Abstract
Chronic myeloid leukemia (CML) represents the first human malignancy successfully treated with a tyrosine kinase inhibitor (TKI; imatinib). However, early relapses and the emergence of imatinib-resistant disease are problematic. Evidence suggests that imatinib and other inhibitors may not effectively eradicate leukemic stem/progenitor cells, and that combination therapy directed to complimentary targets may improve treatment. Abelson helper integration site 1 (Ahi-1)/AHI-1 is a novel oncogene that is highly deregulated in CML stem/progenitor cells where levels of BCR-ABL transcripts are also elevated. Here, we demonstrate that overexpression of Ahi-1/AHI-1 in murine and human hematopoietic cells confer growth advantages in vitro and induce leukemia in vivo, enhancing effects of BCR-ABL. Conversely, RNAi-mediated suppression of AHI-1 in BCR-ABL–transduced lin−CD34+ human cord blood cells and primary CML stem/progenitor cells reduces their growth autonomy in vitro. Interestingly, coexpression of Ahi-1 in BCR-ABL–inducible cells reverses growth deficiencies exhibited by BCR-ABL down-regulation and is associated with sustained phosphorylation of BCR-ABL and enhanced activation of JAK2–STAT5. Moreover, we identified an AHI-1–BCR-ABL–JAK2 interaction complex and found that modulation of AHI-1 expression regulates phosphorylation of BCR-ABL and JAK2–STAT5 in CML cells. Importantly, this complex mediates TKI response/resistance of CML stem/progenitor cells. These studies implicate AHI-1 as a potential therapeutic target downstream of BCR-ABL in CML.
Chronic myeloid leukemia (CML) is a clonal, multistep, multilineage myeloproliferative disorder. It is initiated and propagated by a rare population of CML stem cells that have acquired a BCR-ABL fusion gene (1, 2). The BCR-ABL fusion gene encodes a chimeric oncoprotein that displays constitutively elevated tyrosine kinase activity that drives CML pathogenesis (3, 4). These features deregulate cellular proliferation and apoptosis control through effects on multiple intracellular signaling pathways, including the Ras, phosphatidylinositol 3-kinase (PI3K), JAK–STAT, and NF-κB pathways (5, 6). Recently, imatinib mesylate (IM), which is an inhibitor of the BCR-ABL tyrosine kinase (4), has shown promise in treating CML patients (7–9). However, early relapses and IM-resistant disease have emerged as significant clinical problems in some IM-treated CML patients (10, 11). Relapses are frequently associated with mutations in the BCR-ABL kinase domain (10, 12, 13), accounting for 60–90% of relapses (11). Dasatinib (DS) and nilotinib (NL) are more recently produced small molecule inhibitors of the BCR-ABL–encoded kinase with greater potencies than IM and predicted broader effectiveness in patients with IM-resistant disease (14, 15). Recent studies have indicated that CML stem/progenitor cells in chronic phase patients are less responsive to IM and other tyrosine kinase inhibitors (TKIs), and that they are a critical target population for IM resistance (16–18). In addition, CML stem cells are genetically unstable and rapidly generate IM-resistant mutants in vitro (19). Thus, it is critical to identify other therapies targeting CML stem/progenitor cells to prevent acquisition of resistance. There is also an emerging imperative to develop complementary therapies that target downstream molecular events in the CML stem/progenitor cells of those patients who fail to achieve lasting remission with current treatments.
Abelson helper integration site 1 (Ahi-1) is a novel gene that was identified by provirus insertional mutagenesis in v-abl–induced mouse pre–B cell lymphoma as a candidate cooperate oncogene (20). Mouse Ahi-1 encodes a unique protein with a SH3 domain, multiple SH3 binding sites, and a WD40-repeat domain, which are all known to be important mediators of protein–protein interactions, suggesting that the normal Ahi-1 protein has novel signaling activities and that its deregulation could affect specific cellular signaling pathways. Interestingly, the conserved human homologue (AHI-1) has an additional coiled–coil domain in its N-terminal region. Involvement of Ahi-1 in leukemogenesis is suggested by the high frequency of Ahi-1 mutations seen in certain virus-induced mouse leukemias and lymphomas (20, 21). We recently demonstrated that Ahi-1/AHI-1 expression is regulated at multiple stages of hematopoiesis in a fashion that is highly conserved between mice and humans (22). Ahi-1/AHI-1 is expressed at its highest level in the most primitive hematopoietic cells and is rapidly down-regulated as cells begin to differentiate. Interestingly, marked deregulation of AHI-1 expression is seen in several human leukemic cell lines (22, 23), particularly in a CML cell line (K562) and in Philadelphia chromosome–positive (Ph+ BCR-ABL+) primary leukemic cells, but not Ph− cells, especially in highly enriched leukemic stem cells from patients with CML. In addition, levels of BCR-ABL transcripts are highly elevated in the same CML stem cell population (18, 24), suggesting that it may be important to cooperative activities of AHI-1 and BCR-ABL to generate a permanently expanding clone of deregulated stem cells at the early stage of leukemia development.
