes according to their conservation of expression profiles between the P. vivax and P. falciparum IDC. High correlations (PCC 1 to 0.5) include genes with highly conserved IDC expression profiles, low-to-no correlations (PCC 0.5 to −0.2) include genes with a partial shift in their the IDC expression profile, and negative correlations (PCC −0.2 to −1) include genes with a dramatic change in their IDC expression profiles in P. vivax compared with P. falciparum. (B) Examples of the corresponding gene expression profiles for genes with highly conserved expression profiles (dihydrofolate reductase-thymidilate kinase, DHFR-TS, and MSP1), genes with partial shifts in their expression profiles (MSP8 and plasmepsin IV), and genes with a dramatic change of their IDC expression profiles in P. vivax compared with P. falciparum (KAHRP and lactate/malate dehydrogenase). In the plots, the expression values are log2 transformed, and the expression profiles are mean centered. The expression profiles of the P. falciparum genes are represented by raw data (blue circles) and smoothed lines using Loess smoothing method with 3 polynomial degree 3 (blue line) whereas P. vivax expression is represented by raw values only (red circles).
To gain further insights into the impact of the alterations of transcriptional regulation on P. vivax physiology, we analyzed gene expression correlations within the functionally related gene groups that were classified by the Malaria Parasite Metabolic Pathway database (24). All functional groups exhibited high levels of correlations between the IDC transcriptional profiles and median values ranging from 0.6516 (DNA replication) to 0.9033 (translation initiation) (Fig. S5). These data indicate that the IDC timing of the majority of biological functions is conserved between P. vivax and P. falciparum. However, essentially every functionally related gene group contains a small fraction of genes whose transcriptional timing is shifted between the IDCs of P. falciparum and P. vivax. More studies will be required to determine whether these genes evolved into different biological roles or whether they represent an extended functionality of the conserved pathway. For several functional groups, we observed transcriptional shifts for a high proportion of their genes which suggest significant changes in their timing during the IDC (Table S2).
Two H+ pumping mechanisms have been implicated in maintaining the pH within the food vacuole (25), and although there is no difference in the expression profile of the H+-pyrophoshatase, there is a dramatic shift of expression of three major subunits of the vacuolar H+-ATPase suggesting a more gradual acidification of the food vacuole organelle throughout the P. vivax IDC compared with P. falciparum (25). The food vacuole in malaria parasites is the site of hemoglobin degradation, and thus hemoglobin degradation in P. vivax may proceed at a more gradual rate (Table S2). This is corroborated by a similar transcriptional shift of the major hemoglobinases plasmepsinIV and falcipain2 and 3, and an amino acid transporter that is implicated in the removal of unused amino acids released during hemoglobin degradation (26). In P. falciparum, both copies of falcipain2 show strong expression in the ring/trophozoite stage with falcipain3 being transcribed only very weakly throughout the whole-time course (6). In contrast, in P. vivax, both falcipain3 and the nonsyntenic copy of falcipain2 show strong expression in the early schizonts stage, whereas the syntenic copy of falcipain2 shows evenly distributed transcription throughout the time course. Interestingly, in P. falciparum, plasmepsinIV has been expanded by three additional paralogs (plasmepsin I, II, HAP), all of which show peak expression in the early ring or trophozoite stage compared with the peak of expression in early schizonts for the P. vivax gene. This further supports potential differences in the hemoglobin degradation process between these two species. These results may also, in part, explain the recently described differences in chloroquine sensitivity of P. vivax trophozoites (27).
Another marked difference involves the exp1 and exp2 genes and the etramp family, which are linked with the development of the parasitophorous vacuole (PV) that surrounds the intracellular parasite and is formed immediately after invasion in P. falciparum (28). In P. vivax, both Exp genes and four etramp genes are expressed in schizonts. Variation in the development of the PV, because of the delay of exp and etramp gene expression, might contribute to differences in erythrocyte modifications and lead to the observed differences in rigidity and cytoadherence between P. vivax and P. falciparum infected erythrocytes (12). Consistent with the differences in rigidity and cytoadherance properties between the two Plasmodium species is the observation that P. vivax lacks the genes of the ring-infected erythrocyte surface antigen (resa) family whose function is linked with the increased rigidity of P. falciparum infected erythrocytes (29). RESA proteins were found to be transported to the erythrocytic cytoskeleton via distinct compartments of the PV immediately after invasion (30).
