Abstract
Histidine triad nucleotide-binding protein (HINT), a dimeric purine nucleotide-binding protein from rabbit heart, is a member of the HIT (histidine triad) superfamily which includes HINT homologs and FHIT homologs. Crystal structures of HINT-nucleotide complexes demonstrate that the most conserved residues in the superfamily mediate nucleotide binding and that the HIT motif forms part of the phosphate binding loop. Galactose-1-phosphate uridylyltransferase, whose deficiency causes galactosemia, contains tandem HINT domains with the same fold and mode of nucleotide binding as HINT despite no overall sequence similarity. Features of FHIT, a diadenosine polyphosphate hydrolase and candidate tumor suppressor, are predicted from HINT-nucleotide structures.
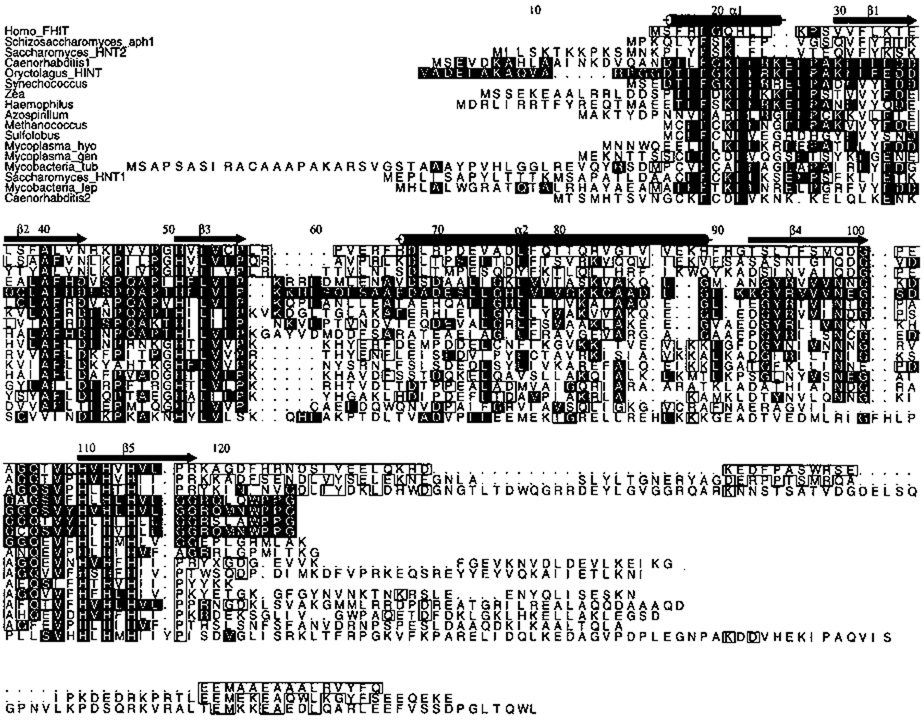
Histidine triad (HIT) proteins are a superfamily of proteins named for a His-ϕ-His-ϕ-His-ϕ-ϕ motif near the C-terminus in which ϕ is a hydrophobic amino acid1. This motif was reported be a binding site for zinc ion2. Primary sequence alignment of HIT proteins (Fig. 1) reveals that there are two branches. One branch contains representatives in all three domains of cellular life, the eukarya, bacteria, and archaea. The second branch appears to be confined to eukarya, with representative sequences in humans and two yeast genera.
Fig. 1.
Sequence alignment of HIT proteins excluding nearly identical proteins, expressed sequence tags and partial sequences. Strict conservation of residues aligning with Phe 19, His 51, Leu 53, and Histidines 110, 112 and 114 was required for inclusion. The top three proteins are members of the FHIT branch. The others are members of the HINT branch. Genera refer their most common species, except Mycobacteria_tub which refers to M. tuberculosis; Mycoplasma_gen, M. genetalium; Mycoplasma_hyo, M. hyorhinis; Mycobacteria_lep, M. leprae. Black boxes designate identity with rabbit (Oryctologus) HINT, white boxes identity with human FHIT at a residue not conserved with HINT. Secondary structure assignments and numbering are those of HINT.
We purified HINT (histidine triad nucleotide-binding protein), a member of the first branch of the HIT superfamily, as a dimeric purine nucleoside and nucleotide-binding protein from rabbit heart cytosol (J.G., N. Liang & J.M.L, in prep.). HINT is nearly identical to proteins that have been given the designation protein kinase C inhibitor-1 (PKCI-1). Bovine PKCI-1 was so named because it was present in brain cytosol fractions that inhibited a mixture of PKC isoforms3. It has not been possible to reproduce this inhibition with HINT from rabbit heart or with purified recombinant HINT (J.G., N. Liang & J.M.L., in prep.). The corresponding human cDNA was cloned in two independent two-hybrid screens. In the first screen, interactions were sought with the product of ATDC4, a cDNA that when overexpressed, suppresses the radiation-sensitivity of a fibroblast cell line from a child with Ataxia-Telangiectasia. In the second screen, interactions were sought with the N-terminal 317 amino acid regulatory domain of PKC isozyme β5. These authors refer to the protein as PKCI-1 (protein kinase C interacting protein) on account of the yeast two-hybrid interaction. The human gene has been mapped to 5q31.26.
The HINT branch of the HIT superfamily contains proteins found in all three domains of cellular life (Fig. 1). Homologs within the eukarya are not restricted to those in plants7, animals8 and fungi9 but represent, additionally, distant eukarya such as Dictyostelium discoidium (Genbank accession number U61986). Within the contiguous core of 102 amino acids that includes all secondary structural elements of HINT, mammalian HINT proteins are nearly identical. HINT proteins from nonmammalian animals and nonanimal eukaryotes represent greater variety. The flatworm Caenorabditis elegans8 contains two HINT-related proteins, one with 68 and the other with 25 identities within the core. Homologs from Zea mays7 and Saccharomyces cerevisiae9 contain, respectively, 54 and 32 identities within this 102 amino acid region.
Remarkably, several predicted proteins from bacteria and archaea have been identified that have greater identity with mammalian HINT than do eukaryotic counterparts. The cyanobacterial Synechococcus has a homolog10 more similar to mammalian HINT than is the Zea protein. The proteobacterial Haemophilus11 and Azospirillum12 genera and the methanogenic archaeon Methanococcus13 have HINT proteins more similar to mammalian HINT than the Saccharomyces HINT protein, Hnt1. The smallest known genome of a free-living organism, that of Mycoplasma genitalium, has 470 predicted coding regions. It encodes a HINT homolog and no homolog of protein kinase C or any other eukaryotic-type protein kinase14.
