Abstract
The cellular aging-associated transcriptional repressor that we previously named as Orpheus was identical to Oct-1, a member of the POU domain family. Oct-1 represses the collagenase gene, one of the cellular aging-associated genes, by interacting with an AT-rich cis-element in the upstream of the gene in preimmortalized cells at earlier population-doubling levels and in immortalized cells. In these stages of cells, considerable fractions of the Oct-1 protein were prominently localized in the nuclear periphery and colocalized with lamin B. During the cellular aging process, however, this subspecies of Oct-1 disappeared from the nuclear periphery. The cells lacking the nuclear peripheral Oct-1 protein exhibited strong collagenase expression and carried typical senescent morphologies. Concomitantly, the binding activity and the amount of nuclear Oct-1 protein were reduced in the aging process and resumed after immortalization. However, the whole cellular amounts of Oct-1 protein were not significantly changed during either process. Thus, the cellular aging-associated genes including the collagenase gene seemed to be derepressed by the dissociation of Oct-1 protein from the nuclear peripheral structure. Oct-1 may form a transcriptional repressive apparatus by anchoring nuclear matrix attachment regions onto the nuclear lamina in the nuclear periphery.
INTRODUCTION
Aging is considered to be one of the phenomena regulated by the network of gene expression (Strehler et al., 1971; Cutler, 1991; Jazwinski, 1996; Smith and Pereira-Smith, 1996). Since Hayflick and Moorhead’s discovery that fibroblasts have a limited proliferative potential in vitro (Hayflick and Moorhead, 1961), the accumulated findings suggest that the phenomenon of cellular or replicative senescence is an intrinsic mechanism of diploid cells (see review, Stanulis-Praeger, 1987; Goldstein, 1990). In other words, the mechanism of cellular aging is genetically controlled as one of the important aspects of organismic aging. From this implication, the growth properties of cultured human diploid cells and numbers of associated cellular events have been analyzed extensively, and several hypotheses have been proposed for the genetic program of aging (Strehler et al., 1971; Zs.-Nagy et al., 1988; Cutler, 1991; Harley et al., 1992; Guarente, 1996). However, neither the pathway of transcriptional regulation in aging nor the transcriptional network controlling the cellular aging is understood so far. To address this issue, it is important to investigate the regulation of cellular aging-associated genes. Levels of the basal expression of several extracellular matrix-associated genes are regulated in a cellular aging-dependent manner. In normal senescent fibroblasts, the expression of interstitial collagenase and stromelysin is prominently up-regulated, while the expression of TIMP-1 (tissue inhibitor of metalloproteinases-1) is down-regulated (Sottile et al., 1989; Millis et al., 1992; Burke et al., 1994). The expression of the fibronectin gene increases (Kumazaki et al., 1991), while that of the α1-collagen gene decreases during cellular senescence (Wistrom and Villeponteau, 1992). Other genes, such as the elongation factor Iα-related gene (Giordano and Foster, 1989), the senescence-associated gene (SAG) (Wistrom and Villeponteau, 1992), and the senescent cell-derived inhibitor 1 gene (sdi1) (Noda et al., 1994), were reported to be significantly induced at the final stage of cellular senescence. Thus, certain common regulatory pathways of transcription are suggested to be involved in the expression of these cellular aging-associated genes. Cellular aging can also be implied to be one of the processes of terminal differentiation or maturation (Bayreuther et al., 1988). By analogy with the myoblast differentiation by the MyoD family (Davis et al., 1987) and the adipocyte differentiation by the C/EBP family (Yeh et al., 1995), a hypothetical master regulator can induce the expression of the cellular aging-associated genes. On the other hand, in yeast, the involvement of transcriptional silencing was reported in the regulation of cellular aging (Kennedy et al., 1995; Smeal et al., 1996). Certain regulatory pathways of transcription may be evoked to induce the aging phenotypes by the release from the transcriptional silencing in aged yeast cells.
In cultured human fibroblasts, the interstitial collagenase gene is one of the best probes to search for the regulatory network of cellular aging-associated genes. Interstitial collagenase, a member of the metalloproteinase family in fibroblasts, plays an important role in the senile atrophy of extracellular matrix. Collagen is degraded in the extracellular matrix by this collagenase in the aged skin and connective tissues. The interstitial collagenase increases significantly in amount and activity in aged fibroblasts (West et al., 1989; Burke et al., 1994). The collagenase gene is dramatically induced in senescent diploid fibroblasts (Sottile et al., 1989; Millis et al., 1992) as well as in precrisis cells of SV40 large T antigen-transformed fibroblast clones (Imai and Takano, 1992). In fibroblasts from the patients with one of the hereditary premature aging syndromes, the Werner syndrome, the collagenase expression is prematurely induced to high levels concomitantly with the accelerated process of cellular aging (Bauer et al., 1986; Millis et al., 1992). In addition, the collagenase expression is almost completely abolished in the immortalized, T antigen-transformed human diploid fibroblasts (Imai and Takano, 1992). These findings suggest that the expression of the collagenase gene seems to be regulated in a cellular aging- and immortalization-dependent manner. We previously reported that the two transcription factors, Pluto and Orpheus, interact with the immortalization-susceptible cis-acting element 2 (ISE2) in the upstream of the collagenase gene and mediate the regulation of the collagenase gene (Imai et al., 1994). Orpheus represses the collagenase expression in preimmortalized cells at earlier population-doubling levels (PDLs). In precrisis cells, the binding of Orpheus is significantly reduced, and, in turn, Pluto becomes prominently bound to ISE2. Pluto is a precrisis cell-specific transcriptional activator for the collagenase gene. In immortalized cells, the Orpheus-mediated transcriptional repression is resumed, and the collagenase expression was completely abolished. Thus, the loss of Orpheus-mediated repression seems to be important in the regulation of the collagenase gene during the process of cellular aging.
In this study, we demonstrated that Orpheus was identical to a member of the POU domain family, Oct-1. Oct-1 interacted with the AT-rich sequence of ISE2 and mediated the repression of the collagenase gene in the cellular aging-associated manner. Consistent with the changes in the binding activity of Orpheus during cellular aging and immortalization, the Oct-1 protein was not detected in the nuclear extract of precrisis cells, but again detected in that of immortalized cells. However, the whole cellular amounts of Oct-1 protein did not change significantly during either process. From the results of immunostaining by anti-Oct-1 antibodies, the loss of Oct-1–mediated repression seemed to be accompanied by the release of Oct-1 protein from the nuclear peripheral region during the cellular aging process. We also found that the Oct-1 protein was densely colocalized with lamin B, one of the important components of the nuclear lamina, in the nuclear periphery. In addition, the cells with no nuclear peripheral localization of Oct-1 showed strong signals of the collagenase expression. These findings open the possibility that the Oct-1–associated structural assembly in the nuclear periphery mediates the cellular aging-associated repression of the collagenase expression. The dissociation of Oct-1 from the transcriptional repressive apparatus may induce the expression of certain cellular aging-associated genes. We discuss the function and regulation of the Oct-1–associated nuclear peripheral structure in cellular aging as an important constituent of the hypothetical molecular counting mechanism of replicative lifespan.