Of the 65 P. falciparum predicted protein kinases (31), 49 syntenic orthologs were found to be transcriptionally regulated through the P. vivax IDC. Expression of 11 of these, however, is considerably delayed in P. vivax. Nine of these belong to the CMGC, NIMA, or CamK classes that are linked to developmental processes and/or regulation of cell proliferation (Table S2). One of these is a well characterized serine/threonine protein kinase PfNek1 (32) whose expression is shifted from early to late schizonts. The Nek1 kinases were found to be involved in the onset of mitosis in other organisms (33). The transcriptional delays of PfNek1 and other protein kinases might signal a delayed onset and progression of P. vivax schizogony compared with P. falciparum. This is consistent with the observed delay in expression of a number of DNA replication factors during the P. vivax IDC (Table S2). Proteins containing the AP2-integrase, DNA-binding domain have been recently implicated in the global regulation of the Plasmodium life cycle (34, 35). All 22 P. falciparum AP2 proteins have syntenic homologues in P. vivax, of which 19 exhibit identical IDC expression profiles between these two species; one homologue (Pv118015) is shifted from the trophozoite stage in P. falciparum to the late schizont stage in P. vivax, and two homologues (Pv086035 and Pv094580) were below the detection threshold of the P. vivax microarray. These data indicate a conserved role of the AP2 proteins in the regulation of the Plasmodium life cycle, but more analyses will be required to determine whether Pv118015, the outlying ortholog, contributes significantly to the transcriptional diversion between the two plasmodium species.
Transcriptional changes between P. vivax and P. falciparum are driven from regulatory elements of individual genes.
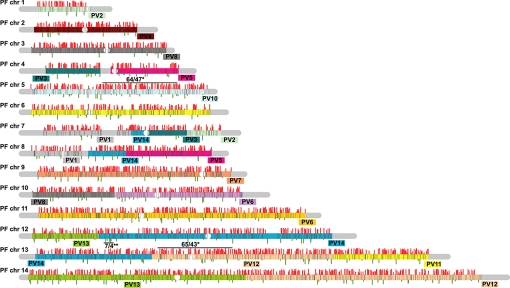
The distribution of transcriptional regulation along Plasmodium chromosomes shows no apparent association with any particular chromosomal region or domain, and differentially expressed genes are evenly scattered along the chromosomes (Fig. 3). This was further confirmed by comparison of transcription correlation values via a sliding window approach along the syntenic block regions with those seen by Pearson correlation presented in Fig. 2A. The two largest highly correlated segments (window of 100 consecutive genes) contain 64 and 65 genes expressed during the P. vivax IDC from which 47 (73%) and 43 (66%) genes are highly correlated, respectively. This is only slightly above the average distribution throughout the genome in which a window of 100 genes contains 43 expressed and 31 (67%) highly correlated genes. A window of 10 genes with 4 negatively correlated genes on chromosome 14 is the region with the lowest level of conservation in IDC transcriptional regulation between P. falciparum and P. vivax (Fig. 3). Our P. falciparum IDC transcriptome analyses revealed an absence of chromosomal domains with transcriptionally coregulated genes (6). Thus, it appears that in Plasmodium parasites, genes are expressed monocistronically, and their tight transcriptional regulation is controlled through regulatory elements associated with each gene individually. These data suggest that the evolutionary diversion of transcriptional regulation between Plasmodium species has taken place at the level of the individual genes, and this diversion is independent of any Plasmodium chromosomes structural features such as isochores.
Fig. 3.
Chromosomal projection of P. vivax–P. falciparum IDC transcriptome correlations. Pearson correlation values for IDC transcriptional profiles were plotted along the P. falciparum chromosomes for all 2,426 P. vivax–P. falciparum positionally conserved gene pairs. All 14 P. falciparum chromosomes are represented by blocks corresponding in size to the size to the individual chromosome. The colored sections correspond to P. vivax syntenic blocks that are projected onto the corresponding regions of the P. falciparum chromosomes. The P. vivax chromosomal origin is indicated for each synteny block (e.g., PV2 – P. vivax chromosome 2). Pearson correlations are proportionally represented by the gray bars above (positive values) or below (negative values) for all 2,426 genes, whereas genes excluded from the analysis are represented by black lines within the chromosome projection boxes. The position of each line reflects a relative location of each gene within the P. falciparum chromosomes. * represents the two regions with the highest proportion of genes with conserved IDC transcriptional regulation: number of expressed genes/number of highly (PCC > 0.5) correlated genes; ** represents the region with the lowest level of IDC correlation: number of expressed genes/number of negatively correlated (PCC < −0.2) genes.