The second branch of the HIT superfamily (Fig. 1), is named for the human FHIT protein which is encoded at the chromosome 3 fragile site15. The FHIT branch consists of two enzymes with demonstrated diadenosine polyphosphate hydrolase activity16, 17 and one protein we predict to have such activity. Diadenosine polyphosphate hydrolases cleave dinucleotide polyphosphates such as A(5′)ppp(5′)A that have adenosine or guanosine nucleosides esterified to a chain of 3 to 5 phosphates18. The phosphate ester linkages are to the 5′-ribose oxygen on both nucleosides such that cleavage of ApppA yields AMP plus ADP, cleavage of AppppG yields AMP plus GTP, and so forth. Phylogenetically, these enzymes constitute a distinct branch of the HIT superfamily that appears to have emerged within eukarya. Proteins within this branch contain 18 conserved residues that differ from those in mammalian HINT. The hydrolases also share a C-terminal extension of 37 to 95 amino acids containing eight invariant amino acids.
The human FHIT gene at 3p14.2 is disrupted in many human tumors15, 19–28. As such, FHIT is a candidate tumor suppressor gene whose loss of function may contribute to the initiation or progression of neoplasias. When FHIT was identified as a member of the HIT protein superfamily15 and subsequently shown to cleave diadenosine polyphosphates17, the molecular basis of its intrinsic nucleotide binding and hydrolase activity were not understood. The existence, demonstrated herein, of two identical purine mononucleotide-binding sites in a HINT dimer suggests the mode of binding of a dinucleotide by FHIT. Sequence alignments and modeling of FHIT from HINT-nucleotide coordinates have suggested the identities of amino acids in FHIT that could be involved in cleavage of diadenosine polyphosphates.
We report here the crystal structures of HINT in its unliganded form, as well as HINT bound to adenosine, GMP and 8-Br-AMP. These structures show that the residues most conserved in the HIT superfamily are involved in binding nucleotides, and that the histidine triad, rather than constituting a zinc binding site2, 5, forms part of the phosphate-binding loop. Because HINT has no PKC inhibitory activity (J.G., N. Liang & J.M.L., in prep.), has bacterial11, 14 and archaeal13 homologs in organisms without protein kinase C, and has been conserved throughout evolution as a nucleotide-binding protein, we argue that classification of HINT as PKCI-1 is unjustified. We propose that this protein and its branch of the HIT superfamily be called HINT.
We also demonstrate that the dimeric structure of HINT, dominated by a 10-stranded anti-parallel β-sheet, can be superimposed upon the monomeric core structure of galactose-1-phosphate uridylyltransferase (GalT) from E. coli29, a highly conserved enzyme whose deficiency in humans causes galactosemia30. Despite a lack of overall sequence similarity, nucleotide binding by GalT is conferred by a subset of the most highly conserved residues in the HIT superfamily. The GalT monomer can be seen as consisting of two repeated HINT domains related by an internal pseudo-two-fold axis. Our analysis suggests that a HINT protein was a common ancestor of FHIT, GalT and other nucleotide-binding proteins. As HINT forms the scaffold for diverse enzymatic activities, it also creates a substructure for dissecting their function.
Biophysical characterization of the HINT dimer
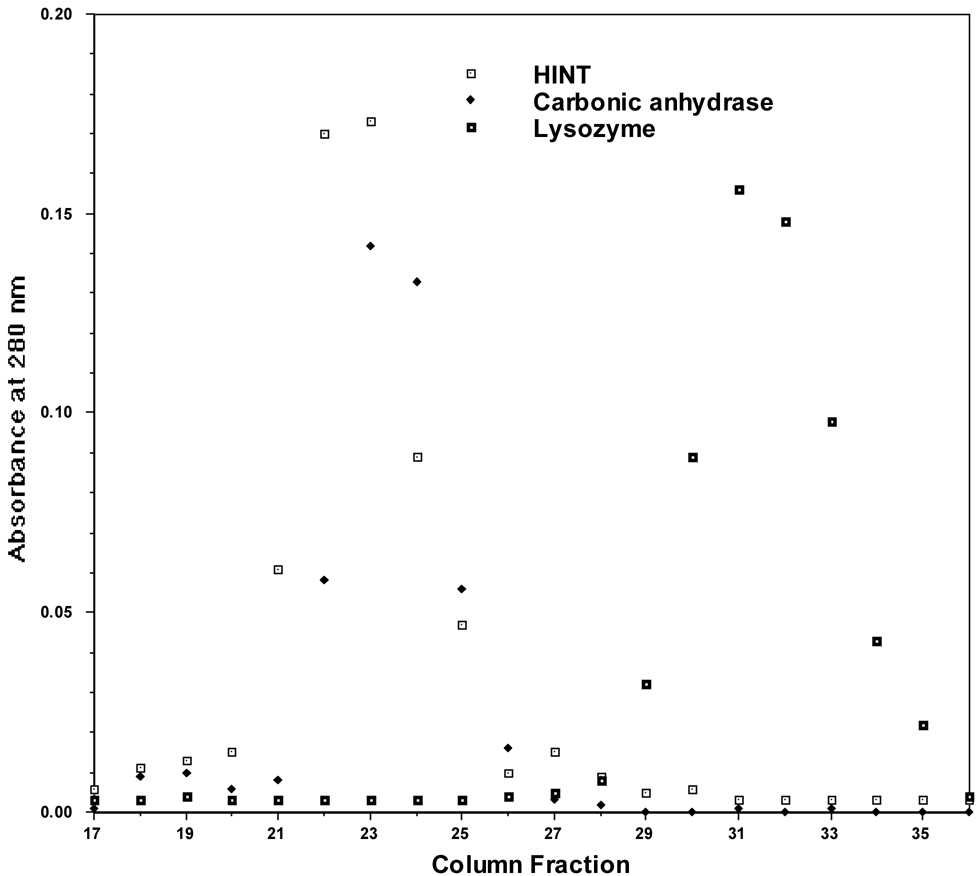
HINT was purified from rabbit heart cytosol by affinity chromatography on a column containing N6-(3-aminopropyl)adenosine-agarose (hereafter, adenosine-agarose) followed by elution with adenosine. The cDNA, isolated from a rabbit heart library with probes made from tryptic peptides of purified HINT (J.G., N. Liang & J.M.L., in prep.), encodes a protein of 13,700 Da (Fig. 1). Recombinant HINT, expressed in E. coli and eluted from adenosine-agarose with adenosine, was subjected to gel filtration on a column calibrated with globular monomeric proteins. HINT coeluted with a protein of 29,000 Da (Fig. 2), indicating that it is dimeric in solution or has an extended monomeric shape.
Fig. 2.
Gel Filtration Chromatography of HINT. A Sephadex G-100 column was calibrated with carbonic anhydrase (MW = 29 kDa) and lysozyme (MW = 14.3 kDa) before HINT was analyzed. 0.5 ml fractions were collected. Peaks were detected by optical density.
Recombinant HINT crystallized in two high symmetry space groups, both of which had specific volumes consistent with a monomer in the asymmetric unit. Because crystallographic symmetry can consist, in part, of authentic molecular symmetry, a monomer in an asymmetric unit can be half of a biological dimer. In fact, the earliest electron density maps of the protein indicated that HINT is globular and consists of two closely associated polypeptide chains related by a crystallographic two-fold axis. The behavior of the protein on gel filtration and a recent structure of unliganded human PKCI-1 in a lower symmetry space group, in which a dimer was found in the asymmetric unit5, confirms that HINT is homodimeric.
