Abstract
The thymic microenvironment is required for T cell development in vivo. However, in vitro studies have shown that when hematopoietic progenitors acquire Notch signaling via Delta-like (Dll)1 or Dll4, they differentiate into the T cell lineage in the absence of a thymic microenvironment. It is not clear, however, whether the thymus supports T cell development specifically by providing Notch signaling. To address this issue, we generated mice with a loxP-flanked allele of Dll4 and induced gene deletion specifically in thymic epithelial cells (TECs). In the thymus of mutant mice, the expression of Dll4 was abrogated on the epithelium, and the proportion of hematopoietic cells bearing the intracellular fragment of Notch1 (ICN1) was markedly decreased. Corresponding to this, CD4 CD8 double-positive or single-positive T cells were not detected in the thymus. Further analysis showed that the double-negative cell fraction was lacking T cell progenitors. The enforced expression of ICN1 in hematopoietic progenitors restored thymic T cell differentiation, even when the TECs were deficient in Dll4. These results indicate that the thymus-specific environment for determining T cell fate indispensably requires Dll4 expression to induce Notch signaling in the thymic immigrant cells.
The thymus is the major necessary site for T cell development. In the physiological condition, hematopoietic progenitor cells (HPCs), which reside in the fetal liver or BM, migrate into the thymus to differentiate into T lineage cells with a diverse and self-compatible TCR repertoire. The importance of the thymus in immunity has been well documented by observations in immunodeficient individuals, in human patients with DiGeorge syndrome, and in mice with the nude (FoxN1) mutation, which lack a thymus and effective T cells. However, it remains unclear how the thymus is essential for T cell development in vivo. T cell development is critically supported by specialized cellular microenvironments composed of the thymic stroma, the three-dimensional structure of epithelial cells, and mesenchymal-originated cells (1). In addition to the cellular components, the thymus is known to produce some growth factors, including IL-7 and stem cell factor, which are likely to contribute to early T cell development. However, these growth factors were not specific for the thymus, as they were also found in other hematopoietic tissues. Thus, recent studies have focused on cellular components of the thymic environment to understand the molecular mechanism that contributes to the specification process for T cell lineage from HPCs migrating into the thymus.
There is accumulating evidence that Notch signaling is critical for the determination of T cell fate from HPCs. Previous studies using Cre-loxP–mediated gene targeting showed that the specific deletion of Notch1 or RBP-J, which codes the signal transducer downstream of Notch1 in HPCs, led to a complete defect of T cell development accompanied by a certain increase of B cells in the thymus (2, 3). In contrast, implantation of HPCs with the enforced expression of the active form of Notch1, intracellular fragment of Notch1 (ICN1), induced the ectopic appearance of CD4 and CD8 double-positive (DP) cells in BM (4). A similar phenotype was observed on the monolayer culture of BM-derived stromal cells in the absence of a thymic environment (5). These results clearly indicated that Notch1-mediated signal is both essential and sufficient for T cell specification at the branch point of T versus B cells. However, it remains to be clarified whether the thymic environment accomplishes its role for T cell development by providing Notch signaling into HPCs in vivo. In fact, both Delta-like (Dll)1 and Dll4 among Notch ligands (NotchL) were detected in the thymus (6–10). Moreover, any stromal cells with the forced expression of NotchL, such as Dll1 and Dll4, are able to induce Notch signaling into HPCs, supporting T lymphopoiesis in vitro (6, 11). Deficiency of both Dll1 and Dll4 in mice is, however, embryonically lethal (12–14), and these mice do not provide information of any potential function of these molecules in T cell development. To overcome this problem, we established mice with a loxP-flanked allele of Dll1 and showed that T cell development is intact in the thymus in a Dll1-null situation (6). Collectively, it is suggested that Dll4 or both Dll1 and Dll4 function as proper ligands to provide Notch signaling required for T cell specification in the thymus.
In this study, we succeeded in establishing mice with a loxP-flanked allele of Dll4 (floxed Dll4), and induced the gene deletion of Dll4 that is specific in thymic epithelial cells (TECs). In these mice, the expression of Dll4 on TECs was abrogated and Notch1-mediated signaling was reduced in the thymus, resulting in the disappearance of T cells and aberrant accumulation of B lineage cells.