Variation of Transcription in Different P. vivax Isolates.
Although P. vivax is generally associated with benign pathology, a wide spectrum of clinical manifestations, including severe and even lethal infections, was reported from many endemic regions including Papua New Guinea, Indonesian Papua, and Sri Lanka (2, 36). This indicates a considerable variability in the host–parasite interactions, virulence, and the growth and transmission rate. Although many recent large-scale genotyping efforts brought some insights in to the P. vivax genetic diversity (37), including gene amplifications (38), little is known about how this translates to intraspecies variability of P. vivax gene expression. Our pairwise comparisons between the three IDC transcriptomes revealed that the timing of gene expression is completely conserved in each isolate with median Pearson correlation ranging from 0.785 ± 0.223 to 0.862 ± 0.207 (Fig. S1). However, significant differences in the total mRNA levels between P. vivax isolates were detected for 249 genes (P value < 0.05) (Fig. 4). Each isolate expresses a distinct group of genes at higher levels compared with the two other isolates, including genes involved in host parasite interactions, regulatory mechanisms, and several enzymatic activities.
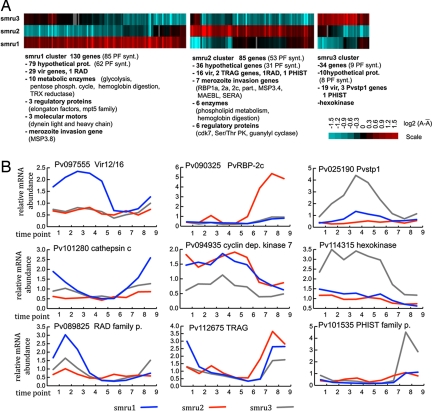
Fig. 4.
Variability of gene expression between P. vivax isolates. (A) mRNA abundance levels were compared between three P. vivax isolates by calculating the total sum of the relative mRNA abundance across the nine time point of the IDC transcriptomes: A = ΣexpTPi, where expTP is the expression ratios measured at each time point i (1 through 9). The heat map represents the log2 transformed mean centered A values for the 249 differentially expressed genes (P value > 0.05) (see Materials and Methods). A total of 130, 85, and 34 genes exhibited increased expression levels in the smru1, smru2, and smru3 isolates, respectively. (B) Representative examples of transcriptional profiles of the deferentially expressed genes including a member of the vir family, rbp 2c, Pvstp1, cathepsin C (high expression in smru1), cyclin dependent kinase 7 (cdk7), hexokinase, and members of the RAD, pvtrag (smru2), and PHIST (smru3) families. The expression profiles represent the gene expression ratios measured by the microarray analyses against the RNA reference pool assembled from the RNA samples from all time points of the three isolates.