MATERIALS AND METHODS
Antibodies
Anti-Oct-1 monoclonal antibodies, YL15 and YL123, were kindly provided by Dr. Masafumi Tanaka of Cold Spring Harbor Laboratory. YL15 and YL123 recognize the POU-domain linker region and the POU homeodomain of Oct-1, respectively (Lai and Herr, 1992). Affinity-purified rabbit polyclonal antibodies against the C-terminal peptide 723–743 of Oct-1 and the C-terminal peptide 444–463 of Oct-2 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), respectively. Anti-lamin B monoclonal and anti-collagenase polyclonal antibodies were purchased from Calbiochem (San Diego, CA) and Quartett (Germany), respectively. An anti-p53 monoclonal antibody, pAb421, was a gift from Dr. Kaoru Segawa.
Cell Culture
A genetically matched pair of parental preimmortalized HuS-L12 and immortalized IML12–4 cell clones were previously established from SV40 T antigen-transformed human diploid fibroblast strain MRC-5 and cultured as described previously (Imai et al., 1993). The preimmortalized cell clone HuS-L12 entered the crisis period at PDL 91–92. HeLa-S3 cells were cultured in minimum essential medium (GIBCO BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (PAA Laboratories Inc.).
Preparation of Nuclear Extracts and Their Partial Purification
Nuclear extracts were prepared from HuS-L12 at various PDLs, IML12–4 and HeLa-S3 as described previously (Imai et al., 1994). For chemical footprint analysis, the nuclear extracts of IML12–4 and HeLa-S3 cells were partially purified with Heparin-Sepharose column (Pharmacia, Piscataway, NJ) and Dignam buffer D. The Orpheus/Oct-1 binding activity was collected by eluting with 0.4 M KCl and diluted to 0.1 M KCl with Dignam buffer D including 25 μg/ml bovine serum albumin (BSA) for further analyses.
Chemical Footprint Analysis
The HindIII-EcoRV fragments of pColSEdel2 or pColSEdel4 were used as the probes for chemical footprint analysis (Imai et al., 1994). Either HindIII or EcoRV site was phosphorylated with [γ-32P]ATP and T4 polynucleotide kinase (Takara Shuzo, Tokyo, Japan). The chemical footprinting was performed with ammonium iron(II) sulfate hexahydrate [Fe(II)] and methidiumpropyl EDTA (MPE, Sigma Chemical, St. Louis, MO) as described previously (Hertzberg and Dervan, 1982; Walker et al., 1994). Briefly, approximately 200–900 pg of radiolabeled DNA fragments and 15–45 U of the Orpheus-binding activity were used for the assay. One unit of the Orpheus-binding activity was defined by electrophoretic mobility shift assay (EMSA) as the specific binding activity that retarded 1% of the radiolabeled probe of the Orpheus-binding site. Binding reaction was carried out as described previously (Imai et al., 1994), after which hydroxyl radical cleavage was performed for 30–40 min after the addition of 5 μM Fe(II)·MPE, 0.0015% H2O2, and 2 mM dithiothreitol at the final concentration. The reaction was terminated by the addition of 20 μl of quenching solution (2 mM thiourea, 90 mM sodium acetate, and 75% ethanol). The product was purified by phenol-chloroform extraction, ethanol-precipitated, and separated with 8% denaturing sequencing gels. The dried sequencing gels were exposed to imaging plates and analyzed by a Bioimage Analyzer BAS2000 system (Fuji Photo Film, Tokyo, Japan) to determine the relative strength of contact between Orpheus and DNA.
EMSA
The binding activity of Orpheus or Oct-1 was detected as described previously, with 5 μg of nuclear extracts of preimmortalized and immortalized cells or with 1 μl of in vitro translated Oct-1, respectively (Imai et al., 1994). Double-stranded oligonucleotide probes of the wild or mutant types were synthesized with automated DNA synthesizers (Sci-Media, Tokyo, Japan) and labeled with [α-32P]dCTP and the Klenow fragment of DNA polymerase I (Takara Shuzo). The sense strand of the wild-type probe for Orpheus was tcgAGGAAATTGTAGTTAAATAATTAGAAAG, and the mutant type was tcgAGGAAGCATGCGTTAACCCGGGAGAAAG. For supershift experiments with anti-Oct-1 polyclonal and monoclonal antibodies, 1 μl of the antibody was added to the reaction mixture after the binding reaction and incubated on ice for 15 min. The dried gels were analyzed by the BAS2000 system and exposed to x ray film.
In Vitro Translation of Oct-1 Protein
The in vitro transcription of Oct-1 RNA was performed with the RNA transcription kit (Stratagene, La Jolla, CA) and pBSoct-1+ DNA (Lai and Herr, 1992) linearized by HindIII digestion. pBSoct-1+ was kindly provided by Dr. Masafumi Tanaka of Cold Spring Harbor Laboratory. The RNA transcripts of Oct-1 were translated in vitro by rabbit reticulocyte lysate in the In Vitro Express translation kit (Stratagene) with 35S-radiolabeled or nonradiolabeled methionine. The precise protocols of transcription and translation were provided by the manufacturer. The Oct-1 protein synthesized with radiolabeled methionine was detected at the molecular mass of 94 kDa by SDS-PAGE.
Site-directed Mutagenesis and Plasmid Construction
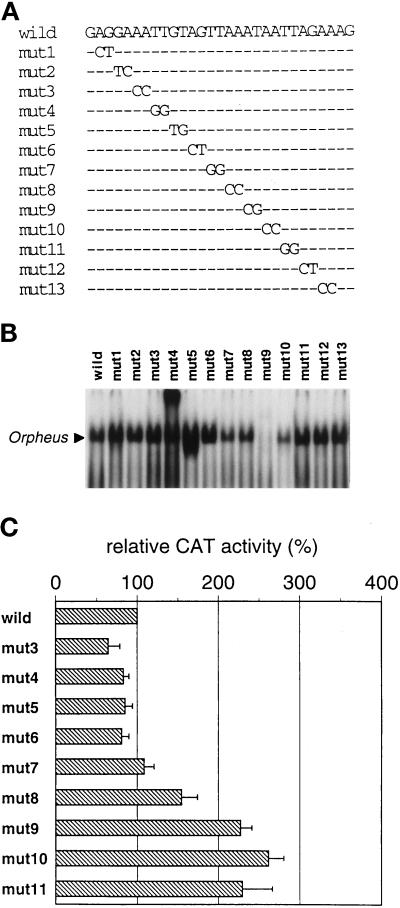
The site-directed mutagenesis was done by the Mutan-Express Km kit (Takara Shuzo) of the oligonucleotide-directed dual amber method (Hashimoto-Gotoh et al., 1995) to introduce sequential 2-base pair (bp) mutations into ISE2. The PCR-amplified HindIII-HindIII fragment carrying ISEs and the synthetic 29 mer oligonucleotide for each mutant (see Figure 2A) were used as the template and the primers for site-directed mutagenesis, respectively. The precise protocols were provided by the manufacturer. The resulting mutants were sequenced to confirm the mutations. The HindIII-EcoRV fragments carrying site-directed mutations were obtained by restriction enzyme digestion, and the corresponding fragment of pColSEdel2 was replaced with these mutant fragments. By using these site-directed mutants, chloramphenicol acetyltransferase (CAT) assay was performed as described previously (Imai et al., 1994).
Figure 2.