Structure of the HINT dimer
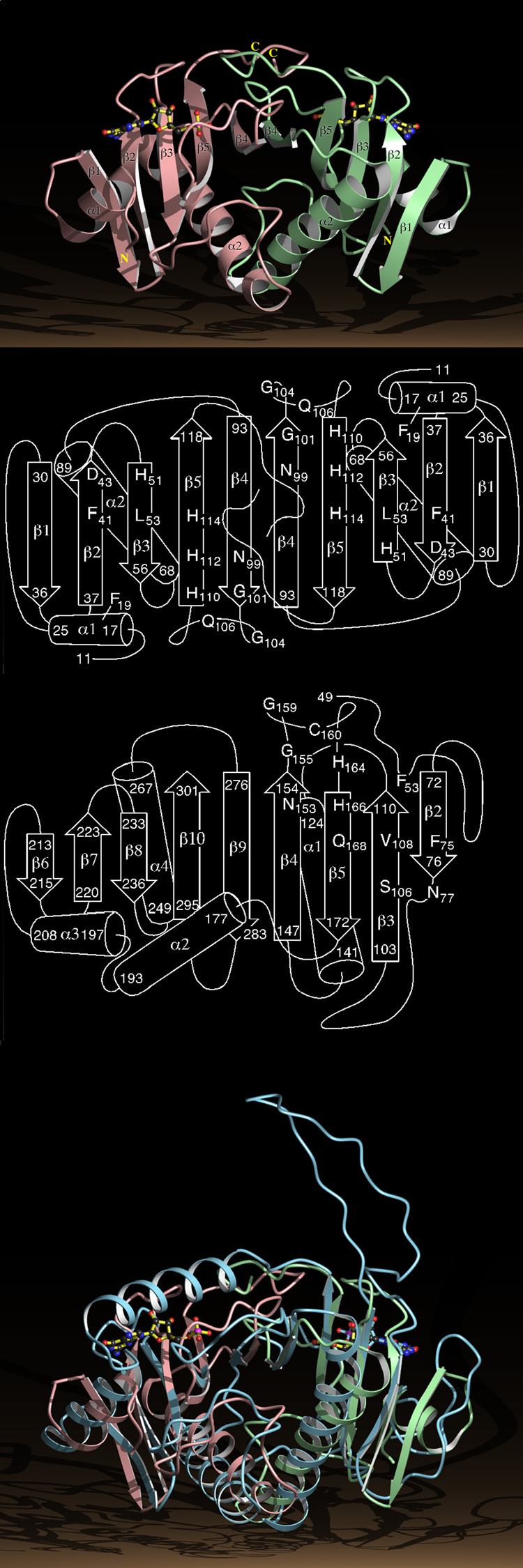
The HINT dimer is shaped like two fused cones with a 35 Å diameter cross-section at their interface and a distance of 55 Å from the tip of one conical monomer to the tip of the other monomer (Fig. 3a). The N-termini, consisting of short helices, α1, are distant from each other, near the tips of the two cones. The C-termini intertwine and are anchored by a bidentate salt-bridge between the guanadino nitrogens of Arg 119 of one monomer and the terminal carboxylic oxygens of Gly 126 of the other monomer. The main structural feature of the HINT dimer is an antiparallel 10-stranded β-sheet with the typical right-handed twist31. The sheet is made up of strands β1, β2, β3, β5, and β4 of one monomer followed by strands β4, β5, β3, β2, and β1 from the other (Fig. 3b). The long helices, α2, of each monomer, which follow β3 in the linear sequence, pack together at the dimer interface and are in the interior of a half-barrel formed by the curved, 10-stranded β-sheet. The structure of the HINT dimer is in agreement with that determined for unliganded PKCI-15.
Fig. 3.
Structure of the HINT Dimer and the GalT Monomer.
a, Ribbon diagram of the HINT-GMP dimer with numbered secondary structural elements. The right hand HINT monomer is in green, the left in pink.
b, Secondary structure of the HINT dimer with the positions of a subset of the conserved residues in the HIT superfamily.
c, Secondary structure of residues 49 to 302 of the GalT core identifying residues that align in three dimensions with HINT.
d, The Cα positions of residues 49 to 302 of the GalT core, represented by a ribbon diagram in blue, superimposed on the HINT-GMP dimer.
This sort of half-barrel has been seen in the nine-stranded anti-parallel β-sheet (Fig. 3c) that forms the core (residues 49 to 302) of a monomer of galactose-1-phosphate uridylyltransferase (GalT) of E. coli29. GalT does not have secondary structural elements that correspond to α1 and β1 of the right hand HINT monomer although the first 22 residues of the GalT core follow the path of these elements. The next 102 residues of the GalT core overlay the remaining four strands and the long helix of HINT. Whereas HINT monomers terminate after β5, GalT continues with a helix that takes its peptide chain to the site of the N-terminus of the second HINT monomer such that residues 198 to 302 of GalT track, in three-dimensions, along the entirety of the second HINT monomer (Figs. 3c and d).
The structural similarity is striking. The most obvious differences between GalT and HINT fail to undermine the similarity between them. First, the N-terminal half of GalT contains an insertion of 21 residues between HINT strands β2 and β3. This insertion occurs at the site of an Ω-shaped β-turn in HINT and has, itself, anti-parallel β-characteristics. Second, GalT is distinct in that the two halves of its sheet are part of the same chain. Nonetheless, three-dimensional least squares superposition done with only secondary structural elements from the N-terminal half of GalT to one HINT monomer aligns the whole GalT core to both HINT monomers. Thus, the GalT core contains a previously unrecognized pseudo-two-fold rotation axis that relates two tandem HINT domains. The HINT dimer possesses a known protein fold, the GalT half-barrel29.
Nucleoside and nucleotide binding by HINT
Ligand-binding studies demonstrate that HINT binds purine nucleosides and nucleotides including adenosine, 8-Br-AMP and GMP, despite the fact that sequence comparisons with other nucleotide-binding proteins did not reveal previously recognized nucleotide-binding motifs (J.G., N. Liang & J.M.L., in prep.). HINT protein crystallizes in the presence and absence of these compounds and the crystals diffract to atomic resolution (Table 1). Electron density maps of the cocrystals clearly show the presence of nucleotides in a cleft on top of the β-sheet (FFig. 4a).
Table 1.
