Abstract
Activation-induced cytidine deaminase (AID) is a mutator enzyme that initiates somatic mutation and class switch recombination in B lymphocytes by introducing uracil:guanine mismatches into DNA. Repair pathways process these mismatches to produce point mutations in the Ig variable region or double-stranded DNA breaks in the switch region DNA. However, AID can also produce off-target DNA damage, including mutations in oncogenes. Therefore, stringent regulation of AID is required for maintaining genomic stability during maturation of the antibody response. It has been proposed that AID phosphorylation at serine 38 (S38) regulates its activity, but this has not been tested in vivo. Using a combination of mass spectrometry and immunochemical approaches, we found that in addition to S38, AID is also phosphorylated at position threonine 140 (T140). Mutation of either S38 or T140 to alanine does not impact catalytic activity, but interferes with class switching and somatic hypermutation in vivo. This effect is particularly pronounced in haploinsufficient mice where AID levels are limited. Although S38 is equally important for both processes, T140 phosphorylation preferentially affects somatic mutation, suggesting that posttranslational modification might contribute to the choice between hypermutation and class switching.
The immune system produces a diverse array of antibodies by a series of DNA transactions that require programmed DNA damage. Ig transcription units are assembled by random joining of variable (V), diversity (D), and joining (J) gene segments by a site-specific recombination reaction mediated by RAG1 and RAG2 proteins in developing B cells (1, 2). Later, during immune responses, antigen-specific clones of B cells are selectively expanded in structures called germinal centers (GCs), where they undergo Ig somatic mutation and class switch recombination (CSR) (3, 4). Somatic hypermutation (SHM) introduces nontemplate nucleotide substitutions into the Ig variable gene (5), which can alter the binding affinity of the antibody molecule. If the affinity is enhanced, the resulting B cell clone is selectively expanded, ultimately resulting in affinity maturation of the antibody response (3, 4). CSR is a region-specific deletional recombination reaction that replaces one antibody-constant region for another, thereby altering antibody effector function (6–11). Class switching does not involve the variable regions, and therefore switched antibodies retain their antigenic specificity. Although somatic mutation and class switching are fundamentally different DNA transactions, they are initiated in the nucleus by the same enzyme, activation-induced cytidine deaminase (AID) (12–14), which introduces uracil:guanine mismatches in transcribed single-stranded (ss) Ig DNA (15–19).
In addition to its effects on Igs, AID produces off-target DNA damage, including point mutations in oncogenes such as bcl6 or c-myc (20–23), and it also induces double-stranded DNA breaks in Igs that serve as substrates for chromosome translocations (24–26). DNA damage by AID is minimized in part because AID expression is restricted to activated B cells within GCs (13) by a requirement for PAX5 and E47 transcription factors (27, 28).
AID levels are limiting for CSR and hypermutation (29–31), and they are known to be regulated by microRNA-155, which controls the half-life of AID mRNA (30, 32). In addition, the concentration of AID in the nucleus is limited by a combination of active export and selective nuclear degradation (33–36). Finally, biochemical and tissue culture experiments indicate that a fraction of AID is posttranslationally modified by phosphorylation of serine 38 (S38) and that this modification may also regulate AID activity (29, 37, 38).
S38 is thought to be a target of c-AMP–dependent protein kinase A (PKA) because S38 is part of a PKA consensus site and can be phosphorylated in vitro by PKA (37). Furthermore, coimmunoprecipitation experiments showed that AID is physically associated with PKA (37, 38). In biochemical assays phosphorylation at S38 is essential for AID to associate with replication protein A (RPA), a single stranded DNA binding protein, and the interaction is required for AID to access actively transcribed DNA (37). Consistent with this observation, AID phosphorylated at S38 is enriched in chromatin (29).
Although the role of S38 phosphorylation has not been tested in vivo, mutating S38 to alanine (AIDS38A) results in catalytically intact AID, which may have an altered substrate preference in biochemical assays in vitro (29, 37, 39). Expression of AIDS38A has also been reported to variably decrease CSR assayed in AID-deficient B cells in vitro (10–80% of wild type) (29, 37, 38, 40) and SHM and gene conversion in chicken DT40 B cells (15% of wild type) (41). Further confounding the question of the function of S38 phosphorylation is the fact that zebrafish AID lacks a serine at this position, yet retains activity in CSR and gene conversion (41–43).
In this study, we report on a novel site of AID phosphorylation at threonine 140 (T140) and examine the impact of S38 and T140 phosphorylation on CSR and SHM in vivo.
RESULTS
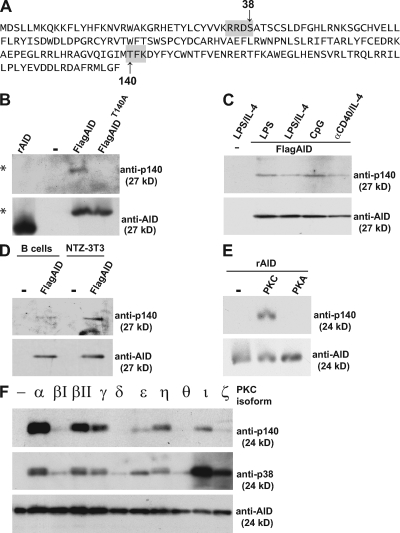
To examine posttranslational modification of AID, we purified the protein from B cells cultured with LPS and IL-4 and subjected the material to mass spectrometry. Analysis of purified AID confirmed phosphorylation at peptides containing S38 (p38), and tyrosine 184 (p184) (29, 37) and revealed additional phosphorylation at T140 (p140; Fig. 1 A).
Figure 1.