Fernandez-Becerra et al.(39) has shown that in natural infections, P. vivax isolates express “large and mostly nonoverlapping” vir gene repertoires. This phenomenon was attributed mainly to the extensive polymorphisms detected in the vir gene family. To assess the level of polymorphisms among the three isolates, we carried out comparative genomic hybridization in a pairwise manner. Of the 262 total, 177 vir-specific oligonucleotide elements present on the P. vivax microarray exhibited equal hybridization signal (P value < 0.05) between any given isolate pair. Of these 177, 153 showed significant expression in all three isolates. Interestingly, 89 (of the 153) vir-genes exhibit identical expression levels whereas 26, 16, and 19 show a significantly increased mRNA abundance in one of the three isolates, respectively (Fig. 4). This suggests that besides the vir gene polymorphisms, transcriptional regulation contributes to the variability of the vir gene repertoire expressed by different P. vivax isolates. Variable transcription is also observed in the pvtrag gene family and the newly identified PHIST and RAD families (Fig. 4). Both the pvtrag (17) and the PHIST (40) gene families have been previously linked with immune evasion and parasite virulence, suggesting that differential transcription of their gene members might be a contributing factor to the P. vivax host–parasite interactions. The second most dominant functionality among the differentially expressed genes represents erythrocyte invasion that involves the rbp, sera, clag, maebl, msp3, and msp7 families (Fig. 4). Transcriptional variations in the rbp and clag genes were linked to switches in invasion phenotypes in other plasmodium species (41, 42), and their variable expression was observed in natural infections (43). In P. vivax, RBP has been shown to specifically recognize reticulocyte-specific receptors as compared with other plasmodium species where members of this conserved rbp homologues gene family have been shown to recognize receptors on mature red blood cells (RBCs) as well (42, 44). The variation in the expression of rbp observed here could therefore reflect parasite-specific adaptation to different host-cell receptors or host-immune status. At this stage, it will require further investigation to determine whether the observed transcriptional differences of both immune evasion and merozoite invasion genes seen here are because of genetic differences in the parasite isolates or the ability of the parasite to adapt to variations in immune status or available red cell receptors in the host. In addition, the recent study showed that in a natural infection, P. falciparum can acquire distinct transcriptional states that are defined by expression of distinct patterns of genes involved in fundamental catabolism and stress response (45). Although, in our studies, we do not observe such extensive transcriptional changes. Variations in a number of genes encoding for enzymes and regulatory proteins suggest that P. vivax has the ability to alter its metabolism and thus adapt to the growth conditions in its host.
Conclusions
There is mounting evidence that the transcriptional cascade of the P. falciparum IDC is highly conserved between different strains obtained from in vitro and in vivo growth (7, 45, 46). Moreover, growth perturbations of P. falciparum parasites induce only miniscule changes in the global transcriptional pattern (47). In P. vivax, we also fail to detect any differences in the timing of gene expression during the IDC between different isolates. These findings are consistent with predictions that the timing of gene expression during the IDC of Plasmodium species does not fluctuate between different strains or isolates (46). Thus, it is reasonable to speculate that many (if not all) of the observed interspecies variations represent conserved transcriptional differences that reflect evolution of the individual Plasmodium species.
Similar to P. falciparum, the intraerythrocytic development of P. vivax is characterized by extensive transcriptional regulation in which each biological function is timed to a specific section of the IDC. Although most of the P. vivax genes exhibit an identical expression profile across the IDC compared with their P. falciparum syntenic orthologs, there are partial shifts as well as dramatic alterations in the mRNA abundance profiles for 22% and 11% genes, respectively. In several cases, these changes represent substantial alteration of the timing for an entire biological function including hemoglobin degradation, host parasite interaction, protein export to the host-cell cytoplasm, and DNA replication. In contrast to the conserved portion of the P. vivax genome, the nonsyntenic P. vivax genes are activated predominantly at the schizont/ring interphase which suggests that the species differences derive mainly from events occurring at red cell invasion and early intraerythrocytic development. Moreover, expression of the majority of vir and pvtrag gene families throughout the IDC indicate major differences in antigenic presentation between P. falciparum and P. vivax. The even distribution of the differentially expressed genes along the P. vivax-P. falciparum chromosomal synteny blocks suggests that the diversion of transcriptional control between these two species occurred at the regulatory elements of individual genes. The intraisolate diversity of gene expression indicates an extensive capacity of P. vivax to adapt to its host and modify its virulence. This fundamental, biological information about P. vivax should facilitate the development of innovative control measures against this remarkably resilient parasite.
Materials and Methods
Sample Collection and RNA Isolation.
Adult patients from Northwestern Thailand (Shoklo Malaria Research Unit, Mahidol University, Thailand) with symptomatic vivax malaria gave informed consent to donate 10 ml of blood. The P. vivax infected blood samples were collected from three preselected patients before receiving standard chloroquine treatment. For more details, see SI Text.
Target DNA Preparations and Microarray Analysis.