Data Collection and Phasing Statistics
| Data Set | Space Group | Resolution (Å) | Reflections Measured/Unique | Complete (%) | Rsyma (%) | MFIDb (%) | |Fh|/|E|c | |
|---|---|---|---|---|---|---|---|---|
| ligand | derivative | |||||||
| adenosine | none | P3(1)21 | 25.82 - 1.90 | 50,474 / 9,074 | 84 | 3 | reference | |
| adenosine | TMLA d | P3(1)21 | 20.85 - 3.00 | 8,581 / 1,899 | 72 | 5 | 20 | 1.7 |
| adenosine | MA e | P3(1)21 | 17.15 - 3.38 | 7,560 / 1,770 | 94 | 9 | 23 | 1.1 |
| adenosine | DMA f | P3(1)21 | 25.82 - 3.00 | 8,520 / 2,621 | 98 | 6 | 12 | 1.3 |
| GMP | none | P3(1)21 | 27.74 - 2.10 | 49,822 / 7,463 | 99 | 3 | n/a | n/a |
| 8-Br-AMP | none | P4(3)2(1)2 | 35.13 - 2.31 | 45,935 / 5,578 | 98 | 5 | n/a | n/a |
| none | none | P4(3)2(1)2 | 35.14 - 2.15 | 42,366 / 6,302 | 90 | 5 | n/a | n/a |
Rsym = ∑ |I − Iavg| ÷ ∑I. I = intensity.
MFID (Mean Fractional Isomorphous Difference) = ∑ |Fph − Fp| ÷ ∑ Fp. Fph = structure factor from derivative data set, Fp = structure factor from adenosine data set.
Fh = calculated structure factor of heavy atom. E = residual lack of closure error44.
TMLA = trimethyllead acetate.
MA = mercuric acetate.
DMA = dimercuric acetate.
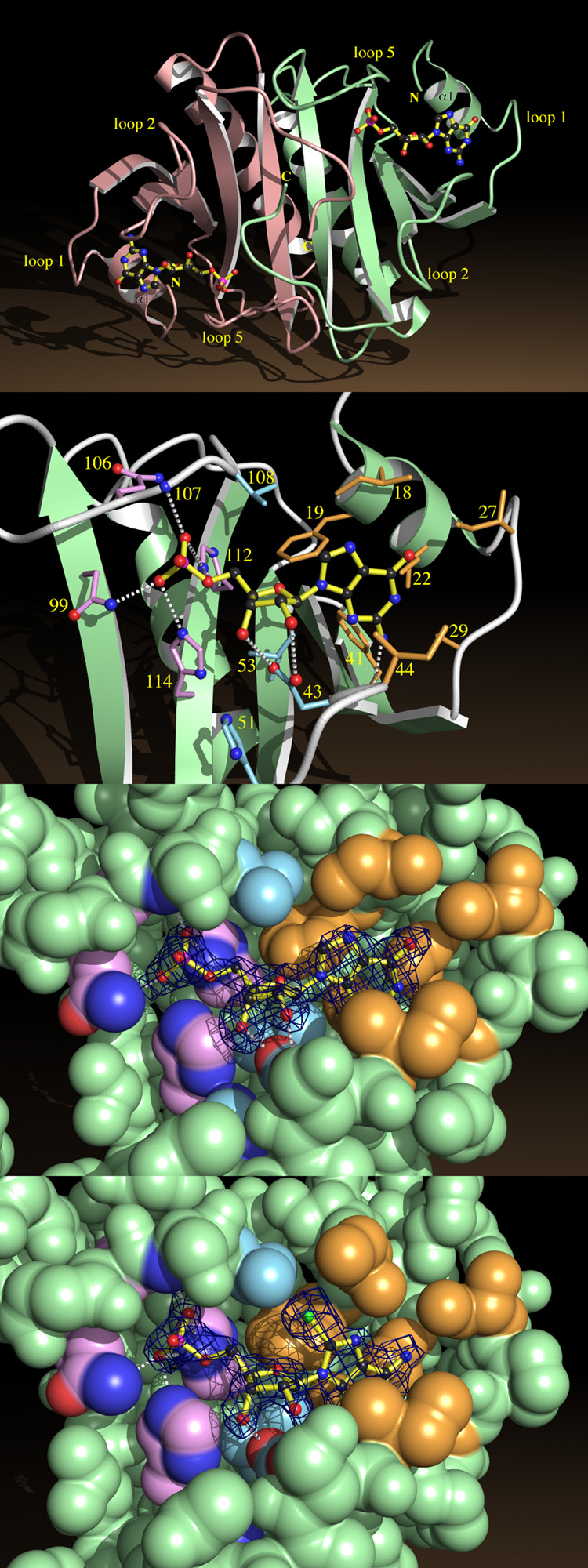
Fig. 4.
Nucleotide Binding by HINT.
a, HINT-GMP dimer shown from above the β-sheet, indicating helices α1 and loops 1, 2 and 5.
b, A close-up of GMP recognition by HINT including nonhydrogen atoms of GMP and side chains from fifteen conserved residues. Side chains surrounding the guanine base are beige, those surrounding ribose are blue, and those surrounding phosphate are purple. Potential hydrogen bonds between GMP and protein are indicated.
c, Coordinates corresponding to GMP in the HINT-GMP structure were removed and the protein model was subjected to simulated annealing at 3000 °C50. A difference electron density map was calculated with coefficients (Fobs - Fcalc) and contoured at 2.4 σ over the omitted GMP coordinates. Agreement between the map and model of GMP demonstrates that phases calculated from the protein model are robust and that GMP binds in this manner. The nucleotide-binding site is represented by its van der Waals surface, color coded as above.
d, A simulated annealing omit map prepared, as above, for HINT-8-Br-AMP.
Loops 1, 2 and 5 and helix α1 form three sides of a cradle that holds the nucleotide on top of the sheet (Fig. 4b). Helix α1 and loop 1 are at the head of the cradle where the base of the nucleotide binds. Loops 2 and 5 are on either side, making contacts with the base and sugar (loop 2) and the sugar and phosphate (loop 5). The cradle is open on top and at its foot where the phosphate-end of the nucleotide is bound. The nucleotide-binding cleft is not at the dimer interface: each monomer binds one nucleotide in an identical manner. Looking down from above (Fig. 4a), one sees two nucleotide cradles, foot to foot, with their 5′-phosphates facing each other. Though the nucleotide-binding cleft accommodates adenosine, the absence of the 5′-phosphate in the HINT-adenosine complex appears to result in a disordered base (see Methods).
The binding pocket for the base is composed of four isoleucines (18, 22, 27 and 44) and two phenylalanines (19 and 41). Buried in this nonpolar environment, a hydrogen bond (~3 Å) between the N2 amino group of guanine and the carbonyl oxygen of His 42 is likely to be strong. Ribose is recognized by a mixture of polar and nonpolar interactions conferred by side chains of Asp 43, His 51, Leu 53 and Val 108. The 5′-phosphate interacts with the side chains of Asn 99, Gln 106, His 112 and His 114 (Figs. 4b, c and d). The 14 residues that make direct contact with nucleotides include five of the six amino acids conserved in every member of the HIT superfamily (Fig. 1) and nine residues that are highly conserved in the HIT superfamily. Within the 102 amino acid core of HINT, apart from conserved Pro and Gly residues, 27 residues are identical in nine or more HIT proteins. The fact that a majority of the most highly conserved residues in the HIT superfamily make direct contacts with nucleotides in HINT provides strong evidence that the conserved function of HIT proteins must depend on binding nucleotides.