AID is phosphorylated at position T140. (A) Amino acid sequence of AID showing the location of S38 and T140 within consensus PKA and PKC sites, respectively (gray boxes). (B) Anti-p140 or -AID immunoblot of recombinant AID (rAID) purified from E. coli or FLAG-tagged AID or AID-T140A immunoprecipitated from retroviral infected AID−/− B cells (* denotes 27-kD FLAG-tagged AID). (C) Anti-p140 and -AID immunoblot of FLAG-tagged AID purified from B cells stimulated with LPS, LPS and IL-4, CpG, or anti-CD40 and IL-4. (D) Anti-p140 and -AID immunoblot of FLAG-tagged AID purified from B cells or NTZ-3T3 cells. (E) Anti-p140 and -AID immunoblot of rAID untreated (-) or treated with PKC or PKA in vitro. (F) Anti-p140, -p38, and -AID immunoblot of rAID untreated (-) or in vitro phosphorylated with the indicated PKC isoforms.
To confirm that AID is phosphorylated at T140 in vivo, we produced AID-p140 phosphospecific antibodies (anti-p140). Anti-p140 was reactive with AID purified from B cells stimulated with LPS and IL-4, but was not reactive with AID-T140A (Fig. 1 B). The level of anti-p140 reactivity in cultured B cells differed depending on the stimulus with the highest levels found with CpG (Fig. 1 C). However, phosphorylation at AID-T140 was not B cell specific and could also be detected in NIH-3T3 NTZ cells expressing AID (Fig. 1 D). Finally, anti-p140 did not react with recombinant AID produced in bacteria, but was reactive with AID phosphorylated in vitro with protein kinase C (PKC) catalytic subunit and not PKA (Fig. 1 E). The PKC family is composed of at least 10 serine/threonine kinases, and anti-p140 and -p38 was reactive to varying degrees with recombinant AID after in vitro phosphorylation with the different isoforms (Fig. 1 F). We conclude that AID is phosphorylated at position T140 and that this site of posttranslational modification resembles S38 in that it is not B cell specific.
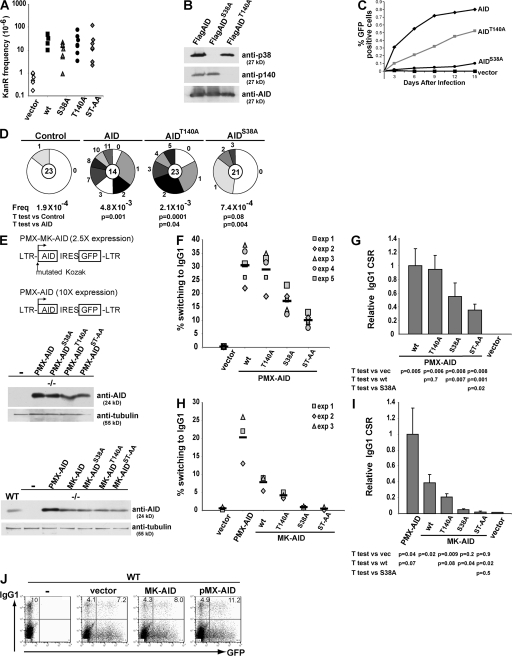
To determine whether T140 is essential for catalytic activity, we compared wild-type and AID-T140A for their ability to revert an inactivating point mutation in a kanamycin resistance encoding plasmid in Escherichia coli (19). Reversion of the point mutation (CCAP94 to CTAL94) by cytidine deamination confers kanamycin resistance, which is assayed by colony formation (19). AID-T140A, AID-S38A, and the double mutant (AID-ST/AA) were indistinguishable from wild type in this assay (Fig. 2 A) (29). Thus, these mutations do not alter AID catalytic activity as assayed in E. coli.
Figure 2.
Comparison of AID, AIDT140A, and AIDS38A hypermutation and CSR activity in B and non–B cells. (A) The graph shows a log plot of numbers of kanamycin-resistant (KanR) colonies after induction of AID, AIDS38A, AIDT140A, AIDST-AA, or empty vector expression. (B) Anti-p140, -p38, or -AID immunoblot of FLAG-tagged AID, AIDS38A, or AIDT140A purified from B cells stimulated with LPS and IL-4. (C) Accumulation of GFP-expressing 3T3-NTZ cells after transduction with AID-, AIDS38A-, or AIDT140A-expressing PMX-MK retroviruses. The x axis indicates the number of days after transduction, and the y axis indicates the percentage of GFP-positive cells measured by flow cytometry. (D) Number of mutations in the GFP gene cloned from 3T3-NTZ cells 11 d after transduction with retroviruses encoding AID, AIDS38A, AIDT140A, or control. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated in the periphery of the charts. The total number of independent sequences analyzed is indicated in the center of each chart. Statistical significance was determined by a two-tailed Student's t test assuming unequal variance and comparing AID expressing with AIDS38A- and AIDT140A-expressing cells. P values are indicated. The numbers of point mutations were as follows: 4 mutations/21,643 bp mutations for vector; 59 mutations/13,174 bp for AID; 15 mutations/19,761 bp for AIDS38A; and 45 mutations/21,643 bp for AIDT140A. (E) Schematic of retroviral constructs used (top); anti-AID immunoblot of AID−/− B cells infected with PMX-AID, or -AIDS38A, or -AIDT140A, or –AIDST-AA (middle); or wild-type B cells (WT) or AID−/− B cells infected with PMX-AID, MK-AID, or -AIDS38A, or -AIDT140A, or –AIDST-AA (bottom). Anti-tubulin was used as a loading control. (F) Graph of flow cytometric analysis of IgG1 expression in AID−/− B cells transduced with PMX-AID, -AIDS38A, -AIDT140A, -AIDST-AA, or control vector and cultured in LPS plus IL-4 for 3 d after infection. The percentage of IgG1+ cells is indicated for five independent experiments, and the mean is indicated by a solid line. (G) Relative efficiency of isotype switching to IgG1. A comparison of PMX-AID versus AIDS38A, AIDT140A, AIDST-AA, or vector control. Bars represent the mean and SD from the five independent experiments in F. (H) Graph of flow cytometric analysis of IgG1 expression in AID−/− B cells transduced with PMX-AID, MK-AID, -AIDS38A, -AIDT140A, -AIDST-AA, or control vector and cultured in LPS plus IL-4 for 3 d after infection. The percentage of IgG1+ cells is indicated for three independent experiments, and the mean is indicated by a solid line. (I) Relative efficiency of isotype switching to IgG1. Bars represent the mean and SD from H. (J) Flow cytometric analysis of IgG1 expression in wild-type B cells alone or transduced with MK-AID, PMX-AID, or control vector and cultured in LPS plus IL-4 for 3 d after infection. The percentage of GFP− or GFP+ that were IgG1+ is indicated on the top left or right of each graph, respectively. Graphs are representative of three independent experiments.