RESULTS AND DISCUSSION
Abrogation of Dll4 on TECs reduces the Notch1-mediated signal in thymocytes
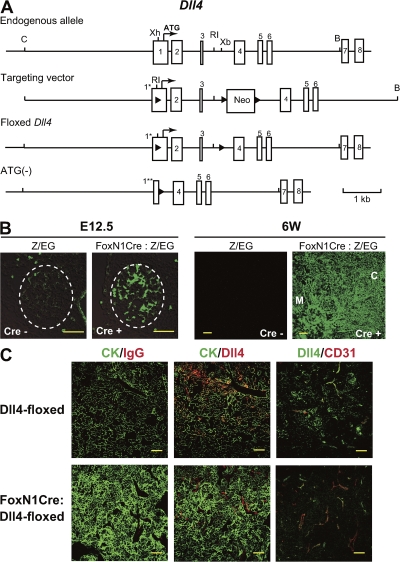
Dll4 is the third member of the Dll family characteristically expressed in the endothelium and thymus (8–10, 15). Immunohistochemical analysis with Dll4-specific antibody revealed that Dll4 was dominantly expressed on the thymic epithelium with cytokeratin (Fig. 1 A), especially in the cortex (K8+K5− region), which is where early T cell development occurs (Fig. 1 B). In contrast, it was found on endothelial cells, but not on the epithelium in gut or salivary gland (Fig. 1 C). These findings suggested that the distinctive expression of Dll4 on the thymic epithelium contributes to the formation of a thymus-specific environment. To examine the physiological significance of Dll4 on the thymic epithelium for T cell development, Dll4-targeting mice, with the Dll4 gene flanked by a loxP sequence (designated Dll4-floxed), were established (Fig. 2 A). The floxed allele of Dll4 gene was removed by Cre recombinase, resulting in the loss of the translational initiation site. These mice were bred with FoxN1-Cre mice, where the Cre gene is driven by the regulatory element of FoxN1 gene (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20080134/DC1). The specific activity of Cre recombinase in FoxN1-Cre mice was observed as the ectopic expression of GFP in the thymic epithelium by crossing with the reporter transgenic mice (16) at the embryonic or adult stage (Fig. 2 B). In mice homozygous for floxed Dll4 with FoxN1-Cre transgene (FoxN1Cre:Dll4-floxed), the expression of Dll4 was completely abrogated on TECs (Fig. 2 C, bottom middle), but was still intact on endothelial cells bearing CD31 (Fig. 2 C, bottom right). These results indicated that the gene deletion was specifically induced in epithelial cells in the thymus. Moreover, this deletion of the Dll4 gene did not affect the expression of Dll1 in the thymus (Fig. S2). Interestingly, cortical and medullary regions were almost indistinguishable in the thymus of mutant mice (unpublished data).
Figure 1.
Dll4 was expressed on thymic epithelium. The expression of Dll4 on the epithelium was detected in thymus (A and B, Thymus), but not gut (C, Gut) or salivary gland (C, Sal. G.) by immunofluorescence histochemistry. Staining was done with anti–pan-cytokeratin (CK; A, green), anti-K5 (K5; B, green), anti-K8 (K8; B, red), anti-Dll4 (Dll4; A, red; B, blue), or control IgG (IgG; A, red; B, blue) antibodies and analyzed by confocal laser microscopy. (A and B, right) High-magnification images of the red square in the middle images. (B, left) Hemotoxylin and eosin staining of serial section. Bars: (A, left and middle) 50 μm; (A, right) 10 μm; (B, left) 200 μm; (B, right) 10 μm; (C) 20 μm.
Figure 2.