The remaining highly conserved residues fall into two classes. Ten are involved in monomer or dimer packing interactions. Some of these, notably His 110 and Leu 116, the first and last residues of the His-ϕ-His-ϕ-His-ϕ-ϕ motif, are buried in the next shell outside the nucleotide-binding site and stabilize it indirectly. Three amino acids, Lys 21, Glu 26 and Glu 34, are charged surface residues neither in contact with the nucleotide nor involved in stabilization of the protein. These residues are candidates for involvement in conserved protein-protein interactions.
Comparison of HINT structures
The nucleotide-binding pocket of HINT is preformed in the absence of a ligand. Minor side chain adjustments accommodate guanine or adenine bases when GMP or 8-Br-AMP are bound. There are main chain adjustments on the order of 1 Å in the nucleotide-free structure, involving Pro 28, Asp 35, Asp 36 and Ser 107. Rather than collapse the nucleotide-binding cradle in the absence of nucleotide, these adjustments slightly peel its sides open. The absence of large conformational changes upon ligand binding is reminiscent of FKBP12 binding to FK506 and to rapamycin. In that system NMR32–34 and crystallographic35 methods revealed that, apart from small differences in the positions and mobilities of loops, the protein conformation was unchanged by ligand binding. Nonetheless, ligand binding allows FKBP12 complexes to interact with target proteins because the ligands become parts of new protein-interaction surfaces36. Similarly, proteins may bind to HINT at its filled or unfilled nucleotide-binding sites. The adenosine-agarose resin used for purification of HINT linked agarose to the 6-amino group of adenosine via a spacer arm. This edge of the base protrudes into the solvent (Fig. 4d) such that the agarose bead would sit on top of the bound nucleotide. A protein could be bound to HINT in a similar manner, making hydrogen bonds to the N7 and N6 groups of adenine (or the N7 and O6 groups of guanine). This mode of binding would potentially explain the conservation of two of the three charged residues on the surface of HINT, Lys 21 and Glu 26, as these residues are on the same face of HINT as the nucleotide.
Conservation of nucleotide-binding between HINT and GalT
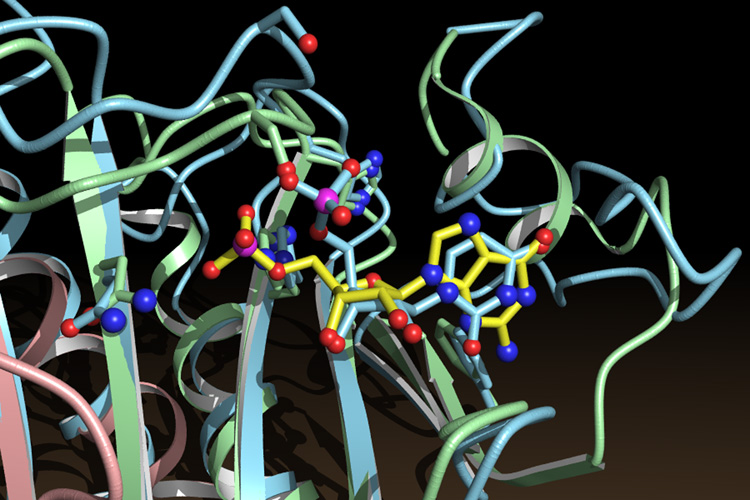
Overall structural similarity between GalT and HINT, though striking, did not by itself lead to the conclusion that GalT and HINT are related. However, superposition of the structures allowed us to examine the modes of nucleotide binding by the two proteins. Remarkably, when the structure of the GalT protein is superimposed on that of HINT, the UMP bound in GalT superimposes upon the nucleotide in the right-hand HINT monomer (Fig. 5). The UMP of GalT lies in a cradle above the β-sheet with the plane of its pyrimidine ring in the plane of the purine. Ribose atoms are superimposed within 0.3 to 0.9 Å. Upon overall protein superposition, the phosphate-binding signature sequence of GalT, including an Asn, a Ser, two Gly and two His residues, now sits atop the phosphate-binding sequence of HINT. Despite a lack of overall sequence similarity, six of 14 consecutive amino acids, from Asn 99 to His 112 of HINT, are identical to the colinear sequence of GalT and have nearly identical side chain conformations. Around the ribose and the uracil base, conservation is maintained via structural similarity rather than sequence identity. Specific polar interactions with ribose are found in GalT at positions that align with HINT. Most of the hydrophobic interactions with ribose and the base are also found at corresponding positions in GalT. The simplest conclusion is that GalT and HINT are related. We propose that ancestral GalT arose via tandem duplication of a HINT coding sequence, followed by divergence of sequences. Because it is apparent that FHIT is also a divergent HIT protein and, like GalT, is an enzyme that performs nucleotide chemistry, the genetics, enzymology and structural biology of GalT may contain information relevant to the activity and mechanism of FHIT.
Fig. 5.
Superposition of the nucleotide-binding sites of HINT and GalT. GMP (yellow bonds) belongs to the HINT protein (green and pink). UMP (blue bonds) belongs to the GalT protein (blue).
Limited sequence similarity between the diadenosine tetraphosphate phosphorylases, Apa1 and Apa2 of S. cerevisiae and the FHIT-related aph1 diadenosine tetraphosphate hydrolase of S. pombe has been noted by Barnes and coworkers16. Unlike hydrolases which use water to cleave P-O bonds, phosphorylases are mechanistically distinct diadenosine polyphosphate cleaving enzymes that transfer inorganic phosphate to AMP to generate ADP and ATP from AppppA via a nucleotidylated enzyme intermediate37, 38. The GalT reaction proceeds through a covalent UMP complex with His 16639. Diadenosine tetraphosphate phosphorylases do not have all of the features of HIT proteins but, like, GalT have a His-X-His-X-Gln-ϕ-ϕ motif in place of the His-ϕ-His-ϕ-His-ϕ-ϕ of the histidine triad. In view of the similar phosphoryl group transfer chemistry performed by GalT and diadenosine tetraphosphate phosphorylases, it seems plausible that the Apa1 and Apa2 phosphorylases may be related to HINT and FHIT as well.
The histidine triad
The histidine triad bound zinc ion in vitro and this motif was interpreted as constituting a physiological zinc-binding site2. We show that the HIT motif is a portion of a nucleotide-binding site. Two of the heavy atom compounds used in the structure determination, namely trimethyllead acetate and dimercuric acetate, bound in this position (see Methods). Metal binding at this site (~3 Å from Asp 43 and 5 to 6 Å from His 51, Ser 107, His 112 and His 114) would be competitive with binding the ribose portion of a nucleoside or nucleotide. We find no evidence for metal ion binding to HINT either in solution or in crystal structures. In contrast, Lima and coworkers reported structures called zinc-PKCI-1, apo-PKCI-1 and apo-PKCI-1 + 0.1 mM zinc. Although no zinc density was found in the “zinc” or “apo + zinc” structures and no differences found between proteins in these structures, an alternative conformation for zinc-PKCI-1 was proposed5. The HIT proteins that are dinucleoside polyphosphate hydrolases require millimolar Mn2+, Mg2+ or Ca2+ for optimum activity and are inhibited by Ni2+, Zn2+ and Cd2+ 16, 17. Metal ions the size of Ni2+ or greater may inhibit FHIT-related hydrolases by binding to the histidine triad in a manner that excludes a catalytically competent complex with nucleotide.