Sequential site-directed mutagenesis of ISE2 in EMSA and CAT assay. (A) The sequences of site-directed mutants used for EMSA and CAT assay are shown. Only mutated nucleotides are presented and other unchanged nucleotides are indicated as dashes. (B) EMSA for the Orpheus-binding activity with the site-directed mutant probes. The types of the probes are indicated at the top. In the lanes of mut4 and mut5, unidentified extra bands were detected by the introduction of the mutations. (C) Transcriptional activities of the collagenase upstream regions carrying the site-directed mutations shown in panel A. The CAT activity in IML12–4 cells transfected with the wild-type construct was assigned a value of 100%. Mean values and standard deviations of CAT activity were calculated from three independent transfection experiments.
Immunostaining
Preimmortalized HuS-L12 and immortalized IML12–4 cells were seeded on glass coverslips. The coverslip was rinsed with phosphate-buffered saline (PBS), and cells were fixed at −20°C for 15 min with a 1:1 mixture of acetone and ethanol. The fixed cells were incubated in a humidified plastic container at 37°C for 1 h with appropriately diluted primary antibodies. After washing 10 times in PBS, cells were incubated under a similar condition with affinity-purified fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) and/or Texas red-conjugated goat anti-rabbit IgG antibodies. The coverslip was rinsed 10 times in PBS and mounted on a slideglass. The images of immunostaining were analyzed by a confocal laser microscopy MRC-600 (Bio-Rad, Tokyo, Japan).
Western Blot Analysis
Thirty micrograms of whole cell extract and 10 μg of nuclear extract per lane from preimmortalized and immortalized cells were separated with 8% SDS-polyacrylamide gel. The electrophoresed proteins were transferred to nitrocellulose membrane (Schleicher & Schuell, Keene, NH). The membrane was washed in Tris-buffered saline (TBS) and immersed for 2 h in TBS with 4% nonfat milk and 0.5% Tween 20. After washing three times in TBS with 0.5% Tween 20, the membrane was incubated at 37°C in TBS with 0.5% Tween 20, 5% BSA, and 5 μl of the anti-Oct-1 monoclonal antibody, YL15. The membrane was washed three times and then incubated for 90 min at room temperature in TBS-BSA-Tween solution with 2 μCi of the 125I-labeled goat anti-mouse IgG antibody (ICN Pharmaceuticals, Cleveland, OH). The membrane was washed three times in TBS-Tween solution and once in TBS solution, and exposed to x ray film (Eastman Kodak, Rochester, NY) for 2–3 d.
RESULTS
Cellular Aging-associated Repressor Orpheus Recognizes the AT-rich Sequence of ISE2 for Its Binding and Repressive Function
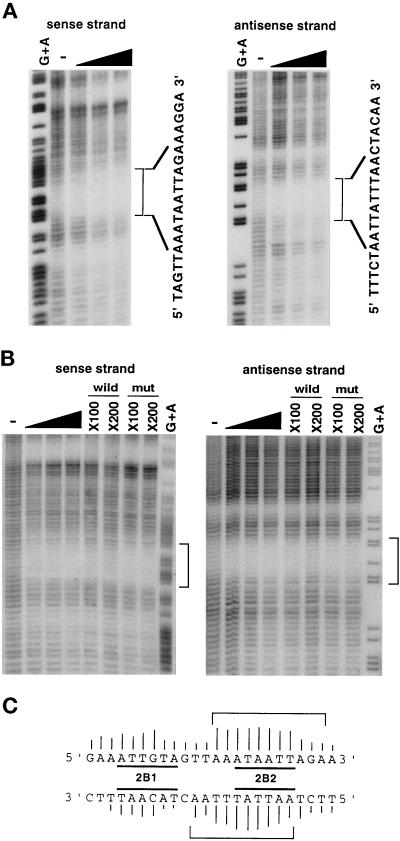
We first determined the Orpheus-binding site more precisely by chemical footprint analysis, using the partially purified nuclear extracts of IML12–4 and HeLa-S3 cells. The nuclear extracts of HeLa-S3 cells contained the binding activities of Orpheus indistinguishable from those of the original Orpheus in the extracts of IML12–4 cells in gel filtration (our unpublished results). The results from the experiments with the extracts of both cells indicated that Orpheus strongly recognized the sequence AAATAATT of ISE2 (Figure 1A and 1B). The protection of this sequence was inhibited by the addition of the wild-type nonradioactive probe, but not by that of the mutant-type probe. As schematically shown in Figure 1C, there were slight differences in the footprint patterns on the sense and the antisense strands at the recognition site. On the sense strand, Orpheus recognized mainly AAATAATT and weakly its 3′-sequence AGAA. On the antisense strand, the recognition sequence of Orpheus also extended more to the 3′-portion. These footprints completely covered up the 2B2 site whose six-base substitution enhanced the transcriptional activity of the collagenase upstream region preferentially in immortalized cells (Imai et al., 1994).
Figure 1.
Chemical footprint analysis for the Orpheus-binding site of ISE2. (A) The footprint patterns on the sense and antisense strands of ISE2 with the partially purified nuclear extract of IML12–4. Fifteen, 30, and 45 U of the Orpheus-binding activity were used in this assay, as indicated by black wedges. The specific footprints of Orpheus are indicated by square brackets with the nucleotide sequences of ISE2. Lanes of G+A and − represent the sequence ladders produced by GA-specific Maxam-Gilbert sequencing and hydroxyl radical cleavage reactions for the intact DNA fragments, respectively. (B) The similar footprint patterns were observed with the partially purified nuclear extract of HeLa-S3, as indicated by square brackets. The footprints of Orpheus were inhibited by the addition of indicated molar excess of the wild-type oligonucleotides, but not by that of mutant-type oligonucleotides. (C) The Orpheus contact sites on the sequence of ISE2 are shown by square brackets. The height of bars on each nucleotide shows the relative strength of the contact between protein and the nucleotide. The 2B1 and 2B2 sites are previously determined as the negative regulatory elements by substitutive mutagenesis (Imai et al., 1994).
Next, we determined nucleotide pairs essential for the binding of Orpheus by introducing sequential two-base substitutions to the recognition site in ISE2. By the substitution of the third and fourth bases AT to CG, the binding of Orpheus was almost completely abolished in EMSA (mut9 in Figure 2, A and B). By the substitution of the adjacent 3′-AA to CC (mut10), the binding of Orpheus was reduced approximately to 30% of the wild type. The substitutions of four bases located at 5′ of the essential AT slightly reduced the binding of Orpheus (mut7 and mut8). The substitution of 3′-TT to GG did not affect the binding of Orpheus (mut11), although Orpheus covered these bases in the footprint pattern. We examined the transcriptional activity of the collagenase upstream region that carried these site-directed mutations. The CAT activity of transfectants with each mutant of mut9, 10, and 11 was approximately 2.5-fold of that with the wild type (Figure 2C). The base substitutions in these three mutants completely overlapped the 2B2 site (see Figure 1C). Because the six-base substitution of the 2B2 site enhanced the CAT activity three- to fourfold in immortalized cells (Imai et al., 1994), these six bases were suggested to be important for the repression by Orpheus. The substitution in mut8 slightly enhanced the CAT activity, but that in mut7 showed no significant effect. These results suggested that the core ATAA is essential for the binding of Orpheus to its recognition sequence and also for its repressive function. Interestingly, the 3′-TT of the recognition site was important for the function but not for the binding of Orpheus.