Specific abrogation of Dll4 on thymic epithelium. (A) Targeted insertion of loxP sequences flanking a part of exon 1 and whole exons 2 and 3 of the Dll4 gene. Numbers indicate exon number. 1*, first exon modified with loxP sequence; 1**, part of exon 1 after gene deletion; ATG, translational initiation codon of Dll4; triangles, loxP sequences; open boxes, Dll4 exons; B, BglII; C, ClaI; RI, EcoRI; Xb, XbaI; Xh, XhoI. (B) Cre activity was targeted to thymic epithelia using mice, designated FoxN1-Cre, in which this recombinase is under the transcriptional control of the FoxN1 locus. The timing and specificity of Cre-mediated recombination was visualized by the expression of enhanced GFP (eGFP) in thymus tissue sections of E12.5 (left) and 6-wk-old (right) crosses of FoxN1-Cre mice with the transgenic Z/EG reporter mice. The thymus anlage in the left image is outlined with a dashed line. C, cortex; M, medulla. (C) The expression of Dll4 on thymic epithelial, but not endothelial, cells was abrogated in Dll4lox/loxFoxN1-Cre (FoxN1Cre:Dll4-floxed) mice. Epithelial and endothelial cells in the thymus were identified by the expression of cytokeratin (CK, green, left and middle) and CD31 (red, right), respectively, with Dll4 (red, middle; green, right). Bars: (B and C) 50 μm.
In the thymus of the Dll4-floxed fetus, as well as the WT fetus, substantial thymocytes (∼30%) bore the ICN1, which was generated by the proteolysis of Notch1 as a signal transducer of Notch signaling, in cytoplasm or nucleus (Fig. 3 A, arrowheads). However, this proportion of thymocytes with ICN1 was markedly decreased in the mutant mice without Dll4 (Fig. 3 B). The specificity of this intracellular staining was confirmed by the reduction of the ratio of double-negative (DN) cells possessing the intracellular fragment after the fetal thymus organ culture (FTOC) with γ-secretase inhibitor (GSI; Fig. 3 B), which blocks the proteolysis of Notch1. These results indicated that the intrathymic immigrants are provided Notch signaling after migrating into the thymus, which is dependent on the Dll4 expressed on epithelial cells.
Figure 3.
Notch signaling was decreased in the thymus with Dll4-null epithelial cells. (A) The cleaved Notch1 fragment was found in fetal thymocytes of E15.5 Dll4lox/lox (Dll4-floxed) embryos. The cells with cleaved Notch1 are indicated with arrowheads. Bar, 10 μm. (B) Frequencies of cells with cleaved Notch1 (ICN1+; mean percentage from five fields in a slide with >100 cells from each embryo ± SD) were counted in E15.5 WT (n = 3), Dll4lox/loxFoxN1Cre (FoxN1Cre:Dll4-floxed, n = 6), or Dll4lox/lox (Dll4-floxed, n = 6) mice. The cultured DN cells were prepared after the FTOC of E14.5 WT fetal thymus for 4 d with γ-secretase inhibitor (GSI, n = 3) or without γ-secretase inhibitor (DMSO, n = 3). Asterisk indicates unpaired Student's t test; P < 0.001.
No T cells, but aberrant B cell accumulation, in the thymus with Dll4-null epithelial cells
Without the Dll4 expression in the TECs (FoxN1Cre:Dll4-floxed), the number of cells obtained from the thymus was significantly decreased (1/3–1/25) compared with that from the thymus in littermates (Dll4-floxed) during the developmental process (Fig. 4 A). Flow-cytometric analysis of 4-wk-old mouse thymus revealed that almost all cells were composed of CD4 and CD8 DN cells expressing neither Thy1, TCR αβ, nor γδ with CD3 in FoxN1Cre:Dll4-floxed mice, in comparison to the substantial presence of DP or CD4 and CD8 single-positive T cells expressing Thy1 and TCR–CD3 complex in Dll4-floxed mice (Fig. 4 B). These DN cells were mainly immature B cells expressing CD19 and, partially, surface IgM, and showed identical phenotype to that seen in BM, but not in the periphery (Fig. 4 B). If DN cells are subdivided into four fractions based on CD44 and CD25 expression, the DN2 and DN3 stages are defined as being CD25 highly positive with TCR gene recombinations. Although more than half of the DN cells belonged to DN2/3 in the control thymus, no CD25-positive cells were detected in FoxN1Cre:Dll4-floxed mice (Fig. 4 C, middle). It was recently reported that the DN1 stage was further distinguished into DN1a, b, and c by the expression of CD24 and c-kit (Fig. 4 C, right), showing that the DN1a+b fraction contained the earliest T cell progenitors (ETPs) and that DN1c possessed differentiation potential for B cell lineage (17). In the thymus of FoxN1Cre:Dll4-floxed mice, few DN1a+b cells, but a substantial amount of DN1c cells, were detected (Fig. 4 C). Similar phenotype was observed in the fetal thymus, where NK cells normally developed, and in the newborn thymus (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20080134/DC1). In peripheral lymphoid tissues such as spleen and lymph node of the mutant mice, only a few mature T cells were detected (Fig. S4). These findings first indicated that the Notch signaling induced by Dll4 on the thymic epithelium is essential for the intrathymic immigrant cells to differentiate into T lineage cells, and that the immigrants tend to differentiate into B lineage cells without the Dll4-mediated Notch signaling. They also indicated that the maintenance of DN1a+b immediately after HPCs have migrated, and further differentiation in the thymus, require the Notch signaling provided by Dll4.