A model of FHIT and implications for catalysis
Identification of two purine mononucleotide binding sites in HINT requires interpretation in light of the identification of FHIT17 and aph116 as HINT-related diadenosine polyphosphate hydrolases. Sequence conservation of these enzymes with HINT at HINT’s dimer interface and the mobility of aph1 on gel filtration40 are consistent with proteins of the FHIT branch functioning as dimers. However, though the α-phosphates of HINT’s bound mononucleotides face each other, the distance between the phosphorous atoms is ~22 Å. If a FHIT dimer uses the two mononucleotide binding sites identified in HINT to bind a diadenosine polyphosphate substrate, it would be expected to exhibit a substantially more closed conformation than a HINT dimer. FHIT-product complexes, containing two mononucleotides, may bear a closer resemblance to HINT-nucleotide complexes.
Asymmetrical diadenosine polyphosphate hydrolases such as FHIT are AMP-yielding enzymes that produce non-identical products17. In the crystal structures of HINT-GMP and HINT-8-Br-AMP, we looked for interactions with the α-phosphate that would be present or possibly strengthened in a FHIT-diadenosine polyphosphate complex. Donation of hydrogen bonds by Asn 99, His 112 and His 114 to the α-phosphate in HINT structures suggests that homologous interactions conferred by Gln 83, His 96 and His 98 in FHIT could stabilize the negative charge on the α-phosphate of a dinucleoside substrate. This interpretation is consistent with mutagenesis of FHIT that showed that substitutions of histidines 96 and 98 but not 35 and 94 dramatically reduced catalytic activity17. Modeling indicates that the residue corresponding to Gln 106 of HINT (Gln 90 of FHIT) could be positioned to interact with the α-β bridging oxygen of a substrate.
Because FHIT is inactivated in many tumors15, 19–27, it is interesting to consider a role for FHIT and diadenosine polyphosphates in cell growth regulation. FHIT substrates, ApppA and AppppA, have long been considered alarmones in bacterial systems which are produced in times of cellular stress41. In mammalian cells, evidence has been presented that ApppA is produced in response to interferon administration42 and AppppA is produced coincident with contact-inhibition of growth43. By analogy to Ras, FHIT-substrate complexes may signal to the cell that these compounds have been made and hydrolysis of substrates may terminate signaling. In contrast to Ras, in which substrate complexes usually signal for mitogenesis, FHIT substrate complexes would be expected to signal for cell-cycle arrest. Recent work demonstrated that FHIT cleaves ApppA with a 1000-fold higher kcat than it cleaves AppppA17. If FHIT signaling depends on binding substrates rather than turning them over, complexes with slowly hydrolyzed substrates could sustain signaling more potently than complexes with good substrates. Continued genetic, biochemical and crystallographic investigations should clarify these issues.
Methods
Crystallization and data collection
Cloning and overexpression of HINT will be described (J.G., N. Liang & J.M.L., in prep.). In the course of purification, recombinant HINT protein was eluted from adenosine-agarose with 0.2 mM adenosine, GMP or 8-Br-AMP in 150 mM NaCl, 20 mM Tris Cl, pH 7.4. Nucleoside-free protein was prepared by dialysis against the same buffer. Proteins were concentrated to 15 mg ml−1 and crystallized by vapor diffusion against 30% polyethylene glycol 8000, 0.1 M sodium acetate, 0.1 M sodium cacodylate, pH 6.5. Trigonal crystals belong to space group P3121 and tetragonal crystals to space group P43212. Diffraction data were collected at 4 °C on Rigaku R-AXIS II image plates using 0.3 mm collimated monochromatized Cu Kα radiation from a Rigaku RU200 rotating anode generator (50 kV, 150 mA). Data were processed with XDS44. Unit cell dimensions refined to a = b = 50.85 Å, c = 81.83 Å for trigonal crystals and a = b = 40.34 Å, c = 143.03 Å for tetragonal nucleotide-free HINT crystals and a = b = 40.32 Å, c = 143.08 Å for tetragonal HINT-8-Br-AMP crystals (Table 1).
Structure determination
Crystals of the HINT-adenosine and HINT-GMP complexes are trigonal and isomorphous. Crystals of HINT-8-Br-AMP and nucleotide-free HINT are tetragonal and nearly isomorphous with each other. The HINT protein structure was solved by multiple isomorphous replacement (MIR) using the HINT-adenosine complex as the parent and three heavy atom derivatives. HINT-adenosine crystals were soaked in 0.1 mM mercuric acetate or dimercuric acetate for 1 hour or were grown in the presence of a grain of trimethyllead acetate. A single lead site per asymmetric unit was located by SHELX45 using data to 4 Å resolution. The heavy atom position and occupancy were refined using the PROTSYS package (G.A.P., unpublished). Single isomorphous replacement phases were calculated from these parameters. The isomorphous difference Patterson map for the mercuric acetate derivative indicated two sites which were located with an isomorphous difference Fourier map calculated with lead phases and (mercuric acid - parent) amplitudes. Phases calculated using both the mercuric acetate and trimethyllead acetate derivatives were used to locate a single dimercuric acetate site near the lead site. Heavy atom positions and MIR phases were refined iteratively using data to 3 Å resolution with the program TENEYCK46 (Table 1). The solvent-flattened47 electron density map showed a clearly defined protein boundary and the central β-sheet. A polyalanine chain consisting of four β-strands was built into this map using the molecular graphics program O48. Improvement in the map was obtained by combining MIR phases with those calculated from the partial atomic model using the program COMBINE49. The amino acid sequence of HINT was built into the COMBINE map and the model was refined using positional and B-factor refinement and simulated annealing with the program X-PLOR50 (Table 2). The lead and dimercuric acetate ions were coordinated to Asp 43 and His 114. Mercuric acetate had labeled the sulfur atoms of Cys 84 and Cys 38. The latter mercurial was additionally coordinated to Asp 35 and His 76. After 110 amino acids of the HINT model were built and refined, nonprotein electron density was apparent in difference electron density maps calculated with coefficients (2Fobs - Fcalc) and (Fobs - Fcalc) with data from the HINT-adenosine and HINT-GMP complexes. Whereas the density in the HINT-adenosine maps was less extensive than one would expect for an entire adenosine molecule, the guanine and the ribose-5′-phosphate were immediately apparent in HINT-GMP maps. The protein model of a HINT monomer from the trigonal HINT-adenosine structure was used to solve the structure of nucleoside-free HINT by the method of molecular replacement using the program AMoRe51. Using data to 4 Å resolution, a rotation and translation solution was found for space group P43212 (R factor = 28.9%) but not for its enantiomorph P41212 or for the 6 other primitive space groups in the 422 point group. The resulting model was rebuilt and refined. Difference electron density maps, examined for evidence of nucleoside or metal ion binding, only showed peaks of electron density characteristic of water molecules. On the other hand, similar maps for the HINT-8-Br-AMP complex revealed, in addition to electron densities for water molecules, electron density corresponding to 8-Br-AMP. Superposition of the HINT-GMP model on the 8-Br-AMP electron density indicated that the two nucleotides are bound in the same manner, both in the anti conformation. The 8-Br-AMP electron density lacked density corresponding to the 2-amino group of guanine and contained density at high contour corresponding to bromine at the 8 position. Each model was individually rebuilt with addition of no more than 70 waters, and refined, excluding no data from stated resolution limits (Table 2). Eight percent of the data were reserved for free R-factor analysis: a cycle of refinement was rejected if it did not reduce the free R-factor52. The adenine ring of adenosine was disordered, thus the nucleoside was modeled only as ribose. No electron density was observed for residues 1 to 11 of the 126 amino acid predicted sequence of HINT. Because of side chain disorder, Arg 12 was modeled as Ala. Refinement and model statistics are provided in Table 2. Coordinates will be deposited in the Protein Data Bank and may be obtained by ftp (asterix.jci.tju.edu/pub/HINT).