Orpheus Is Identical to a Member of the POU Domain Family, Oct-1
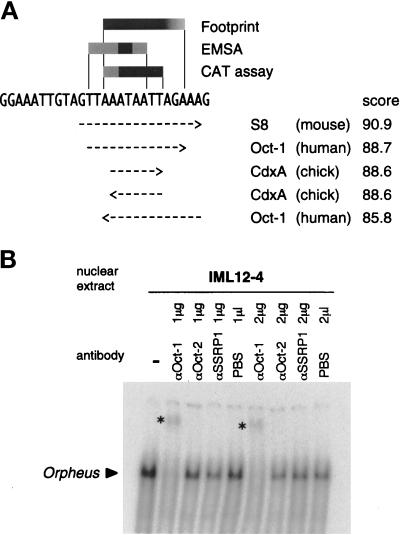
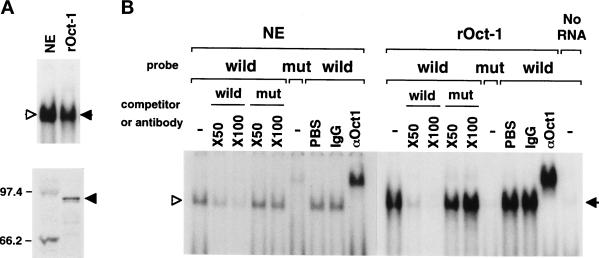
The summaries of chemical footprint analysis, EMSA, and CAT assays on the site-directed mutants are shown in Figure 3A. All these results consistently indicated that Orpheus represses the collagenase transcription by interacting with AAATAATT. By searching for sequences homologous to this sequence in the TRANSFAC database of transcription factor-binding sites (Figure 3A), several homeodomain proteins including Oct-1, a member of the POU domain family, were suggested to bind to this sequence. Consistently, the band of Orpheus in EMSA was specifically supershifted by anti-Oct-1 polyclonal and monoclonal antibodies (Figures 3B and 4B). Thus, Orpheus seemed to be Oct-1 or a protein closely related to Oct-1. The in vitro translated Oct-1 protein was able to bind to the Orpheus-binding sequence in EMSA (Figure 4A). The electrophoretic mobility, the binding specificity of the in vitro translated Oct-1, and the profiles of supershift by the anti-Oct-1 antibody were indistinguishable from those of Orpheus in the extract of immortalized T antigen-transformed fibroblasts (Figure 4, A and B). From these results, we concluded that the transcription factor we named as Orpheus was identical to Oct-1.
Figure 3.
Identification of Orpheus as a member of the POU domain family, Oct-1. (A) The search results of the Orpheus-binding sequence in the TRANSFAC database of transcription factor-binding sites. The summaries of chemical footprint analysis, EMSA, and CAT assay are schematically shown on the nucleotide sequence of ISE2. The arrows indicating rightward or leftward represent the binding sequences of transcription factors matching to the sense or antisense strand of ISE2, respectively. The matching scores were calculated from the nucleotide matrices of transcription factor-binding sites in the database. (B) Supershift experiment for the Orpheus-binding activity with the nuclear extract of IML12–4 cells and antibodies indicated on top. The anti-SSRP1 polyclonal antibody specifically recognizes the HMG-box protein SSRP1. The bands of Orpheus supershifted by an anti-Oct-1 polyclonal antibody are indicated by asterisks.
Figure 4.
The binding activity and specificity of Orpheus (NE, open triangle) and the in vitro translated Oct-1 (rOct-1, closed triangle). (A) The in vitro translated Oct-1 protein shows the binding activity indistinguishable from that of Orpheus in the nucelar extract of IML12–4 cells (top panel). The Oct-1 protein synthesized in vitro with radiolabeled methionine was detected at 94 kDa by SDS-PAGE (bottom panel). Molecular markers of 97.4 and 66.2 kDa are indicated at left. (B) The competition and supershift experiments were performed with the indicated molar excess of wild and mutant oligonucleotides and the anti-Oct-1 YL15 monoclonal antibody, respectively. Very faint binding of Oct-1 was detected in the rabbit reticulocyte lysate with no exogenous RNA.
The Oct-1 Protein Disappears from the Nuclear Periphery during Cellular Aging and Is Resumed in Immortalization
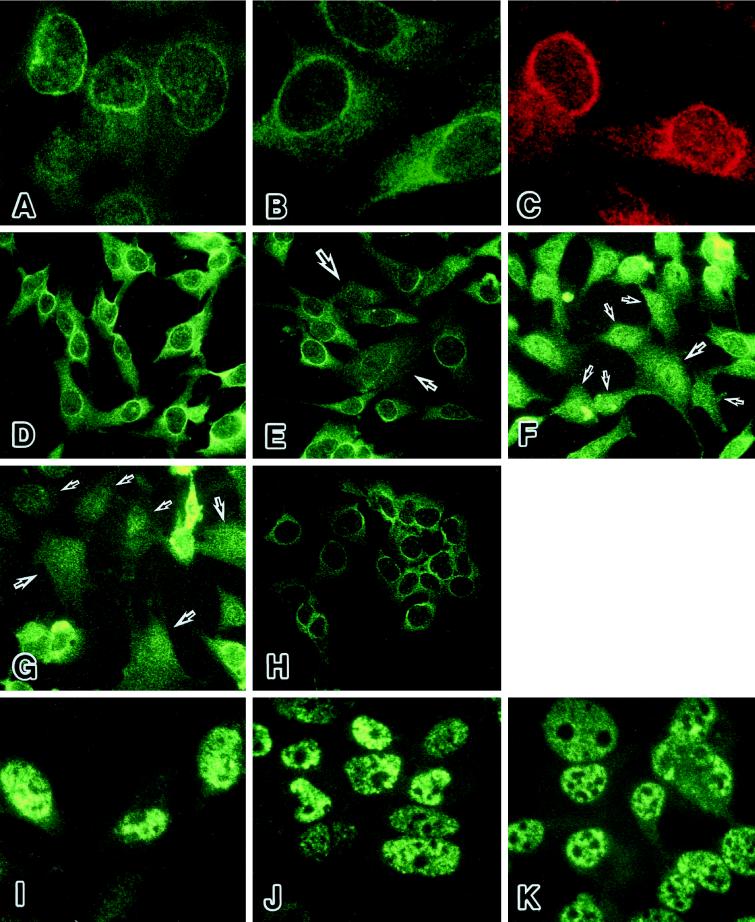
Oct-1 is reported to be located in the compartment of nuclear matrix and the soluble nuclear compartment (van Wijnen et al., 1993; Kim et al., 1996). We pursued a possibility that the release from the Oct-1-mediated repression accompanies certain changes in the localization of Oct-1 protein during the cellular aging process. We immunostained preimmortalized HuS-L12 cells at an earlier PDL by the anti-Oct-1 monoclonal antibody, YL15, which recognizes the unique POU-domain linker region of Oct-1. The nuclear periphery was strongly stained by this antibody (Figure 5A). The intranuclear compartment was also stained in fuzzy speckled patterns, and the cytoplasmic region was stained diffusely. A small number of cells with strong signals in the cytoplasmic region were observed in the population. Two other anti-Oct-1 antibodies, a monoclonal antibody YL123, which recognizes the POU homeodomain of Oct-1, and a polyclonal antibody against the C-terminal amino acids of Oct-1, showed quite similar profiles of staining (Figure 5, B and C). In preimmortalized cells at PDL 66, the collagenase expression was undetectable, and almost all the cells were prominently stained in the nuclear periphery by the anti-Oct-1 YL15 antibody (Figure 5D). However, a small number of cells with punctuated or no staining in the nuclear periphery were detected in the population of cells at PDL 73, although the majority of cells showed the ring-formed staining in the nuclear periphery (Figure 5E). This type of cells, with punctuated or no staining in the nuclear periphery, showed enlarged cell shape characteristic of the senescent cells. At PDL 84, a high level of the collagenase expression was detectable, and many more clusters of cells exhibited no staining in the nuclear periphery (Figure 5F). At PDL 91, most of the cells showed the senescent phenotype and exhibited no staining in the nuclear periphery (Figure 5G). Similar results were obtained by immunostaining with the monoclonal antibody YL123 and the polyclonal antibody against the C terminus of Oct-1 (our unpublished results). These changes in the subnuclear localization of Oct-1 were not due to certain degenerative alteration of nuclei during the cellular aging process, because the nuclear distribution of p53 protein was unchanged in HuS-L12 cells at earlier and later PDLs (Figure 5, I and J). In immortalized cells, both Oct-1–mediated collagenase repression and strong Oct-1 staining in the nuclear periphery were resumed to similar levels as in the preimmortalized cells at PDL 66 (Figure 5H). The results obtained by the three independent anti-Oct-1 antibodies were similar in the immortalized cells (our unpublished results). The staining profile of p53 was unchanged in immortalized cells, compared with those of preimmortalized cells (Figure 5K). The cellular aging- and immortalization-dependent changes of Oct-1 localization seemed to be parallel to the switching of derepressed and repressed states on the collagenase gene during both processes.