To determine whether there is interdependence between S38 and T140 phosphorylation, we expressed AID, AID-S38A, or AID-T140A in stimulated AID−/− B cells and performed Western blotting experiments with anti-p140 and -p38 antibodies. We found that AID-S38A was normally phosphorylated at position T140 and, conversely, that AID-T140A was normally phosphorylated at S38. We conclude that the two sites of phosphorylation are not interdependent (Fig. 2 B).
AID is phosphorylated in 3T3 fibroblasts (Fig. 1 D) and mutates indicator substrates when it is expressed in these cells. Mutator activity can be measured by reversion of a green fluorescent protein indicator containing a premature stop codon (NIH-3T3 NTZ cells [44]). When assayed by FACS or sequencing, AID-S38A and -T140A showed <15 and <50% of wild-type levels, respectively (Fig. 2, C and D) (29). We conclude that T140 is required for normal levels of mutation in NIH-3T3 NTZ cells in vitro.
AID can be assayed for its ability to induce CSR in B cells in vitro by retroviral complementation of AID-deficient B cells stimulated with LPS and IL-4 (45). Because AID expression levels may impact on the results of the assay, we used two retroviral vectors that directed different levels of AID expression (Fig. 2 E). B cell retroviral infection with PMX-AID (25) or MK-AID, which contains a mutant Kozak sequence, results in AID levels that are 10- or 2.5-fold higher than physiological levels, respectively (Fig. 2 E). Consistent with the high levels of AID expression, B cells infected with PMX-AID switched to IgG1 at higher efficiency than cells infected with MK-AID (Fig. 2, F and H). AID mutants showed varying degrees of activity in this assay (Fig. 2, F and H). High levels of AID-T140A expression reconstituted nearly 95% of control levels of class switching (Fig. 2, F and G). In contrast, lower levels of AID-T140A expression resulted in 50% of control levels of switching (Fig. 2, H and I). As previously reported, AID-S38A mutants varied in activity depending on expression levels ranging from nearly undetectable to 60% of control levels (Fig. 2, G and I) (29, 37, 38, 40). High-level expression of the double mutant, AID-ST/AA, resulted in a modest drop in class switching compared with AID-S38A, 35 versus 55% of control levels of switching, respectively, which is consistent with an additive effect of the two mutations (Fig. 2, F and G). We conclude that phosphorylation of S38 and T140 are important for class switching in LPS- and IL-4–stimulated B cells. Furthermore, higher levels of AID expression drive higher levels of switching to IgG1 and diminish the requirement for S38 or T140 phosphorylation. Therefore, the precise contribution of these posttranslational modifications to AID function is difficult to evaluate in this assay.
To determine if AID protein levels are a rate-limiting factor in CSR, we augmented AID levels in wild-type LPS- and IL-4–stimulated B cells with retrovirally expressed AID and measured isotype switching to IgG1. Compared with wild type alone or wild type transduced with empty PMX retrovirus, cells supplemented with AID from the PMX-AID retrovirus displayed a >40% increase in isotype switching (Fig. 2 J). We conclude that AID levels are rate limiting and that superphysiological levels can drive higher rates of isotype switching.
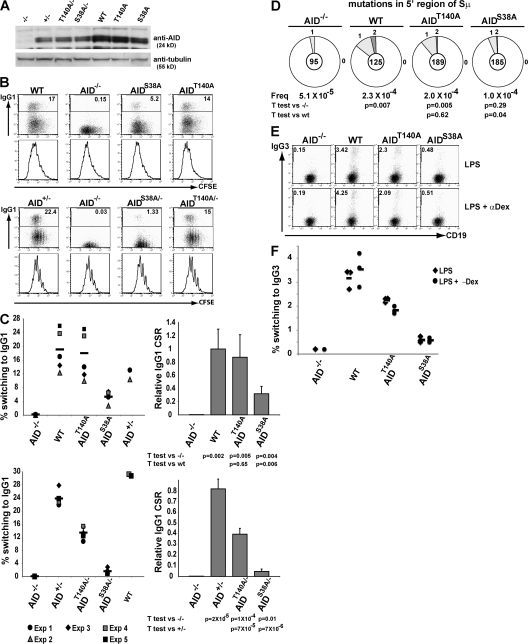
To examine the physiological function of AID phosphorylation at positions S38 and T140, we produced mice that carry S38A or T140A mutations in AID, AIDS38A, and AIDT140A, respectively (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20081319/DC1). AIDS38A and AIDT140A mutant B cells developed normally (not depicted) and were assayed for CSR to IgG1 after stimulation with LPS and IL-4 in culture. Cell division was monitored by labeling cells with CFSE and tracking dye dilution by flow cytometry. AID protein levels expressed by wild-type, AIDS38A, and AIDT140A B cells were indistinguishable when measured by Western blotting (Fig. 3 A), and heterozygous AID+/−, AIDS38A/−, and AIDT140A/− expressed half as much AID as their homozygous counterparts (Fig. 3 A). AIDS38A and AIDT140A B cells divided normally in response to LPS and IL-4, but were impaired in switch recombination to IgG1 (Fig. 3, B and C). AIDS38A mutant B cells showed 32% the level of CSR to IgG1 of wild-type controls, whereas AIDT140 produced a milder defect resulting in 87% of wild-type CSR (Fig. 3, B and C). In both cases, the defect was exacerbated in haploinsufficient mice. Although AID+/− B cells displayed IgG1 switching, 80% of wild-type B cells, AIDS38A/−, and AIDT140A/− showed 4 and 45% the level of wild-type B cells, respectively (Fig. 3, B and C). This relative decrease was consistent within each experiment, despite overall switching rates varying from experiment to experiment (Fig. 3, B and C). We also measured mutation within the region 5′ of the Sμ switch repeats. Similar to CSR, AIDS38A and AIDT140A had mutation rates of 25 and 85%, respectively, compared with WT (Fig. 3 D). Because different stimulation conditions may result in differential levels of AID phosphorylation (Fig. 1 C), we measured isotype switching to IgG3. After stimulation with LPS or LPS and anti-dextran in culture, AIDS38A B cells showed 19 or 16% the level of CSR to IgG3 of wild-type controls, whereas AIDT140A produced a milder defect resulting in 71 or 52% wild-type CSR (Fig. 3, E and F). We conclude that both AID-S38 and -T140 phosphorylation are required for physiological levels of CSR, but neither is essential for this reaction.