Figure 4.
No T cells, but substantial B cells, appear in the thymus with Dll4-null epithelium. (A) Thymic cellularity (mean ± SD) of fetus (E15.5; left, n = 3; right, n = 5), neonate (N.B.; left, n = 5; right, n = 3), 4-wk-old (4W, n = 4), or 8-wk-old (8W, n = 3) Dll4lox/loxFoxN1-Cre (FoxN1Cre:Dll4-floxed, shaded columns) mice compared with Dll4lox/lox (Dll4-floxed, open columns) mice. Asterisks indicate unpaired Student's t test; P < 0.001. (B) Flow cytometry of thymocytes from young adult (4W) Dll4lox/lox mice with (bottom, FoxN1Cre:Dll4-floxed) or without (top, Dll4-floxed) FoxN1-Cre transgene. Thymocytes were stained with monoclonal antibodies to surface molecules as shown. Numbers in the profiles indicate the relative percentages, in CD4−CD8− cells (right, CD4+CD8 vs. Thy1.2), for each corresponding quadrant or fraction. (C) The ETP population was not observed in Dll4lox/loxFoxN1-Cre (4W) mice. Thymocytes from Dll4lox/lox (top) or Dll4lox/loxFoxN1-Cre mice (bottom) were stained for lineage markers (Lin.: CD4, CD8, CD3, B220, CD19, Gr1, CD11b, TER119), CD44, CD25, CD24, and c-kit. Profiles are shown with the gate (Lin.−, middle; Lin.−CD44+CD25− as DN1, right). Numbers on the plots represent the frequency of cells lying in the indicated regions within the gate. The ETP population is identified as Lin.−CD44+CD25−CD24− or lowc-kit+ cells (DN1a+b). DN1c and DN1–3 populations are also shown in the plots. Data are representative of three experiments.
HPCs with an active form of Notch1 give rise to T cell lineage in Dll4-deficient thymus
Next, we examined whether the Dll4-deficient thymus retains any functions for a T cell development niche other than Dll4. For this, ICN1, which bypasses Notch signaling without any Notch–NotchL interaction, was expressed into WT HPCs of fetal liver by gene introduction with retrovirus vector, and these cells were cultured in thymic lobes obtained from Dll4-floxed mice with or without FoxN1-Cre transgene for FTOC. After a 13-d culture, all mock- or ICN1-introduced cells differentiated into T lineage cells, in part to DP stage or DN stage with CD25 expression, and they also bore Thy1 (unpublished data) in the lobe derived from Dll4-floxed fetus (Fig. 5 A, top). On the other hand, the mock-introduced cells mainly differentiated into CD19-positive B lineage cells in the lobe from FoxN1Cre:Dll4-floxed fetus (Fig. 5 A, bottom left), but even when without Dll4 on the epithelium, the ICN1-introduced cells differentiated into T lineage cells (Fig. 5 A, bottom right), as seen in the lobe with intact Dll4. Also, the ICN1-introduced HPCs could migrate and differentiate into DP cells in the thymus after intravenous injection into the irradiated FoxN1Cre:Dll4-floxed mice (Fig. 5 B), although the ICN1-transduced cells stopped their differentiation at DP stage (18). These results indicated that the depletion of Dll4 on the epithelium does not deprive the thymus of environment potential for T cell development other than the Dll4, and, if the intrathymic immigrants already have Notch signaling, the subsequent differentiation for the T cell lineage is possible in the thymus without Dll4.