Figure 5.
The localization of Oct-1 protein in nuclear periphery during cellular senescence and immortalization. (A-C) The nuclear periphery of HuS-L12 cells at PDL 66 or 73 was prominently stained by three independent anti-Oct-1 antibodies. (A) YL15, a monoclonal antibody against the unique POU-domain linker region of Oct-1. (B) YL123, a monoclonal antibody against the POU homeodomain of Oct-1. (C) A polyclonal antibody against the C terminus of Oct-1. (D–H) The prominent signals of Oct-1 protein disappeared from the nuclear periphery during cellular aging and were resumed in immortalization. The Oct-1 protein was stained with YL15 in HuS-L12 cells at PDL 66 (D), 73 (E), 84 (F), and 91 (G) and in immortalized IML12–4 cells (H). Arrows indicate the cells that lack the Oct-1–associated nuclear peripheral structure. (I–J) The staining patterns of p53 were mostly unchanged as a control. The p53 protein was stained with pAb421 in HuS-L12 cells at PDL 66 (I) and 91 (J) and in immortalized IML12–4 cells (K).
Collagenase Is Highly Expressed in the Cells That Lack the Oct-1-associated Nuclear Peripheral Structure
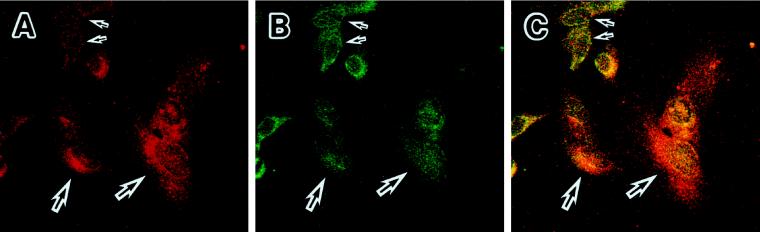
To confirm the relationship between the localization of Oct-1 and the collagenase expression, we double-stained the preimmortalized cells at PDLs 66 and 91 with the anti-collagenase polyclonal and the anti-Oct-1 monoclonal antibodies. At PDL 66, a very rare fraction of cells exhibited staining signals of collagenase, and most of the cells showed no signals of collagenase (our unpublished results). At PDL 91, many cells were strongly stained with the anti-collagenase antibody. The most strongly stained cells exhibited the enlarged, senescent morphology (Figure 6A, large arrows). In these cells, Oct-1 completely disappeared from the nuclear periphery (Figure 6B). On the contrary, faint signals of the collagenase expression were observed in the cells with detectable signals of Oct-1 in the nuclear periphery (Figure 6, A and 6B, small arrows). Although the cells with the intermediate profiles of both staining signals were also observed, this tendency was confirmed when these two images of collagenase and Oct-1 signals were merged (Figure 6C). These results suggested that the Oct-1–associated structural assembly in the nuclear periphery was involved in the mechanism of collagenase repression in cellular aging and immortalization.
Figure 6.
The relationship between the collagenase expression (red) and the localization of Oct-1 (green). Precrisis HuS-L12 cells at PDL 91 were double immunostained with an anti-collagenase polyclonal antibody (A) and YL15 (B). These two images of staining are merged in panel C. Large and small arrows represent cells with no staining of Oct-1 in the nuclear periphery and strong signals of collagenase in the cytoplasm and those with the opposite characteristics, respectively.
Oct-1 Is Colocalized with Lamin B, a Component of the Nuclear Lamina, in the Nuclear Periphery
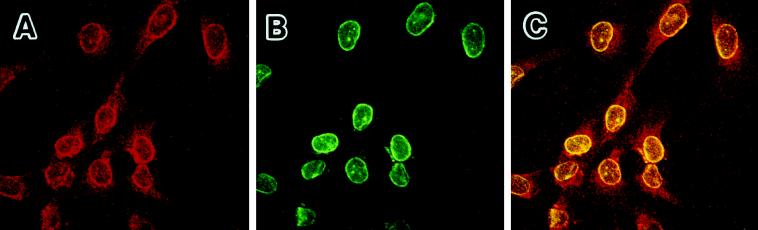
From the prominent immunostaining profiles of Oct-1 in the nuclear periphery, one may imply a possibility that Oct-1 mediates the repression of collagenase by attaching to the structure of nuclear lamina. The nuclear lamina carries a fibrous meshwork structure that forms the backing of the inner nuclear membrane. Two types of components are classified in the nuclear lamina: lamin A/C and lamin B (Moir et al., 1995). Lamin B, especially, is associated with the nuclear matrix attachment regions (MARs), the peripheral chromatin, and the sites of DNA replication (Ludérus et al., 1992; Belmont et al., 1993; Moir et al., 1994). Thus, we examined the colocalization of Oct-1 and lamin B. We double-stained the preimmortalized cells at PDL 66 with anti-Oct-1 polyclonal and anti-lamin B monoclonal antibodies. The prominent staining of Oct-1 was again observed in the nuclear periphery with the anti-Oct-1 polyclonal antibody (Figure 7A, red). The lamin B showed profiles of strong shell-like staining in the nuclear lamina, as reported by others (Meier and Georgatos, 1994; Moir et al., 1994) (Figure 7B, green). In certain fractions of cells, brightly stained dots were also observed in the nuclei. When these two images of staining were merged, the colocalization of Oct-1 and lamin B was clearly demonstrated in the nuclear periphery (Figure 7C, yellow).
Figure 7.
The colocalization of Oct-1 (red) with lamin B (green) in the nuclear periphery. HuS-L12 cells at PDL 66 were double immunostained with the anti-Oct-1 polyclonal antibody (A) and an anti-lamin B monoclonal antibody (B). These two images of staining are merged, and the colocalization of Oct-1 with lamin B is represented by yellow (C).