Figure 3.
CSR in AIDS38A and AIDT140A mice. (A) Anti-AID immunoblot of AID−/−, AID+/−, AIDT140A/−, AIDS38A/−, wild-type (WT), AIDT140A, and AIDS38A B cells stimulated with LPS and IL-4. Anti-tubulin immunoblot was used as a loading control. (B) Flow cytometric analysis of IgG1 expression and CFSE dye dilution by WT, AID−/−, AIDT140A, and AIDS38A (top) or AID+/−, AID−/−, AIDT140A/−, or AIDS38A/− B cells (bottom) stimulated with LPS and IL-4. The percentage of IgG1+ cells is indicated on the top right of each graph. (C) Isotype switching to IgG1 by WT, AIDT140A, and AIDS38A (top) or AIDT140A/−, AIDS38A/−, and AID+/− cells (bottom) as in B for the indicated number of independent experiments. Solid lines represent means. Bar graphs represent the mean relative efficiency and SD of IgG1 CSR compared with WT. (D) Number of mutations in the 5′ of Sμ region cloned from AID−/−, WT, AIDT140A, and AIDS38A B cells stimulated with LPS and IL-4 sorted for IgM expression and five cell divisions. Pie charts and statistical analysis, as in Fig. 2 D, represent summary of two experiments. The numbers of point mutations were as follows: 3 mutations/59,185 bp mutations for AID−/−; 18 mutations/77,875 bp for WT; 11 mutations/115,255 bp for AIDS38A; and 24 mutations/117,747 bp for AIDT140A. (E) Flow cytometric analysis of IgG3 expression in AID−/−, WT, AIDT140A, and AIDS38A B cells stimulated with LPS or LPS and anti-dextran for 6 d. The percentage of IgG3+ cell is indicated on the top left of each graph. (F) Results of E from three independent experiments. Solid line represents the mean.
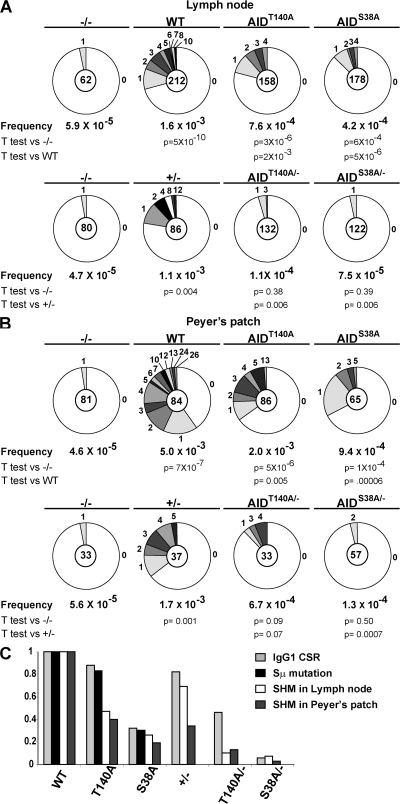
To examine the role of AID phosphorylation in SHM, we cloned and sequenced the DNA region downstream of IgJH4 from purified lymph node GC B cells (46). The effect of AIDS38A on somatic mutation was similar to CSR, resulting in 30% of wild-type activity (Fig. 4 A). In contrast, AIDT140A had more profound effects on somatic mutation than CSR, resulting in 45% of wild-type activity, which was not significantly different from AIDS38A (Fig. 4 A). Similar results were obtained from Peyer's patch B cells, where mutation rates for AIDS38A and AIDT140A were 20 and 40% of controls, respectively (Fig. 4 B). Haploinsufficiency by itself resulted in a mild decrease in hypermutation with 70% hypermutation activity in AID+/− versus wild type (Fig. 4 A). Haploinsufficiency magnified the defect in hypermutation of AIDS38A/− and AIDT140A/−, with neither statistically rising above the background levels of mutation found in AID−/− lymph node GC B cells (Fig. 4 A). Similar, but slightly less pronounced, effects were found in chronically stimulated Peyer's patch GC B cells (Fig. 4 B). We conclude that both AID-S38 and -T140 phosphorylation are required for optimal somatic mutation and that the T140 has a more profound effect on this reaction than on CSR (Fig. 4 C).
Figure 4.
SHM in GC B cells from AIDS38A and AIDT140A mice. (A) GC B cells were purified from the lymph nodes of 5 immunized AID−/−, AID+/−, AIDT140A/−, AIDS38A/−, WT, AIDT140A, or AIDS38A mice. Pie charts indicated the number of mutations in the intronic region 3′ of JH4. Pie charts and statistical analysis as in 2D. The numbers of point mutations were as follows: 2 mutations/33,666 bp mutations for AID−/−; 184 mutations/115,116 bp for WT; 41 mutations/96,654 bp for AIDS38A; and 65 mutations/85,794 bp for AIDT140A (top). 2 mutations/43,440 bp mutations for AID−/−; 51 mutations/46,689 bp for AID+/−; 5 mutations/66,246 bp for AIDS38A/−; and 8 mutations/71,676 bp for AIDT140A/− (bottom). (B) GC B cells purified from Peyer's patch analyzed the same as A. The numbers of point mutations were as follows: 2 mutations/43,983 bp mutations for AID−/−; 227 mutations/45,612 bp for WT; 33 mutations/35,295 bp for AIDS38A; and 95 mutations/46,698 bp for AIDT140A (top). 1 mutation/179,191 bp mutations for AID−/−; 34 mutations/20,091 bp for AID+/−; 4 mutations/31,494 bp for AIDS38A/−; and 12 mutations/17,919 bp for AIDT140A/− (bottom). (C) Summary of efficiency of both SHM and isotype switching to IgG1 relative to wild-type by AIDT140A/−, AIDS38A/−, AIDT140A,and AIDS38A/AID+/− cells. Bars represent the means from and statistics are reported in Figs. 3 C, 3 D, and 4 [A and B].