Figure 5.
Hematopoietic progenitors with active form of Notch1 give rise to T cell lineage in Dll4-deficient thymus. Hematopoietic progenitors (Lin.−c-kit+ cells) derived from E15.5 fetal liver were infected with the retrovirus encoding the intracellular region of Notch1 (ICN1) or empty vector (Mock). Infected cells were monitored by expression of GFP. These cells were cultured into deoxyguanosine-treated thymic lobes of Dll4lox/loxFoxN1-Cre (FoxN1Cre:Dll4-floxed) or Dll4lox/lox (Dll4-floxed) fetuses for 13 d as FTOC and analyzed for the expression of CD4, CD8, CD25, and CD19 (A). Alternatively, these cells were injected intravenously into irradiated Dll4lox/loxFoxN1-Cre (FoxN1Cre:Dll4-floxed) or Dll4lox/lox (Dll4-floxed) neonate mice. After 3 wk, thymocytes were analyzed for expression of CD4, CD8, and CD19 (B). Profiles are shown with the gate (GFP+ or GFP+CD4−CD8−, DN-gated in A). Numbers in the profiles indicate the relative percentages for each corresponding quadrant.
Among the NotchLs, Jagged1 and Jagged2 are dispensable for T cell development as a physiological ligand for Notch1 because inactivation of these genes either did not affect or did cause a minor defect in the T cell appearance in vivo, respectively (19, 20). In contrast, other NotchLs, Dll1 and Dll4, were shown to retain the potential to induce T lymphopoiesis in vitro (6, 11). Therefore, it has been strongly suggested that Dll1 and Dll4 are candidates for critical NotchLs in the thymus. However, T cell development was intact when Dll1-floxed mice were crossed with FoxN1Cre mice (unpublished data), as previously demonstrated (6). The difference in the contribution of Dll1 and Dll4 for T cell development may be explained by a quantitative or qualitative mechanism. Although the expression of Dll1 was detected in the thymus, there was less of it than of Dll4 (Fig. S2) (10), suggesting that Dll4 dominantly functions as a Notch1 ligand in the thymus. Alternatively, there may be a preferential combination between Notch receptors and ligands in the physiological condition. A recent study suggested an advantage of the interaction between Notch1 and Dll4, compared with that between Notch1 and Dll1, for promoting T cell development (21). This would be consistent with the results of this study.
Without the expression of Dll4 on the thymic epithelium, aberrant accumulation of B cells during thymic ontogeny was observed. In TCR-β−/−δ−/− mice, in which no T cells appear, a few B cells occurred in the thymus, but the cell number was lower than that in normal thymus (22). A small accumulation of B cells was found in hCD3ε transgenic mice that do not develop T cells, but that cell number was much lower (<1/10) than that in Dll4-deficient thymus (23). Substantial B cell accumulation was also detected in the mice receiving some modifications of Notch signaling after the thymus has normally developed (2, 3, 24, 25). Combining the in vitro observation that Notch signaling blocked B cell development (4, 5, 26), it is suggested that B cell accumulation in Dll4-deficient thymus is not simply a reflection of the complete impairment of T cell development, but is caused by the reduced Notch signaling.
Hematopoietic progenitors without Notch signaling give rise to B lineage cells and lose the potential to differentiate into T lineage cells in the thymus (2, 3). There were reports showing the presence of multi- or bipotent stem cells for T or B cell lineages in the thymus (10, 27). Collectively, it was suggested that all thymic lymphocytes originate from such multipotent stem cells that can migrate into the thymus. On the other hand, some reports indicated that only lineage-committed precursors, and not the stem cells, were detected in the thymus (17, 28, 29). These possibilities may be acceptable in this study showing that Notch signaling provided by Dll4 in the thymus induces the development of T cells, but blocks B cell development, from the committed precursors or the stem cells, and that aberrant accumulation of B lineage cells instead of T lineage cells can occur without Dll4-mediated Notch signaling. This is consistent with a recent study showing that transcriptional repressor, LRF, contributes to the determination of B cell fate by repressing Notch signaling, and suggests that abundant Notch signaling induced in the thymus overcomes this repression (30). Another study indicated that continuous Notch signaling is necessary to determine the fate to T cell lineage (31). Collectively, it is likely that the necessity of the thymus in T cell development is dependent on the expression of Dll4 in the thymic epithelium, which can provide abundant and continuous Notch signaling sufficient for the specification to T cell lineage from immigrant cells.