The Amounts of Nuclear Oct-1 Protein Vary during Cellular Aging and Immortalization, but Not Those in the Whole Cell Extracts
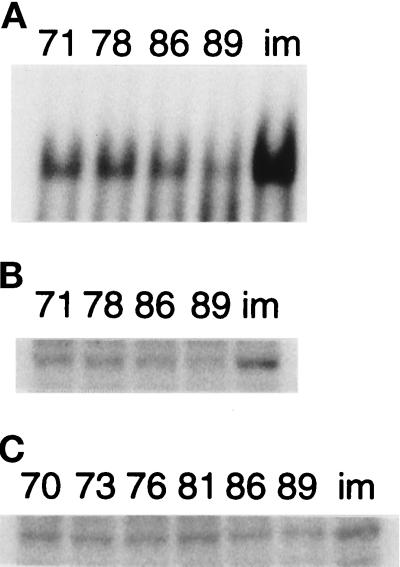
To confirm the immunocytological changes of Oct-1 protein in cellular aging and immortalization, we examined the binding activity and the amount of Oct-1 protein in both processes. The binding activity of Oct-1 to its binding site of ISE2 was gradually reduced during the cellular aging process and almost completely abolished at PDL 89. In immortalized cells, the higher level of Oct-1–binding activity was resumed, compared with that in the preimmortalized cells at PDL 71 (Figure 8A). Concomitantly with these changes of binding activity, the amounts of Oct-1 protein in the nuclear extract varied in cellular aging and immortalization (Figure 8B). These results were consistent with the transcriptional changes of the collagenase gene and the immunocytological changes of Oct-1 protein. In the whole cell extracts, however, the changes in the amounts of Oct-1 protein did not seem to be parallel to those in the nuclear extracts (Figure 8C). The significant amount of Oct-1 protein was observed even in the extract at PDL 89. These results showed that both the localization of Oct-1 protein and the transcriptional status of the collagenase gene were concomitantly regulated in a cellular aging- and immortalization-dependent manner.
Figure 8.
The amounts of nuclear and cellular Oct-1 protein during cellular aging and immortalization. (A) The Oct-1-DNA complexes were detected in EMSA with the nuclear extracts of HuS-L12 at indicated PDLs and IML12–4 (im). (B) The amounts of nuclear Oct-1 protein were determined in the same nuclear extracts as panel A by Western blotting. (C) The total amounts of cellular Oct-1 protein were determined in the whole cell extracts of HuS-L12 at indicated PDLs and IML12–4.
DISCUSSION
Oct-1 Functions as a Cellular Aging-associated Transcriptional Repressor
From the characteristics of the Orpheus-binding activity, we demonstrated clearly that Orpheus was identical to Oct-1. The sequence AAATAATT of ISE2 was determined as the precise Orpheus-binding site that contributed to the binding activity and the repressor function of Orpheus. This Orpheus-binding site was found to be consistent with a potential Oct-1– binding sequence, and the Orpheus–DNA complex was completely supershifted by the anti-Oct-1 monoclonal YL15 and polyclonal antibodies. The in vitro translated Oct-1 protein exhibited the binding properties indistinguishable from those of Orpheus. We also detected binding activity quite similar to that of Orpheus in HeLa-S3 cells that were known to express the high level of Oct-1 protein (Sturm et al., 1988). This Oct-1–binding sequence of ISE2 is not canonical, but Oct-1 is reported to recognize degenerate AT-rich sequences (Verrijzer et al., 1992; Bendall et al., 1993). By the analysis of x ray crystallography, the POU homeodomain of Oct-1 recognizes the 3′-half-portion AAAT of the canonical octamer ATGCAAAT (Klemm et al., 1994). The 5′-half-portion of the Oct-1–binding site of ISE2 was also AAAT, and the POU homeodomain may recognize this portion. In fact, by mutations of AT in AAAT, the Oct-1 binding was completely abolished in both cases of the canonical octamer and the binding site of ISE2 (our unpublished results). Although the core ATAA of the Oct-1 recognition site in ISE2 is essential for both binding and repressive function of Oct-1, the most 3′-TT of this site contributed to the repressive function but not for the binding of Oct-1 (see Figure 2, mut11). Although this reason is unclear, the flexibility of the DNA recognition of Oct-1 may be involved in this inconsistency. The mutation in mut11, which itself does not affect the Oct-1 binding, may induce certain structural alteration of Oct-1 that affects its repressive activity. Taken together, we concluded that Orpheus, i.e., Oct-1, repressed the collagenase expression by interacting with the sequence AAATAATT of ISE2.
The Function and Regulation of Oct-1–associated Structural Assembly
We found that the significant fraction of Oct-1 protein was prominently localized in the nuclear periphery of the preimmortalized cells at earlier PDLs. This subspecies of Oct-1 protein was also demonstrated to be colocalized with lamin B, a structural component of nuclear lamina, in the nuclear periphery. Although the direct physical interaction between Oct-1 and lamin B remained unclear, these results suggested that the Oct-1 protein was possibly involved in certain structural assembly associated with the nuclear lamina. Accompanying the loss of Oct-1–mediated collagenase repression, Oct-1 disappeared from the nuclear periphery during the cellular aging process. The specific localization of nuclear Oct-1 protein was resumed after immortalization, and the collagenase expression was strongly repressed again. We also showed indirect evidence for the relationship between the localization in the nuclear periphery and the repressive function of Oct-1. The double immunostain of collagenase and Oct-1 revealed that the strong collagenase expression was observed in the cells that carried no staining of Oct-1 in the nuclear periphery. These results suggested that Oct-1 was involved in the formation of a nuclear apparatus to mediate silencing of a certain group(s) of genes. This hypothetical apparatus may be associated with the structure of nuclear lamina or nuclear matrix, and the release of Oct-1 from the structural assembly may cause the derepression of the silenced genes. We could not detect the repressive function of Oct-1 in transient transfection assays with the Oct-1 expression vector and various CAT reporter constructs carrying the ISEs (our unpublished results). The repressive function of Oct-1 may be inert in this case, because the Oct-1 protein overexpressed by the exogenous Oct-1 gene did not reside appropriately in the apparatus of the nuclear periphery, and the cells overexpressing Oct-1 seemed to become dead (our unpublished results).
During cellular aging and immortalization, the binding activity of Oct-1 changed coincidentally with the amounts of nuclear Oct-1 protein. These findings also supported that the loss of Oct-1-mediated repression was evoked by the loss of Oct-1 protein from the nuclear periphery. However, we found that the whole cellular amounts of Oct-1 protein did not change significantly during these cellular processes. Although the other possibilities cannot be excluded, the Oct-1 protein may be prevented from translocating to the correct nuclear peripheral compartment in precrisis cells. We detected a motif of the potential nuclear localization signal at the N terminus of the POU homeodomain in Oct-1. This amino acid sequence RRRKKR is juxtaposed to Ser385. This serine residue is phosphorylated in an M-phase–specific manner, and the phosphorylation leads to the loss of DNA-binding activity in Oct-1 (Roberts et al., 1991; Segil et al., 1991). In the case of SWI5 and v-Jun, the phosphorylation of serine residues close to their nuclear localization signals inhibits their translocation to the nucleus (Moll et al., 1991; Chida and Vogt, 1992). The localization and/or the binding activity of Oct-1 protein may be regulated by phosphorylation in the process of cellular aging.