Table 2.
Refinement Statistics
| RMS Deviation from Ideality | ||||||
|---|---|---|---|---|---|---|
| Model | Waters | R factora | Free R factorb | Resolution (Å) | Bond Lengths (Å) | Bond Angles (°) |
| HINT-adenosinec | 70 | 0.200 | 0.264 | 10 - 1.90 | 0.010 | 1.74 |
| HINT-GMP | 66 | 0.194 | 0.254 | 10 - 2.10 | 0.011 | 1.72 |
| HINT-8-Br-AMP | 51 | 0.163 | 0.265 | 10 - 2.31 | 0.011 | 1.71 |
| HINT | 51 | 0.176 | 0.263 | 10 - 2.15 | 0.010 | 1.62 |
R factor = ∑ |Fobs - Fcalc| ÷ ∑ Fobs.
Free R factor = R factor of test set reflections not used in refinement50.
Adenosine modeled as ribose.
Molecular graphics
Fig. 1 was made with Alscript53. Fig. 3a, 3d, Fig. 4 and Fig. 5 were made with Molscript54, Rayscript (http://www.rose.brandeis.edu/users/peisach/rayscript/) and Rayshade (http://www-graphics.stanford.edu/~cek/rayshade/rayshade.html).
ACKNOWLEDGMENTS
We thank David Harrison, Geoff Stamper and Robert F. Williams for technical assistance and Kay Huebner, Carlo Croce, Mike Blackburn, Deborah Andrew, Perry Frey and Larry Barnes for helpful discussions. Work in the laboratories of J.M.L., D.R. and G.A.P. was supported by grants from the National Institutes of Health and, in part, by a grant from the Lucille P. Markey Charitable Trust. C.B. was a Fellow of the Leukemia Society of America.
References
- 1.Seraphin B. The HIT protein family: a new family of proteins present in prokaryotes yeast and mammals. DNA Sequence. 1992;3:177–179. doi: 10.3109/10425179209034013. [DOI] [PubMed] [Google Scholar]
- 2.Mozier NM, Walsh MP, Pearson JD. Characterization of a novel zinc binding site of protein kinase C inhbitor-1. FEBS Lett. 1991;279:14–18. doi: 10.1016/0014-5793(91)80238-x. [DOI] [PubMed] [Google Scholar]
- 3.McDonald JR, Walsh MP. Ca2+-binding proteins from bovine brain including a potent inhibitor of protein kinase C. Biochem. J. 1985;232:559–567. doi: 10.1042/bj2320559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brzoska PM, et al. The product of the ataxia-telangiectasia group d complementing gene, atdc interacts with a protein kinase c substrate and inhibitor. Proc. Natl. Acad. Sci. USA. 1995;92:7824–7828. doi: 10.1073/pnas.92.17.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima C, Klein MG, Weinstein IB, Hendrickson WA. Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins. Proc. Natl. Acad. Sci. USA. 1996;93:5357–5362. doi: 10.1073/pnas.93.11.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brzoska PM, et al. Cloning, mapping, and in vivo localization of a human member of the PKCI-1 protein family (PRKCNH1) Genomics. 1996;36:151–156. doi: 10.1006/geno.1996.0435. [DOI] [PubMed] [Google Scholar]
- 7.Simpson GG, Clark G, Brown JW. Isolation of a maize cDNA encoding a protein with extensive similarity to an inhibitor of protein kinase C and a cyanobacterial open reading frame. Biochim. Biophys. Acta. 1994;1222:306–308. doi: 10.1016/0167-4889(94)90183-x. [DOI] [PubMed] [Google Scholar]
- 8.Wilson R, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 9.Frohlich KU, et al. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 1991;114:443–453. doi: 10.1083/jcb.114.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustos SA, Schaefer MR, Golden SS. Different and rapid responses of four cyanobacterial psbA transcripts to changes in light intensity. J. Bacteriol. 1990;172:1998–2004. doi: 10.1128/jb.172.4.1998-2004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Fani R, et al. Cloning of histidine genes of Azospirillum brasilense: organization of the ABFH gene cluster and nucleotide sequence of the hisB gene. Mol. Gen. Genet. 1989;216:224–229. doi: 10.1007/BF00334360. [DOI] [PubMed] [Google Scholar]
- 13.Bult CJ, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 14.Fraser CM, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 15.Ohta M, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Garrison PN, Barnes LD. Cloning of the Schizosaccharomyces pombe gene encoding diadenosine 5´,5-P1,P4-tetraphosphate (Ap4A) asymmetrical hydrolase: sequence similarity with the histidine triad (HIT) protein family. Biochem. J. 1995;312:925–932. doi: 10.1042/bj3120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes LD, et al. FHIT, a putative tumor suppressor in humans, is a dinucleoside 5',5'"-P-1,P-3-triphosphate hydrolase. Biochemistry. 1996;35:11529–11535. doi: 10.1021/bi961415t. [DOI] [PubMed] [Google Scholar]
- 18.McLennan AG. Ap4A and Other Dinucleoside Polyphosphatases. Boca Raton, Florida: CRC Press; 1992. [Google Scholar]
- 19.Sozzi G, et al. The FHIT gene at 3p14.2 is abnormal in lung cancer. Cell. 1996;85:17–26. doi: 10.1016/s0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- 20.Sozzi G, et al. Aberrant FHIT transcripts in Merkel cell carcinoma. Cancer Research. 1996;56:2472–2474. [PubMed] [Google Scholar]
- 21.Negrini M, et al. The FHIT gene at 3p14.2 is abnormal in breast carcinomas. Cancer Research. 1996;56:3173–3179. [PubMed] [Google Scholar]
- 22.Virgilio L, et al. FHIT gene alterations in head and neck squamous cell carcinomas. Proc. Natl. Acad. Sci. USA. 1996;93:9770–9775. doi: 10.1073/pnas.93.18.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shridhar R, et al. Frequent breakpoints in the 3p14.2 fragile site, Fra3b, in pancreatic tumors. Cancer Research. 1996;56:4347–4350. [PubMed] [Google Scholar]
- 24.Panagopoulos I, et al. The FHIT and PTPRG genes are deleted in benign proliferative breast disease associated with familial breast cancer and cytogenetic rearrangements of chromosome band 3p14. Cancer Research. 1996;56:4871–4875. [PubMed] [Google Scholar]