During embryonic development, Notch–NotchL interaction between two equipotent progenitors gives rise to Notch signaling to adopt a distinct developmental fate according to the lateral inhibition theory (32). In intrathymic T cell development, however, NotchL is expressed on the mature epithelium of the thymic environment and induces Notch signaling into immature immigrant cells expressing Notch derived from HPCs. Thus, our findings clearly indicate that Notch–NotchL interaction by direct contact between cells with distinct origin and development contributes to the determination of cell fate, and this should benefit our understanding of organogenesis through Notch–NotchL interaction in mature individuals.
MATERIALS AND METHODS
Generation of mice with floxed Dll4.
For construction of a targeting vector containing sequences corresponding to a floxed Dll4 allele, the loxP-flanked neomycin phosphotransferase (NeoR) gene was inserted after exon 3, and a third loxP site was also placed before the translational initiation site in exon 1. After the NeoR gene was depleted in ES cell clones containing the homologous recombination allele of Dll4, one of these clones was injected into blastocysts and chimeric mice were obtained. Mice with floxed Dll4 allele were bred with FoxN1-Cre transgenic mice for removal of the floxed allele. All mice were maintained in specific pathogen–free conditions, and all mouse experiments were approved by the Animal Experimentation Committee (Tokai University, Kanagawa, Japan).
Immunohistochemistry.
Immunohistochemical analysis was performed as previously described (6). Tissue sections of thymus were stained with FITC-anti-pan-cytokeratin (Sigma-Aldrich), biotinylated goat anti-Dll4 (R&D Systems) antibodies, biotinylated goat IgG, nonlabeled anti-K5 (Covance), anti-K8 (TROMA-I; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), or anti-CD31 (eBioscience) antibodies. For the staining of the cleaved Notch1 fragment, thymocytes (5 × 104) were fixed in PBS containing 4% PFA at 4°C for 10 min. Antigen retrieval was accomplished by autoclave treatment in Target Retrieval Solution (Dako) at 105°C for 5 min. These slides were stained with rabbit anti-cleaved murine Notch1 antibody (Cell Signaling Technology) or control rabbit IgG (Dako). The signals were visualized with ABC kit mixture and DAB (Sigma-Aldrich). Five random fields were digitally photographed and printed so that the number of cells with and without cleaved Notch1 could be counted. The percentage of cleaved Notch1-positive cells was calculated as follows: (No. of stained cells with anti-cleaved Notch1) − (No. of stained cells with control rabbit IgG)/(total No. of cells counted) × 100.
Retroviral infections.
Lineage marker–negative, c-kit–positive cells were obtained from fetal liver embryonic day (E)15.5 by cell sorting with JSAN automatic cell sorter (Bay Bioscience) and infected with the retrovirus encoding the intracellular region of Notch1 (ICN1/MIGR1) or empty vector (MIGR1), as previously described (5).
FTOCs.
The retrovirus-infected fetal liver cells were aliquoted at 2,000/cells well in Terasaki plates (Sumitomo Bakelite), and one deoxyguanosine-treated lobe per well was added. The cells and lobes were incubated as hanging drop cultures for 48 h, and then lobes were removed, rinsed, and cultured on insert filters for 11 d.
Transplantation.
The retrovirus-infected fetal liver cells were injected intravenously into irradiated (450 rad) neonate Dll4lox/loxFoxN1-Cre mice. After 3 wk, thymocytes were obtained and analyzed by flow cytometry.
Online supplemental material.