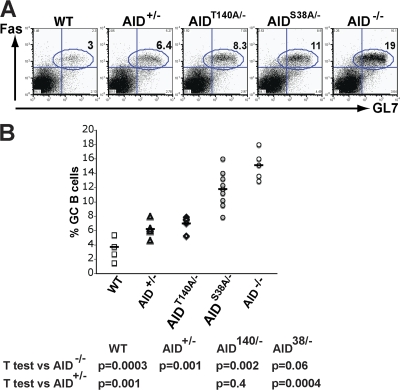
AID-deficient mice and humans have large GCs compared with controls (12, 14). To determine whether this effect is caused by loss of AID protein or its activity, we measured the number of GC B cells in AID−/−, AID+/−, AIDT140A/−, AIDS38A/−, and wild-type mice (Fig. 5, A and B). We found that the amount of AID activity was inversely proportional to the size of the GC response. AID−/− mice showed the highest number of GC B cells, wild-type mice the fewest, and AID+/− haploinsufficient mice were intermediate between the two (Fig. 5, A and B). Decreasing AID activity, but not protein, in AIDS38A/− mice resulted in an increase in the number of GC B cells proportional with the relative decrease in activity when compared with AID+/− controls (Fig. 5, A and B). We conclude that the number of GC B cells in immunized mice is inversely proportional to the amount of AID activity.
Figure 5.
Percentage of GC B cells from AIDS38A and AIDT140A mice. (A) Representative FACS analysis that shows percentage of FAS+GL7+ GC B cells in AID−/−, AID+/−, AIDT140A/−, AIDS38A/−, and wild-type (WT) mice 10 d after immunization. (B) Percentage of GC B cells in the lymph nodes of AID−/−, AID+/−, AIDT140A/−, AIDS38A/−, and WT mice 10 d after immunization from 6–8 mice. Each point represents an immunization experiment from a single mouse.
DISCUSSION
Mice and humans deficient in AID have larger GCs than controls (12, 14). In this study, we observe larger GCs in AID-haploinsufficient mice together with further enlargement in AIDS38A/− mice, indicating that the GC phenotype is directly related to the level of AID activity. AID activity might impact GC size by a variety of mechanisms, including antibody gene or generalized genomic damage. For example, increased AID activity could result in higher rates of detrimental antibody gene mutation, leading to loss of affinity, loss of expression, or development of self-reactive antibodies (3, 4). Conversely, higher rates of off-target genomic damage by AID may lead to mutations that result in cell death (47). Irrespective of the mechanism, the magnitude of the effect of AID levels on GC size emphasizes the importance of AID regulation in vivo.
Phosphorylation of AID at position S38 is believed to facilitate the interaction between AID and RPA, a cofactor that is required for AID to access transcribed DNA in vitro (37). RPA binds to ssDNA and it was proposed that RPA might stabilize ssDNA liberated during transcription to serve as a substrate for AID (48). Consistent with this idea, AID-S38A is inactive on transcribed dsDNA in the absence of RPA, while retaining catalytic activity on ssDNA (37). Nevertheless, AID-S38A is active at between 20 and 35% of wild-type levels in hypermutation and CSR in vivo. One way to reconcile these results would be to posit that association between AID and RPA is only required for a subset of AID substrates that do not stabilize ss regions spontaneously. For example, ssDNA in switch regions is stabilized spontaneously by R-loops, but this does not occur in Ig variable region DNA. This model might predict that the S38A mutation would preferentially interfere with Ig variable region hypermutation (49, 50). However, the AIDS38A mutation affects switching and mutation equally; therefore, the mechanism by which S38 phosphorylation alters AID function is likely to be independent of whether the ssDNA substrate is created by R-loops. An alternative nonexclusive explanation for our results is that S38 phosphorylation enhances AID activity by facilitating its interaction with chromatin (29).
Biochemical studies with AID purified from insect cells assayed on artificial substrates showed that S38A mutation does not alter overall AID activity, but shifts target preference in a way that might result in specific loss of activity on hotspots found in the Ig switch region (39). This in vitro bias is not consistent with our mutation data in transfected fibroblasts or GC B cells because we find lower levels of SHM in all cases and the selective hotspot bias found in the in vitro transcription systems would not affect the rate of SHM (29).
Previous coimmunoprecipitation and in vitro kinase experiments suggest that PKA phosphorylates AID at S38; the importance of this kinase in regulating AID is supported by the finding that inhibition of its activity in B cells inhibits class switching (37, 38). Our experiments indicate that T140 is not a PKA target site, and that S38 can be phosphorylated by either PKA or PKC in vitro (Fig. 1 E). In addition, PKC family members overlap in their preference to phosphorylate T140 or S38 in vitro (Fig. 1 F) and S38 or T140 phosphorylation is nonexclusive (Fig. 2 B). Thus, AID activity may be modulated by phosphorylation on one or both sites, depending on which kinase is activated. The PKC family is composed of at least 10 serine/threonine kinases, many of which are expressed in activated B cells (51, 52). They have been implicated in a wide variety of cellular processes, including growth, differentiation, tolerance, immunity, and tumor development (51). Which family members phosphorylate AID and the precise pathways that regulate phosphorylation in vivo remains to be determined.