The microarray contained 5,727 60-mer oligonucleotides that represented a total of 5,335 predicted genes in the P. vivax genome by at least one unique probe. From these, 5,063 are represented by a single oligonucleotide, whereas 272 genes (typically long ORFs) are represented by two or more oligonucleotide elements. The P. vivax microarray oligonucleotide set was designed by OligoRankPick (15). Sequences of all oligonucleotides and further information are available online at http://zblab.sbs.ntu.edu.sg/vivax/index.html. The P. virax long-oligo array is also available through the Pathogen Functional Genomics Resource Center (http://pfgrc.jcvi.org).
Data Processing and Analysis.
Microarray data processing and analysis (including the Fast Fourier Transform) was carried out as described (6). For more details, see SI Text.
Supplementary Material
Acknowledgments.
This work was supported by Biomedical Research Council Singapore Grants BMRC 04/1/22/19/364 and BMRC 05/1/22/19/398 and National Medical Research Council Singapore Grant NMRC/CPG/016/2005.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807404105/DCSupplemental.
References
- 1.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 2.Price RN, et al. Vivax malaria: Neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 3.Hay SI, et al. The global distribution and population at risk of malaria: Past, present, and future. Lancet Infect Dis. 2004;4:327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krotoski WA, et al. Observations on early and late post-sporozoite tissue stages in primate malaria. IV. Pre-erythrocytic schizonts and/or hypnozoites of Chesson and North Korean strains of Plasmodium vivax in the chimpanzee. Am J Trop Med Hyg. 1986;35:263–274. doi: 10.4269/ajtmh.1986.35.263. [DOI] [PubMed] [Google Scholar]
- 5.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozdech Z, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 8.Grannham PCC. Malaria Parasites and Haemosporidia. Oxford: Blackwell Scientific Publications; 1966. [Google Scholar]
- 9.Mons B. Preferential invasion of malarial merozoites into young red blood cells. Blood Cells. 1990;16:299–312. [PubMed] [Google Scholar]
- 10.Aikawa M. Parasitological review. Plasmodium: The fine structure of malarial parasites. Exp Parasitol. 1971;30:284–320. doi: 10.1016/0014-4894(71)90094-4. [DOI] [PubMed] [Google Scholar]
- 11.Aikawa M, Miller LH, Rabbege J. Caveola–vesicle complexes in the plasmalemma of erythrocytes infected by Plasmodium vivax and P cynomolgi. Unique structures related to Schuffner's dots. Am J Pathol. 1975;79:285–300. [PMC free article] [PubMed] [Google Scholar]
- 12.Suwanarusk R, et al. The deformability of red blood cells parasitized by Plasmodium falciparum and P. vivax. J Infect Dis. 2004;189:190–194. doi: 10.1086/380468. [DOI] [PubMed] [Google Scholar]
- 13.Carlton J, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008 doi: 10.1038/nature07327. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell BM, et al. Simple in vitro assay for determining the sensitivity of Plasmodium vivax isolates from fresh human blood to antimalarials in areas where P. vivax is endemic. Antimicrob Agents Chemother. 2003;47:170–173. doi: 10.1128/AAC.47.1.170-173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu G, et al. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinformatics. 2007;8:350–362. doi: 10.1186/1471-2105-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.del Portillo HA, et al. A superfamily of variant genes encoded in the subtelomeric region of Plasmodium vivax. Nature. 2001;410:839–842. doi: 10.1038/35071118. [DOI] [PubMed] [Google Scholar]
- 17.Jalah R, et al. Identification, expression, localization and serological characterization of a tryptophan-rich antigen from the human malaria parasite Plasmodium vivax. Mol Biochem Parasitol. 2005;142:158–169. doi: 10.1016/j.molbiopara.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Duraisingh MT, et al. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 19.Freitas-Junior LH, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 20.Tham WH, Payne PD, Brown GV, Rogerson SJ. Identification of basic transcriptional elements required for rif gene transcription. Int J Parasitol. 2007;37:605–615. doi: 10.1016/j.ijpara.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Galinski MR, Medina CC, Ingravallo P, Barnwell JW. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992;69:1213–1226. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- 22.Drew DR, Sanders PR, Crabb BS. Plasmodium falciparum merozoite surface protein 8 is a ring-stage membrane protein that localizes to the parasitophorous vacuole of infected erythrocytes. Infect Immun. 2005;73:3912–3922. doi: 10.1128/IAI.73.7.3912-3922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rug M, et al. The role of KAHRP domains in knob formation and cytoadherence of P falciparum-infected human erythrocytes. Blood. 2006;108:370–378. doi: 10.1182/blood-2005-11-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginsburg H. Progress in in silico functional genomics: The malaria Metabolic Pathways database. Trends Parasitol. 2006;22:238–240. doi: 10.1016/j.pt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 25.Saliba KJ, et al. Acidification of the malaria parasite's digestive vacuole by a H+-ATPase and a H+-pyrophosphatase. J Biol Chem. 2003;278:5605–5612. doi: 10.1074/jbc.M208648200. [DOI] [PubMed] [Google Scholar]