Fig. S1 shows the construct of FoxN1-Cre transgenic mice. Fig. S2 shows the expression of Dll1 in Dll4-floxed or FoxN1-Cre:Dll4-floxed mice. Fig. S3 shows flow cytometry of fetal or neonatal thymocytes in Dll4-floxed or FoxN1-Cre:Dll4-floxed mice. Fig. S4 shows flow cytometry of peripheral lymphocytes in young-adult (4W) Dll4-floxed or FoxN1-Cre:Dll4-floxed mice. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20080134/DC1.
Supplementary Material
Acknowledgments
We thank N. Abe, D. Suzuki, and Y. Nakano for experimental support; H. Kawamoto, T. Sato, H. Yagita, and R.H. MacDonald for discussions; and the Developmental Studies Hybridoma Bank maintained by The University of Iowa for anti-K8 mAb.
This work was supported by a Grant-in-Aid for Scientific Research (B), a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and 2007 Tokai University School of Medicine Project Research to K. Hozumi.
The authors have no conflicting financial interests.
References
- 1.Anderson, G., and E.J. Jenkinson. 2001. Lymphostromal interactions in thymic development and function. Nat. Rev. Immunol. 1:31–40. [DOI] [PubMed] [Google Scholar]
- 2.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H.R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. [DOI] [PubMed] [Google Scholar]
- 3.Han, H., K. Tanigaki, N. Yamamoto, K. Kuroda, M. Yoshimoto, T. Nakahata, K. Ikuta, and T. Honjo. 2002. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14:637–645. [DOI] [PubMed] [Google Scholar]
- 4.Pui, J.C., D. Allman, L. Xu, S. DeRocco, F.G. Karnell, S. Bakkour, J.Y. Lee, T. Kadesch, R.R. Hardy, J.C. Aster, and W.S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308. [DOI] [PubMed] [Google Scholar]
- 5.Hozumi, K., N. Abe, S. Chiba, H. Hirai, and S. Habu. 2003. Active form of Notch members can enforce T lymphopoiesis on lymphoid progenitors in the monolayer culture specific for B cell development. J. Immunol. 170:4973–4979. [DOI] [PubMed] [Google Scholar]
- 6.Hozumi, K., N. Negishi, D. Suzuki, N. Abe, Y. Sotomaru, N. Tamaoki, C. Mailhos, D. Ish-Horowicz, S. Habu, and M.J. Owen. 2004. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat. Immunol. 5:638–644. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt, T.M., M. Ciofani, H.T. Petrie, and J.C. Zúñiga-Pflücker. 2004. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J. Exp. Med. 200:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailhos, C., U. Modlich, J. Lewis, A. Harris, R. Bicknell, and D. Ish-Horowicz. 2001. Delta4, an endothelial specific Notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation. 69:135–144. [DOI] [PubMed] [Google Scholar]
- 9.Benedito, R., and A. Duarte. 2005. Expression of Dll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr. Patterns. 5:750–755. [DOI] [PubMed] [Google Scholar]
- 10.Heinzel, K., C. Benz, V.C. Martins, I.D. Haidl, and C.C. Bleul. 2007. Bone marrow-derived hematopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J. Immunol. 178:858–868. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt, T.M., and J.C. Zúñiga-Pflücker. 2002. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity. 17:749–756. [DOI] [PubMed] [Google Scholar]
- 12.Hrabe de Angelis, M., J. Mclntyre II, and A. Gossler. 1997. Maintenance of somite borders in mice requires the Delta homologue Dll1. Nature. 386:717–721. [DOI] [PubMed] [Google Scholar]
- 13.Krebs, L.T., J.R. Shutter, K. Tanigaki, T. Honjo, K.L. Stark, and T. Gridley. 2004. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Gene. Dev. 18:2469–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duarte, A., M. Hirashima, R. Benedito, A. Trindade, P. Diniz, E. Bekman, L. Costa, D. Henrique, and J. Rossant. 2004. Dosage-sensitive requirement for mouse Dll4 in artery development. Gene. Dev. 18:2474–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shutter, J.R., S. Scully, W. Fan, W.G. Richards, J. Kitajewski, G.A. Deblandre, C.R. Kintner, and K.L. Stark. 2000. Dll4, a novel Notch ligand expressed in arterial endothelium. Gene. Dev. 14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 16.Novak, A., C. Guo, W. Yang, A. Nagy, and C.G. Lobe. 2000. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 28:147–155. [PubMed] [Google Scholar]