How T140 phosphorylation modulates AID activity is not known, but this modification differs from S38 in that it preferentially affects somatic mutation. Differential regulation of class switching and somatic mutation is a well-characterized physiological feature of gene diversification in B cells. For example, B cells undergoing switch recombination in response to LPS and IL-4 in vitro do not mutate their Ig variable regions. This cannot be ascribed to a general absence of the factors that mediate hypermutation because the DNA 3′ of the Iμ promoter and the switch regions themselves are mutated by AID in LPS- and IL-4–stimulated B cells. Indeed, the amount of mutation in switch regions in AID-T140A–expressing B cells is directly proportional to the amount of class switching and differs from Ig variable region gene mutation, which is more severely affected (Fig. 4 C). One explanation for the disparity between the effects of AID-T140A on switching and mutation would be that there are specific requirements for AID in the two reactions. This idea is supported by the finding that mutations in the amino or carboxyl terminal regions of AID preferentially affect somatic mutation or class switching (45, 53). Finding that phosphorylation at T140 preferentially affects somatic mutation suggests a mechanism by which AID activity on closely related substrates might be differentially regulated.
AID activity is regulated by several mechanisms, including restricted expression, mRNA stability, nuclear degradation, nuclear export, and multiple phosphorylation sites. Although each of these alone appears to have a small effect on CSR and SHM, the combined effects are dramatic. For example AID haploinsufficiency supports rates of 80 and 70% of wild type in CSR and lymph node SHM, respectively (Fig. 4 C). However, CSR is decreased to 5% of AID+/− controls, and lymph node SHM was at background levels in AIDS38A/− mice (Fig. 4 C). Similarly, AIDT140A/− B cells were severely, but selectively, impaired in lymph node SHM (10% of AID+/− vs. 57% for CSR to IgG1; Fig. 4 C). Peyer's patch B cells supported SHM levels nearly three times higher then lymph node. In this case, SHM rates in AID+/− and AIDT140A/− resemble those of AID and AIDT140A in the lymph node (Fig. 4, A and B). Thus, AID appears to be controlled by multiple potentially overlapping mechanisms. We speculate that this type of combinatorial regulation facilitates fine control of AID levels, which is required because small imbalances in its expression can result in catastrophic effects on genomic stability (25).
MATERIALS AND METHODS
Mice.
To produce AIDS38A and AIDT140A mice, AID S38 or T140 was mutated to alanine using the same gene-targeting strategy (Fig. S1). The long arm of the targeting vector was 5 kb long with 3′ within the intron between AID exons 2 and 3 (Fig. S1). The short arm was a 3.3-kb fragment extending with the 3′ into the intron between exons 3 and 4. A LoxP-flanked neomycin-resistance gene was used for positive selection, and a diphtheria toxin gene was used for negative selection. The targeting construct was linearized and transfected into C57BL/6 embryonic stem cells. The genotype was confirmed by Southern blot and PCR amplification using a primer outside of the targeting construct, followed by sequencing. Mice were crossed to AID−/− C57BL/6 mice (12) to produce AID+/−, AIDS38A/−, or AIDT140A/− mice used for analysis. The presence of mutants were confirmed by RT-PCR and sequencing of AID mRNA. All experiments with mice were done according to protocols approved by the Rockefeller University Institutional Animal Care and Use Committee.
Lymphocyte isolation and culturing and retroviral infection.
Lymphocyte isolation, cultures, retrovirus infection, and analysis were previously described (29). The PMX-MK-AID was the same as the previously described PMX-AID, except the Kozak CCACCATGG was changed to GGTTTATGG. B cells were purified from mouse spleens by depletion with anti-CD43 beads (Miltenyi Biotec) and cultured in 25 μg/ml LPS (Sigma-Aldrich) alone for switching to IgG3 or with 5 ng/ml IL-4 (Sigma-Aldrich) for IgG1or with 2.5 ng/ml anti-α–δ-dextran (Fina BioSolutions) for IgG3. Cells were stained with CSFE and/or with biotinylated anti–mouse IgG1 or IgG3 antibodies and streptavidin APC (BD Biosciences).
PCR and mutation analysis.
For SHM analysis, age-matched 8–16-wk-old mice were immunized by footpad injected with 50 μg of NP-CGG (Biosearch Technologies) precipitated in alum and lymph node, and Peyer's patches were dissected 11 d after immunization. GC B cells were stained with APC anti-CD19, FITC anti-GL7, and PE anti-FAS antibodies (BD Biosciences) and purified by cell sorting. In two independent experiments, DNA samples were pooled from 2 or 3 separate mice. To amplify the intronic region 3′ of JH4, four separate PCR reactions were performed on each sample with PFU polymerase (Stratagene) on genomic DNA from 5,000 equivalent cells. Amplified products were pooled, cloned, and sequenced. The intron region 3′ of JH4 was amplified with a common VHJ558 family primer (5′-GGAATTCGCCTGACATCTGAGGACTCTGC-3′) and (5′-CTGGACTTTCGGTTTGGTG-3′) for 9 cycles at 94°C (30 s), at 55°C (30 s), and at 72°C (90 s), and then (5′-GGTCAAGGAACCTCAGTCA-3′) and (5′-TCTCTAGACAGCAACTAC-3′) for 21 cycles at 94°C (30 s), at 55°C (30 s), and at 72°C (30 s). For 5′μ switch region analysis after 96-h LPS/IL-4 stimulation, IgM+ cells from two mice that CFSE labeling indicated had divided 5 times were sorted. Conditions and primers were previously described (29). The NTZ-3T3 assay and GFP gene mutational analysis were performed as previously described 11 d after retrovirus infection (33, 44). PCR products were cloned into TOPO-TA cloning kit (Invitrogen) and sequenced with T7 primer. E. coli assays were performed exactly as previously described (19).
Protein analysis.