- 26.Martin RE, et al. The “permeome” of the malaria parasite: An overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 2005;6:R26. doi: 10.1186/gb-2005-6-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharrock WW, et al. Plasmodium vivax trophozoites insensitive to chloroquine. Malar J. 2008;7:94–100. doi: 10.1186/1475-2875-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spielmann T, et al. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol Microbiol. 2006;59:779–794. doi: 10.1111/j.1365-2958.2005.04983.x. [DOI] [PubMed] [Google Scholar]
- 29.Mills JP, et al. Effect of plasmodial RESA protein on deformability of human red blood cells harboring Plasmodium falciparum. Proc Natl Acad Sci USA. 2007;104:9213–9217. doi: 10.1073/pnas.0703433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Culvenor JG, Day KP, Anders RF. Plasmodium falciparum ring-infected erythrocyte surface antigen is released from merozoite dense granules after erythrocyte invasion. Infect Immun. 1991;59:1183–1187. doi: 10.1128/iai.59.3.1183-1187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: The kinome of a divergent eukaryote. BMC Genomics. 2004;5:79–97. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorin D, et al. PfPK7, an atypical MEK-related protein kinase, reflects the absence of classical three-component MAPK pathways in the human malaria parasite Plasmodium falciparum. Mol Microbiol. 2005;55:184–196. doi: 10.1111/j.1365-2958.2004.04393.x. [DOI] [PubMed] [Google Scholar]
- 33.Osmani SA, Pu RT, Morris NR. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell. 1988;53:237–244. doi: 10.1016/0092-8674(88)90385-6. [DOI] [PubMed] [Google Scholar]
- 34.Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Silva EK, et al. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci USA. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tjitra E, et al. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: A prospective study in Papua, Indonesia. PLoS Med. 2008;5:e128. doi: 10.1371/journal.pmed.0050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karunaweera ND, et al. Extensive microsatellite diversity in the human malaria parasite Plasmodium vivax. Gene. 2008;410:105–112. doi: 10.1016/j.gene.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 38.Suwanarusk R, et al. Chloroquine resistant Plasmodium vivax: In vitro characterisation and association with molecular polymorphisms. PLoS ONE. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez-Becerra C, et al. Variant proteins of Plasmodium vivax are not clonally expressed in natural infections. Mol Microbiol. 2005;58:648–658. doi: 10.1111/j.1365-2958.2005.04850.x. [DOI] [PubMed] [Google Scholar]
- 40.Sargeant TJ, et al. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortes A, et al. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog. 2007;3:e107. doi: 10.1371/journal.ppat.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyer JK, Amaladoss A, Genesan S, Preiser PR. Variable expression of the 235-kDa rhoptry protein of Plasmodium yoelii mediate host cell adaptation and immune evasion. Mol Microbiol. 2007;65:333–346. doi: 10.1111/j.1365-2958.2007.05786.x. [DOI] [PubMed] [Google Scholar]
- 43.Nery S, et al. Expression of Plasmodium falciparum genes involved in erythrocyte invasion varies among isolates cultured directly from patients. Mol Biochem Parasitol. 2006;149:208–215. doi: 10.1016/j.molbiopara.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaur D, Mayer DC, Miller LH. Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int J Parasitol. 2004;34:1413–1429. doi: 10.1016/j.ijpara.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 45.Daily JP, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450:1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 46.Llinas M, et al. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 2006;34:1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gunasekera AM, et al. Plasmodium falciparum: Genome wide perturbations in transcript profiles among mixed stage cultures after chloroquine treatment. Exp Parasitol. 2007;117:87–92. doi: 10.1016/j.exppara.2007.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.