Possible Involvement of Oct-1–associated Structural Assembly in the Mechanism of Cellular Aging
Oct-1 is a ubiquitously expressed member of the POU domain family and suggested so far to play important roles in transcriptional activation and DNA replication in mammalian cells (Coenjaerts et al., 1994; Herr and Cleary, 1995). Recently, the transcriptional repressive or silencing function of Oct-1 has been reported (Kim et al., 1996; Sterling and Bresnick, 1996). Oct-1 is involved in the silencing of the human thyrotropin β gene (hTSHβ) by interacting with the AT-rich silencer element that functions as a MAR (Kim et al., 1996). A significant fraction of Oct-1 is shown consistently to be associated with the nuclear matrix. Oct-1 may anchor the hTSHβ MAR-silencer region onto the nuclear matrix and mediate the transcriptional silencing through the formation of certain highly ordered chromatin structures. Although direct evidence remains to be obtained, Oct-1 may similarly function as a MAR-binding protein that mediates transcriptional repression in the case of the collagenase gene. 1) The region containing the Oct-1 binding sequence of ISE2 is about 70% AT-rich and consists of an ATC (ATG) sequence that is proposed to be an important characteristic of MAR (Dickinson et al., 1992). 2) In the approximately 30-kbp region upstream of the collagenase gene, we detected many potential binding sites of Oct-1. These multiple Oct-1–binding sites seemed to be clustered in AT-rich MAR-like regions (our unpublished findings). 3) When the prominent localization of Oct-1 in the nuclear periphery was abolished in the process of cellular aging, the strong collagenase expression was observed. 4) Oct-1 in the nuclear periphery was colocalized with lamin B. Lamin B is reported to be one of the MAR-binding components of the nuclear lamina and to be associated with the chromatin structure in the nuclear periphery and the sites of DNA replication during S phase (Ludérus et al., 1992; Belmont et al., 1993; Moir et al., 1994). Thus, Oct-1 may form a transcriptional repressive apparatus by anchoring MARs onto the nuclear lamina in the nuclear periphery. Other molecules interacting with Oct-1 may be involved in the formation of the transcriptional repressive apparatus. In fact, we detected another unidentified factor whose binding site was 5′ adjacent to the Oct-1–binding site in ISE2. This factor regulates the collagenase expression negatively (our unpublished results). Oct-1 may function cooperatively with this factor to repress the collagenase expression.
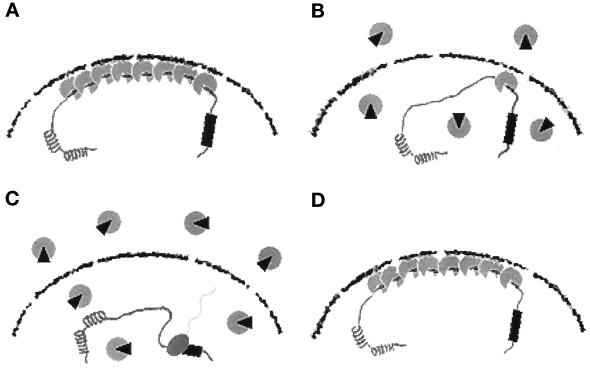
Taken together, we speculate a comprehensive model for the cellular aging-associated regulation of the collagenase gene (Figure 9). In preimmortalized cells at earlier PDLs, Oct-1 is involved in the formation of the repressive apparatus that locates in the nuclear periphery (Figure 9A). This structural assembly may be associated with the nuclear lamina or the nuclear matrix and function as a global mechanism for the negative regulation of transcription. The cellular aging-associated genes, including the collagenase gene, are under the control of this regulatory mechanism. In the process of cellular aging, the Oct-1–mediated repressive apparatus is gradually dissociated by the liberation of Oct-1 from the structural assembly (Figure 9B). Certain modification and/or protein–protein interaction of Oct-1 may be important for this change. When the extent of Oct-1 dissociation reaches a certain threshold, the cellular aging-associated genes are totally released from the transcriptionally repressed states, and the high levels of expression are induced in precrisis cells (Figure 9C). By the derepression of the cellular aging-associated genes, cells cease to proliferate and show various senescent phenotypes. Cells can be immortalized if certain mutational events occur in the global regulatory mechanism of transcription and the repressed state is resumed (Figure 9D). In our previous study, we proposed the hypothesis of “molecular counter” as the central regulatory mechanism of cellular senescence (Imai et al., 1993). This hypothetical molecular counter counts cellular replication cycles and terminates cellular proliferation after a certain limited number of cellular doublings. It is an intriguing possibility that the sequential dissociation of Oct-1 protein from the structural assembly in the nuclear periphery may be involved in this hypothetical molecular counting mechanism of replicative lifespan. Identification of the regulatory proteins that effect the binding and functional ability of Oct-1 will provide much information concerning a possible role of Oct-1 as a device for the molecular counter of cellular doublings.
Figure 9.
A comprehensive model for the cellular aging- and immortalization-associated regulation of the collagenase gene. The four representative stages of the collagenase gene regulation are schematically shown in panels A–D. The Oct-1–associated transcriptional repressive apparatus is presented by gray particles. A gray particle with a small black wedge shows a hypothetical modified form of Oct-1 protein. See text for details. (A) Preimmortalized cells at earlier PDLs. (B) Preimmortalized cells in the process of cellular aging. (C) Precrisis cells. (D) Immortalized cells.
ACKNOWLEDGMENTS
We thank Masafumi Tanaka and Winship Herr of Cold Spring Harbor Laboratory for providing antibodies and plasmids; Kyoji Ohyama of Department of Anatomy, Keio University School of Medicine, for immunostaining; and Hiroaki Kitano of Sony Computer Science Laboratory for his critical discussion. S.I. and T.T. were supported by the Grant-in-Aids for Scientific Research from the Ministry of Education, Science, Sports and Culture in Japan, respectively. S.I. was also supported by Keio University Sakaguchi-Memorial Medical Science Fund. For part of this study, S.I. was awarded the ASCB/Glenn Foundation Award in 1996.