Anti-AID and -p38 antibodies were previously described (29). To produce anti-p140 antibodies, rabbits were immunized with phosphopeptide GVQIGIM(pT)FKDYFYC (AID 133–148) coupled to keyhole limpet hemocyanin. Phosphospecific antibodies were purified by negative selection on unphosphorylated peptide AID 133–148 coupled to Sulfolink gel (Thermo Fisher Scientific), followed by positive selection on the phosphopeptide. Cells were extracted in lysis buffer (20 mM Tris, pH 8.0, 400 mM NaCl, 1% Nonidet P-40, 0.5 mM EDTA, 25 mM NaF, and 1 mM DTT). For immunoprecipitation, 2 mg of extracts were incubated with anti-AID antibody and protein A–Sepharose (GE Healthcare) for 2 h. For FLAG immunoprecipitation, anti-FLAG agarose beads (Sigma-Aldrich) were incubated with extracts for 2 h and AID was eluted with 0.5 μg/ml of FLAG peptide (Sigma-Aldrich) in lysis buffer. Western blots were performed on immunoprecipitated protein or on 50 μg of total cell extracts with the indicated anti-AID antibody; anti-tubulin (Abcam) was used as a loading control. To compare AID levels from retroviral-infected B cells, Western blots were performed on cells sorted for GFP expression. AID purification and mass spectrometry analysis of phosphorylation was performed as previously described on extracts from 109 wild-type B cells purified from mouse spleens by depletion with anti-CD43 beads (Miltenyi Biotec) and cultured in RPMI medium, 10% FBS, 5 ng/ml IL-4 (Sigma-Aldrich), and 25 μg/ml LPS (Sigma-Aldrich) for 72 h (29).
In vitro phosphorylation.
100 ng of purified recombinant AID protein was incubated with PKA as previously described (29) or 0.1 U of PKC catalytic subunit or isoforms (Calbiochem) for 30 min at 30°C in a buffer containing 40 mM MES, pH 6.0, 1 mM EGTA, 10 mM MgCl2, 1 mM DTT, and 0.1 mM ATP.
Online supplemental material.
Fig. S1 shows gene-targeting strategy to produce AIDS38A and AIDT140A mice. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081319/DC1.
Supplementary Material
Acknowledgments
We thank the members of the Nussenzweig laboratory, K. Velizon, and T. Shengelia for cell sorting, and T. Eisenreich for mice breeding.
This work was supported in part by National Institutes of Health grants (to B.T. Chait and M.C. Nussenzweig). M.C. Nussenzweig is a Howard Hughes Medical Institute Investigator.
The authors have no conflicting financial interests.
Abbreviations used: AID, activation-induced cytidine deaminase; CSR, class switch recombination; GC, germinal center; PKA, protein kinase A; PKC, protein kinase C; s38, serine 38; SHM, somatic hypermutation; ss, single-stranded; T140, threonine 140.
References
- 1.Fugmann, S.D., A.I. Lee, P.E. Shockett, I.J. Villey, and D.G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495–527. [DOI] [PubMed] [Google Scholar]
- 2.Jung, D., C. Giallourakis, R. Mostoslavsky, and F.W. Alt. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570. [DOI] [PubMed] [Google Scholar]
- 3.Meffre, E., R. Casellas, and M.C. Nussenzweig. 2000. Antibody regulation of B cell development. Nat. Immunol. 1:379–385. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky, K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. [DOI] [PubMed] [Google Scholar]
- 5.McKean, D., K. Huppi, M. Bell, L. Staudt, W. Gerhard, and M. Weigert. 1984. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 81:3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barreto, V.M., A.R. Ramiro, and M.C. Nussenzweig. 2005. Activation-induced deaminase: controversies and open questions. Trends Immunol. 26:90–96. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri, J., U. Basu, A. Zarrin, C. Yan, S. Franco, T. Perlot, B. Vuong, J. Wang, R. Phan, A. Datta, et al. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94:157–214. [DOI] [PubMed] [Google Scholar]
- 8.Di Noia, J.M., and M. Neuberger. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22. [DOI] [PubMed] [Google Scholar]
- 9.Honjo, T., H. Nagaoka, R. Shinkura, and M. Muramatsu. 2005. AID to overcome the limitations of genomic information. Nat. Immunol. 6:655–661. [DOI] [PubMed] [Google Scholar]
- 10.Li, Z., Z. Luo, D. Ronai, F. Kuang, J. Peled, M. Iglesias-Ussel, and M. Scharff. 2007. Targeting AID to the Ig genes. Adv. Exp. Med. Biol. 596:93–109. [DOI] [PubMed] [Google Scholar]
- 11.Stavnezer, J., and C.E. Schrader. 2006. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 22:23–28. [DOI] [PubMed] [Google Scholar]
- 12.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 13.Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. [DOI] [PubMed] [Google Scholar]
- 14.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 15.Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F.W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. [DOI] [PubMed] [Google Scholar]
- 17.Dickerson, S.K., E. Market, E. Besmer, and F.N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single-stranded DNA. J. Exp. Med. 197:1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham, P., R. Bransteitter, J. Petruska, and M.F. Goodman. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107. [DOI] [PubMed] [Google Scholar]
- 19.Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. [DOI] [PubMed] [Google Scholar]
- 20.Pasqualucci, L., A. Migliazza, N. Fracchiolla, C. William, A. Neri, L. Baldini, R.S. Chaganti, U. Klein, R. Kuppers, K. Rajewsky, and R. Dalla-Favera. 1998. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA. 95:11816–11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, M., J.L. Duke, D.J. Richter, C.G. Vinuesa, C.C. Goodnow, S.H. Kleinstein, and D.G. Schatz. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845. [DOI] [PubMed] [Google Scholar]
- 22.Pasqualucci, L., P. Neumeister, T. Goossens, G. Nanjangud, R.S. Chaganti, R. Kuppers, and R. Dalla-Favera. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346. [DOI] [PubMed] [Google Scholar]
- 23.Shen, H.M., A. Peters, B. Baron, X. Zhu, and U. Storb. 1998. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 280:1750–1752. [DOI] [PubMed] [Google Scholar]