- 25.Mao L, Fan YH, Lotan R, Hong WK. Frequent abnormalities of fhit, a candidate tumor suppressor gene, in head and neck cancer cell lines. Cancer Research. 1996;56:5128–5131. [PubMed] [Google Scholar]
- 26.Man S, Ellis IO, Sibbering M, Blamey RW, Brook JD. High levels of allele loss at the FHIT and ATM genes in non-comedo ductal carcinoma in situ and grade I tubular invasive breast cancers. Cancer Research. 1996;56:5484–5489. [PubMed] [Google Scholar]
- 27.Yanagisawa K, et al. Molecular analysis of the FHIT gene at 3p14.2 in lung cancer cell lines. Cancer Research. 1996;56:5579–5582. [PubMed] [Google Scholar]
- 28.Geurts JMW, Schoenmakers EFPM, Roijer E, Stenman G, Van de Ven WJM. Expression of reciprocal hybrid transcripts of HMGIC and FHIT in a pleomorphic adenoma of the parotid gland. Cancer Research. 1997;57:13–17. [PubMed] [Google Scholar]
- 29.Wedekind JE, Frey PA, Rayment I. Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 Å resolution. Biochemistry. 1995;34:11049–11061. doi: 10.1021/bi00035a010. [DOI] [PubMed] [Google Scholar]
- 30.Levy HL, Hammersen G. Newborn screening for galactosemia and other galactose metabolic defects. J. Pediatr. 1978;92:871–877. doi: 10.1016/s0022-3476(78)80351-5. [DOI] [PubMed] [Google Scholar]
- 31.Branden C, Tooze J. Introduction to Protein Structure. New York: Garland Publishing; 1991. [Google Scholar]
- 32.Rosen MK, Michnick SW, Karplus M, Schreiber SL. Proton and nitrogen sequential assignments and secondary structure determination of the human FK506 and rapamycin binding protein. Biochemistry. 1991;30:4774–4789. doi: 10.1021/bi00233a020. [DOI] [PubMed] [Google Scholar]
- 33.Michnick SW, Rosen MK, Wandless TJ, Karplus M, Schreiber SL. Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin. Science. 1991;252:836–839. doi: 10.1126/science.1709301. [DOI] [PubMed] [Google Scholar]
- 34.Moore JM, Peattie DA, Fitzgibbon MJ, Thomson JA. Solution structure of the major binding protein for the immunosuppressant FK506. Nature. 1991;351:248–250. doi: 10.1038/351248a0. [DOI] [PubMed] [Google Scholar]
- 35.Wilson KP, et al. Comparitive X-ray Structures of the Major Binding Protein for the Immunosuppressant FK506 (Tacrolimus) in Unliganded Form and in Complex with FK506 and Rapamycin. Acta Crystallogr. D. 1995;51:511–521. doi: 10.1107/S0907444994014514. [DOI] [PubMed] [Google Scholar]
- 36.Clardy J. The chemistry of signal transduction. Proc. Natl. Acad. Sci. USA. 1995;92:56–61. doi: 10.1073/pnas.92.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guranowski A, Sillero A. Enzymes cleaving dinucleoside polyphosphates. In: McLennan AG, editor. Ap4A and Other Dinucleoside Polyphosphates. Boca Raton, FL: CRC Press; 1992. pp. 81–133. [Google Scholar]
- 38.Booth JW, Guidotti G. An alleged yeast polyphosphate kinase is actually diadenosine-5', 5"'-P1,P4-tetraphosphate alpha, beta-phosphorylase. J. Biol. Chem. 1995;270:19377–19382. doi: 10.1074/jbc.270.33.19377. [DOI] [PubMed] [Google Scholar]
- 39.Wedekind JE, Frey PA, Rayment I. The structure of nucleotidylated Histidine-166 of Galactose-1-Phosphate Uridylyltransferase provides insight into phosphoryl group transfer. Biochemistry. 1996;35:11560–11569. doi: 10.1021/bi9612677. [DOI] [PubMed] [Google Scholar]
- 40.Robinson AK, de la Pena CE, Barnes LD. Isolation and characterization of diadenosine tetraphosphate (Ap4A) hydrolase from Schizosaccharomyces pombe. Biochemica et Biophysica Acta. 1993;1161:139–148. doi: 10.1016/0167-4838(93)90207-8. [DOI] [PubMed] [Google Scholar]
- 41.Kitzler JW, Farr SB, Ames BN. Intracellular functions of ApnN: prokaryotes. In: McLennan AG, editor. Ap4A and Other Dinucleoside Polyphosphates. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- 42.Vartanian A, Narovlyansky A, Amchenkova A, Turpaev K, Kisselev L. Interferons induce accumulation of diadenosine triphosphate (Ap3A) in human cultured cells. FEBS Lett. 1996;381:32–34. doi: 10.1016/0014-5793(96)00073-7. [DOI] [PubMed] [Google Scholar]
- 43.Segal E, Le Pecq JB. Relationship between cellular diadenosine 5',5'"-P1,P4-tetraphosphate level, cell density, cell growth stimulation and toxic stresses. Exp. Cell Res. 1986;167:119–126. doi: 10.1016/0014-4827(86)90209-0. [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 45.Sheldrick GM. SHELXS86, a Program for the Solution of Crystal Structures. Gottingen, FRG: Univ. of Gottingen; 1985. [Google Scholar]
- 46.Ten Eyck LF, Arnone A. Three-dimensional Fourier Synthesis of Human Deoxyhemoglobin at 2.5 Angstrom Resolution. J. Mol. Biol. 1976;100:3–11. doi: 10.1016/s0022-2836(76)80029-0. [DOI] [PubMed] [Google Scholar]
- 47.Wang BC. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- 48.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 49.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNAse I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 50.Brunger AT. X-PLOR Version 3.1 Manual. New Haven, Connecticut: Yale University; 1993. [Google Scholar]
- 51.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. D. 1994;50:157–163. [Google Scholar]
- 52.Brunger AT. The free R value: a more objective statistic for crystallography. Methods in Enzymol. 1997;277 doi: 10.1016/s0076-6879(97)77021-6. in press. [DOI] [PubMed] [Google Scholar]
- 53.Barton GJ. ALSCRIPT, a tool to format multiple sequence alignments. Protein Engineering. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 54.Kraulis PJ. MolScript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]