- 17.Porritt, H.E., L.L. Rumfelt, S. Tabrizifard, T.M. Schmitt, J.C. Zúñiga-Pflücker, and H.T. Petrie. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 20:735–745. [DOI] [PubMed] [Google Scholar]
- 18.Izon, D.J., J.A. Punt, L. Xu, F.G. Karnell, D. Allman, P.S. Myung, N.J. Boerth, J.C. Pui, G.A. Koretzky, and W.S. Pear. 2001. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity. 14:253–264. [DOI] [PubMed] [Google Scholar]
- 19.Mancini, S.J.C., N. Mantei, A. Dumortier, U. Suter, H.R. MacDonald, and F. Radtke. 2005. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood. 105:2340–2342. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, R., Y. Lan, H.D. Chapman, C. Shawber, C.R. Norton, D.V. Serreze, G. Weinmaster, and T. Gridley. 1998. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Gene. Dev. 12:1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besseyrias, V., E. Fiorini, L.J. Strobl, U. Zimber-Strobl, A. Dumortier, U. Koch, M.L. Arcangeli, S. Ezine, H.R. MacDonald, and F. Radtke. 2007. Hierarchy of Notch-Delta interactions promoting T cell lineage commitment and maturation. J. Exp. Med. 204:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mombaerts, P., A.R. Clarke, M.A. Rudnicki, J. Iacomini, S. Itohara, J.J. Lafaille, L. Wang, Y. Ichikawa, R. Jaenisch, M.L. Hooper, and S. Tonegawa. 1992. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature. 360:225–231. [DOI] [PubMed] [Google Scholar]
- 23.Tokoro, Y., T. Sugawara, H. Yaginuma, H. Nakauchi, and Y. Takahama. 1998. A mouse carrying genetic defect in the choice between T and B lymphocytes. J. Immunol. 161:4591–4598. [PubMed] [Google Scholar]
- 24.Wilson, A., H.R. MacDonald, and F.R. Radtke. 2001. Notch1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch, U., T.A. Lacombe, D. Holland, J.L. Bowman, B.L. Cohen, S.E. Egan, and C.J. Guidos. 2001. Subversion of the T/B lineage decision in the thymus by lunatic fringe-mediated inhibition of Notch-1. Immunity. 15:225–236. [DOI] [PubMed] [Google Scholar]
- 26.Ordentlich, P., A. Lin, C.P. Shen, C. Blaumueller, K. Matsuno, S. Artavanis-Tsakonas, and T. Kadesch. 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18:2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambandam, A., I. Maillard, V.P. Zediak, L. Xu, R.M. Gerstein, J.C. Aster, W.S. Pear, and A. Bhandoola. 2005. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat. Immunol. 6:663–670. [DOI] [PubMed] [Google Scholar]
- 28.Masuda, K., M. Itoi, T. Amagai, N. Minato, Y. Katsura, and H. Kawamoto. 2005. Thymic anlage is colonized by progenitors restricted to T, NK, and Dendritic cells lineages. J. Immunol. 174:2525–2532. [DOI] [PubMed] [Google Scholar]
- 29.Harman, B.C., W.E. Jenkinson, S.M. Parnell, S.W. Rossi, E.J. Jenkinson, and G. Anderson. 2005. T/B lineage choice occurs prior to intrathymic Notch signaling. Blood. 106:886–892. [DOI] [PubMed] [Google Scholar]
- 30.Maeda, T., T. Merghoub, R.M. Hobbs, L. Dong, M. Maeda, J. Zakrzewski, M.R. van den Brink, A. Zelent, H. Shigematsu, K. Akashi, et al. 2007. Regulation of B versus T lymphoid fate decision by the proto-oncogene LRF. Science. 316:860–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taghon, T.N., E. David, J.C. Zúñiga-Pflücker, and E.V. Rothenberg. 2005. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Gene. Dev. 19:965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis, J. 1998. Notch signaling and the control of cell fate choices in vertebrates. Semin. Cell Dev. Biol. 9:583–589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.