- 24.Dorsett, Y., D. Robbiani, M. Jankovic, B. Reina-San-Martin, T. Eisenreich, and M. Nussenzweig. 2007. A role for AID in chromosome translocations between c-myc and the IgH variable region. J. Exp. Med. 204:2225–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramiro, A.R., M. Jankovic, E. Callen, S. Difilippantonio, H.T. Chen, K.M. McBride, T.R. Eisenreich, J. Chen, R.A. Dickins, S.W. Lowe, et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramiro, A.R., M. Jankovic, T. Eisenreich, S. Difilippantonio, S. Chen-Kiang, M. Muramatsu, T. Honjo, A. Nussenzweig, and M.C. Nussenzweig. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438. [DOI] [PubMed] [Google Scholar]
- 27.Gonda, H., M. Sugai, Y. Nambu, T. Katakai, Y. Agata, K.J. Mori, Y. Yokota, and A. Shimizu. 2003. The balance between Pax5 and Id2 activities is the key to AID gene expression. J. Exp. Med. 198:1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayegh, C.E., M.W. Quong, Y. Agata, and C. Murre. 2003. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 4:586–593. [DOI] [PubMed] [Google Scholar]
- 29.McBride, K.M., A. Gazumyan, E.M. Woo, V.M. Barreto, D.F. Robbiani, B.T. Chait, and M.C. Nussenzweig. 2006. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl. Acad. Sci. USA. 103:8798–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorsett, Y., K.M. McBride, M. Jankovic, A. Gazumyan, T.H. Thai, D.F. Robbiani, M. Di Virgilio, B.R. San-Martin, G. Heidkamp, T.A. Schwickert, et al. 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 28:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takizawa, M., H. Tolarova, Z. Li, W. Dubois, S. Lim, E. Callen, S. Franco, M. Mosaico, L. Feigenbaum, F.W. Alt, et al. 2008. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J. Exp. Med. 205:1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng, G., P. Hakimpour, P. Landgraf, A. Rice, T. Tuschl, R. Casellas, and F.N. Papavasiliou. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 28:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride, K.M., V. Barreto, A.R. Ramiro, P. Stavropoulos, and M.C. Nussenzweig. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito, S., H. Nagaoka, R. Shinkura, N. Begum, M. Muramatsu, M. Nakata, and T. Honjo. 2004. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA. 101:1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brar, S.S., M. Watson, and M. Diaz. 2004. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 279:26395–26401. [DOI] [PubMed] [Google Scholar]
- 36.Aoufouchi, S., A. Faili, C. Zober, O. D'Orlando, S. Weller, J.C. Weill, and C.A. Reynaud. 2008. Proteasomal degradation restricts the nuclear lifespan of AID. J. Exp. Med. 205:1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basu, U., J. Chaudhuri, C. Alpert, S. Dutt, S. Ranganath, G. Li, J.P. Schrum, J.P. Manis, and F.W. Alt. 2005. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 438:508–511. [DOI] [PubMed] [Google Scholar]
- 38.Pasqualucci, L., Y. Kitaura, H. Gu, and R. Dalla-Favera. 2006. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl. Acad. Sci. USA. 103:395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pham, P., M.B. Smolka, P. Calabrese, A. Landolph, K. Zhang, H. Zhou, and M.F. Goodman. 2008. Impact of phosphorylation and phosphorylation-null mutants on the activity and deamination specificity of activation-induced cytidine deaminase. J. Biol. Chem. 283:17428–17439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinkura, R., I.M. Okazaki, T. Muto, N.A. Begum, and T. Honjo. 2007. Regulation of AID function in vivo. Adv. Exp. Med. Biol. 596:71–81. [DOI] [PubMed] [Google Scholar]
- 41.Chatterji, M., S. Unniraman, K.M. McBride, and D.G. Schatz. 2007. Role of activation-induced deaminase protein kinase A phosphorylation sites in Ig gene conversion and somatic hypermutation. J. Immunol. 179:5274–5280. [DOI] [PubMed] [Google Scholar]
- 42.Barreto, V.M., Q. Pan-Hammarstrom, Y. Zhao, L. Hammarstrom, Z. Misulovin, and M.C. Nussenzweig. 2005. AID from bony fish catalyzes class switch recombination. J. Exp. Med. 202:733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakae, K., B.G. Magor, H. Saunders, H. Nagaoka, A. Kawamura, K. Kinoshita, T. Honjo, and M. Muramatsu. 2006. Evolution of class switch recombination function in fish activation-induced cytidine deaminase, AID. Int. Immunol. 18:41–47. [DOI] [PubMed] [Google Scholar]
- 44.Yoshikawa, K., I.M. Okazaki, T. Eto, K. Kinoshita, M. Muramatsu, H. Nagaoka, and T. Honjo. 2002. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 296:2033–2036. [DOI] [PubMed] [Google Scholar]
- 45.Barreto, V., B. Reina-San-Martin, A.R. Ramiro, K.M. McBride, and M.C. Nussenzweig. 2003. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell. 12:501–508. [DOI] [PubMed] [Google Scholar]
- 46.Jolly, C.J., N. Klix, and M.S. Neuberger. 1997. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 25:1913–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jankovic, M., A. Nussenzweig, and M.C. Nussenzweig. 2007. Antigen receptor diversification and chromosome translocations. Nat. Immunol. 8:801–808. [DOI] [PubMed] [Google Scholar]
- 48.Chaudhuri, J., C. Khuong, and F.W. Alt. 2004. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 430:992–998. [DOI] [PubMed] [Google Scholar]
- 49.Roy, D., K. Yu, and M.R. Lieber. 2008. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 28:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinkura, R., M. Tian, M. Smith, K. Chua, Y. Fujiwara, and F.W. Alt. 2003. The influence of transcriptional orientation on endogenous switch region function. Nat. Immunol. 4:435–441. [DOI] [PubMed] [Google Scholar]
- 51.Guo, B., T.T. Su, and D.J. Rawlings. 2004. Protein kinase C family functions in B-cell activation. Curr. Opin. Immunol. 16:367–373. [DOI] [PubMed] [Google Scholar]
- 52.Saijo, K., I. Mecklenbrauker, C. Schmedt, and A. Tarakhovsky. 2003. B cell immunity regulated by the protein kinase C family. Ann. N. Y. Acad. Sci. 987:125–134. [DOI] [PubMed] [Google Scholar]
- 53.Shinkura, R., S. Ito, N.A. Begum, H. Nagaoka, M. Muramatsu, K. Kinoshita, Y. Sakakibara, H. Hijikata, and T. Honjo. 2004. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 5:707–712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.