REFERENCES
- Bauer EA, Silverman N, Busiek DF, Kronberger A, Deuel TF. Diminished response of Werner’s syndrome fibroblasts to growth factors PDGF and FGF. Science. 1986;234:1240–1243. doi: 10.1126/science.3022382. [DOI] [PubMed] [Google Scholar]
- Bayreuther K, Rodemann HP, Hommel R, Dittmann K, Albiez M, Francz PI. Human skin fibroblasts in vitro differentiate along a terminal cell lineage. Proc Natl Acad Sci USA. 1988;85:5112–5116. doi: 10.1073/pnas.85.14.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123:1671–1685. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall AJ, Sturm RA, Danoy PAC, Molloy PL. Broad binding-site specificity and affinity properties of octamer 1 and brain octamer-binding proteins. Eur J Biochem. 1993;217:799–811. doi: 10.1111/j.1432-1033.1993.tb18308.x. [DOI] [PubMed] [Google Scholar]
- Burke EM, Horton WE, Pearson JD, Crow MT, Martin GR. Altered transcriptional regulation of human interstitial collagenase in cultured skin fibroblasts from older donors. Exp Gerontol. 1994;29:37–53. doi: 10.1016/0531-5565(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Chida K, Vogt PK. Nuclear translocation of viral Jun but not of cellular Jun is cell cycle dependent. Proc Natl Acad Sci USA. 1992;89:4290–4294. doi: 10.1073/pnas.89.10.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenjaerts FEJ, van Oosterhout JAWM, van der Vliet PC. The Oct-1 POU domain stimulates adenovirus DNA replication by a direct interaction between the viral precursor terminal protein-DNA polymerase complex and the POU homeodomain. EMBO J. 1994;13:5401–5409. doi: 10.1002/j.1460-2075.1994.tb06875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler RG. Human longevity and aging: possible role of reactive oxygen species. Ann NY Acad Sci. 1991;621:1–28. doi: 10.1111/j.1749-6632.1991.tb16965.x. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lasser AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Giordano T, Foster DN. Identification of a highly abundant cDNA isolated from senescent WI-38 cells. Exp Cell Res. 1989;185:399–406. doi: 10.1016/0014-4827(89)90310-8. [DOI] [PubMed] [Google Scholar]
- Goldstein S. Replicative senescence: the human fibroblast comes of age. Science. 1990;249:1129–1133. doi: 10.1126/science.2204114. [DOI] [PubMed] [Google Scholar]
- Guarente L. Do changes in chromosomes cause aging? Cell. 1996;86:9–12. doi: 10.1016/s0092-8674(00)80072-0. [DOI] [PubMed] [Google Scholar]
- Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T, Mizuno T, Ogasahara Y, Nakagawa M. An oligodeoxyribonucleotide-directed dual amber method for site-directed mutagenesis. Gene. 1995;152:271–275. doi: 10.1016/0378-1119(94)00750-m. [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Herr W, Cleary MA. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes & Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- Hertzberg RP, Dervan PB. Cleavage of double helical DNA by (methidiumpropyl-EDTA)iron(II) J Am Chem Soc. 1982;104:313–315. [Google Scholar]
- Imai S, Fujino T, Nishibayashi S, Manabe T, Takano T. Immortalization-susceptible elements and their binding factors mediate rejuvenation of regulation of the type I collagenase gene in simian virus 40 large T antigen-transformed immortal human fibroblasts. Mol Cell Biol. 1994;14:7182–7194. doi: 10.1128/mcb.14.11.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Saito F, Ikeuchi T, Segawa K, Takano T. Escape from in vitro aging in SV40 large T antigen-transformed human diploid cells: a key event responsible for immortalization occurs during crisis. Mech Ageing Dev. 1993;69:149–158. doi: 10.1016/0047-6374(93)90079-7. [DOI] [PubMed] [Google Scholar]
- Imai S, Takano T. Loss of collagenase gene expression in immortalized clones of SV40 T antigen-transformed human diploid fibroblasts. Biochem Biophys Res Commun. 1992;189:148–153. doi: 10.1016/0006-291x(92)91537-z. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. Longevity, genes, and aging. Science. 1996;273:54–59. doi: 10.1126/science.273.5271.54. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kim MK, Lesoon-Wood LA, Weintraub BD, Chung JH. A soluble transcription factor, Oct-1, is also found in the insoluble nuclear matrix and posesses silencing activity in its alanine-rich domain. Mol Cell Biol. 1996;16:4366–4377. doi: 10.1128/mcb.16.8.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- Kumazaki T, Robetorye RS, Robetorye SC, Smith JR. Fibronectin expression increases during in vitro cellular senescence: correlation with increased cell area. Exp Cell Res. 1991;195:13–19. doi: 10.1016/0014-4827(91)90494-f. [DOI] [PubMed] [Google Scholar]
- Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludérus MEE, de Graaf A, Mattia E, den Blaauwen J L, Grande M A, de Jong L, van Driel R. Binding of matrix attachment regions to lamin B1. Cell. 1992;70:949–959. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- Meier J, Georgatos SD. Type B lamins remain associated with the integral nuclear envelop protein p58 during mitosis: implications for nuclear reassembly. EMBO J. 1994;13:1888–1898. doi: 10.1002/j.1460-2075.1994.tb06458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millis AJT, Hoyle M, McCue HM, Martini H. Differential expression of metalloproteinase and tissue inhibitor of metalloproteinase genes in aged human fibroblasts. Exp Cell Res. 1992;201:373–379. doi: 10.1016/0014-4827(92)90286-h. [DOI] [PubMed] [Google Scholar]
- Moir RD, Montag-Lowy M, Goldman RD. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Goldman RD. The dynamic properties and possible functions of nuclear lamins. Int Rev Cytol. 1995;162B:141–182. doi: 10.1016/s0074-7696(08)62616-9. [DOI] [PubMed] [Google Scholar]
- Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- Roberts SB, Segil N, Heinz N. Differential phosphorylation of the transcription factor Oct-1 during the cell cycle. Science. 1991;253:1022–1026. doi: 10.1126/science.1887216. [DOI] [PubMed] [Google Scholar]
- Segil N, Roberts SB, Heinz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- Smith JR, Pereira-Smith OM. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273:63–67. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- Sottile J, Mann DM, Diemer V, Millis AJT. Regulation of collagenase and collagenase mRNA production in early- and late-passage human diploid fibroblasts. J Cell Physiol. 1989;138:281–290. doi: 10.1002/jcp.1041380209. [DOI] [PubMed] [Google Scholar]
- Stanulis-Praeger BM. Cellular senescence revisited: a review. Mech Ageing Dev. 1987;38:1–48. doi: 10.1016/0047-6374(87)90109-6. [DOI] [PubMed] [Google Scholar]
- Sterling K, Bresnick E. Oct-1 transcription factor is a negative regulator of rat CYP1A1 expression via an octamer sequence in its negative regulatory element. Mol Pharmacol. 1996;49:329–337. [PubMed] [Google Scholar]
- Strehler B, Hirsch G, Gusseck D, Johnson R, Bick M. Codon-restriction theory of aging and development. J Theor Biol. 1971;33:429–474. doi: 10.1016/0022-5193(71)90091-9. [DOI] [PubMed] [Google Scholar]
- Sturm RA, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes & Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, Bidwell JP, Fey EG, Penman S, Lian JB, Stein JL, Stein G S. Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP-1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Alkema MJ, van Weperen WW, van Leeuwen HC, Strating MJJ, van der Vliet PC. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S, Hayes S, O’Hare P. Site-specific conformational alteration of the Oct-1 POU domain-DNA complex as the basis for differential recognition by Vmw65 (VP16) Cell. 1994;79:841–852. doi: 10.1016/0092-8674(94)90073-6. [DOI] [PubMed] [Google Scholar]
- West MD, Pereira-Smith OM, Smith JR. Replicative senescence of human skin fibroblasts correlates with a loss of regulation and overexpression of collagenase activity. Exp Cell Res. 1989;184:138–147. doi: 10.1016/0014-4827(89)90372-8. [DOI] [PubMed] [Google Scholar]
- Wistrom C, Villeponteau B. Cloning and expression of SAG: a novel marker of cellular senescence. Exp Cell Res. 1992;199:355–362. doi: 10.1016/0014-4827(92)90445-e. [DOI] [PubMed] [Google Scholar]
- Yeh W-C, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- Zs.-Nagy I, Cutler RG, Semsei I. Dysdifferentiation hypothesis of aging and cancer: a comparison with the membrane hypothesis of aging. Ann NY Acad Sci. 1988;521:215–225. doi: 10.1111/j.1749-6632.1988.tb35280.x. [DOI] [PubMed] [Google Scholar]