In this study, biological and molecular functions of Ahi-1/AHI-1 and its cooperative activities with BCR-ABL were extensively investigated in primitive mouse and human hematopoietic cells using several overexpression, suppression, and inducible model systems. We found that overexpression of Ahi-1 alone in primitive hematopoietic cells confers a proliferative advantage in vitro and induces a lethal leukemia in vivo; these effects are enhanced by BCR-ABL. Stable suppression of AHI-1 by small interfering RNA in BCR-ABL–transduced primitive human cord blood (CB) cells and primitive leukemic cells from CML patients reduces their growth autonomy in vitro. The regulatory role of Ahi-1/AHI-1 in mediating BCR-ABL transforming activities can be further explained by demonstration of a direct physical interaction between AHI-1 and BCR-ABL at endogenous levels in CML cells. This is associated with JAK2 and results in modulation of sensitivity to TKIs and differing levels of BCR-ABL tyrosine phosphorylation and JAK2–STAT5 activity in BCR-ABL+ CML cells where Ahi-1/AHI-1 expression is either coexpressed or inhibited.
RESULTS
Overexpression of Ahi-1 alone can transform IL-3–dependent BaF3 cells in vitro and in vivo, and these effects can be enhanced by BCR-ABL
To investigate the transforming potential of Ahi-1 in hematopoietic cells, we cloned full-length Ahi-1 cDNA into a MSCV-internal ribosomal entry site (IRES)-YFP (MIY) vector and overexpressed it in an IL-3–dependent cell line (BaF3). Quantitative real-time RT-PCR analysis (Q-RT-PCR) showed that Ahi-1 transcript levels were greatly increased in Ahi-1–transduced BaF3 clonal cell lines compared with control cells (Fig. S1 A available at http://www.jem.org/cgi/content/full/jem.20072316/DC1). Ahi-1–transduced cells have increased proliferative activity in the presence of IL-3, and this effect was markedly enhanced when IL-3 was not added (3–4-fold; P < 0.01; Fig. S1 C). Ahi-1–transduced cells also had greater cell viability in the presence of a low concentration of IL-3 (2 ng/ml) compared with control cells (twofold; Fig. S1 D). Interestingly, overexpressing both Ahi-1 and BCR-ABL in BaF3 cells further enhanced these perturbations, compared with cells transduced with either BCR-ABL or Ahi-1 alone (1.5–3.5-fold in the absence or presence of IL-3; P < 0.05; Fig. S1, C and D). Western blot analysis of protein from two individual clonal lines revealed that both protein expression and tyrosine kinase activity of p210BCR-ABL were highly increased in cells cotransduced with Ahi-1 and BCR-ABL, compared with BCR-ABL–transduced cells (Fig. S1 E). We also detected higher levels of Ahi-1 protein expression in the same dually transduced cells than in those transduced with Ahi-1 alone. More interestingly, endogenous Ahi-1 expression is increased in cells transduced with BCR-ABL alone compared with control cells with expression levels being similar to those detected in Ahi-1–transduced BaF3 cells (Fig. S1 E).
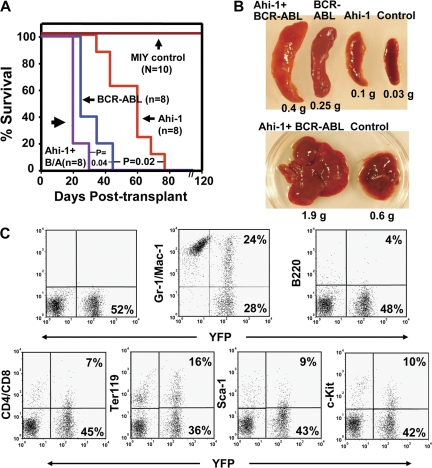
To investigate effects of overexpression of Ahi-1 on the ability of transduced cells to induce leukemias in vivo, we injected transduced cells into sublethally irradiated NOD/SCID–β2 microglobulin (β2m)−/− mice. Strikingly, mice injected intravenously with Ahi-1–transduced BaF3 cells had a lethal leukemia within 70 d (5 × 106 cells/per mouse; Fig. 1 A). Disease latency was shortened to 40 d with BCR-ABL–transduced cells alone. Leukemogenic activity was further increased by introduction of cotransduced Ahi-1 and BCR-ABL cells, producing a latency of 26 d (P < 0.05; Fig. 1 A). Mice injected with either parental BaF3 cells or vector control cells had no evidence of disease after 120 d. Leukemic mice injected with either Ahi-1 or BCR-ABL–transduced cells developed splenomegaly and hepatomegaly, with 50–90% of YFP+/Ahi-1+, GFP+/BCR-ABL+, or both YFP+GFP+ cells detectable in these tissues (Fig. 1 B). As expected, larger spleens and livers were observed in mice injected with both Ahi-1 and BCR-ABL–transduced cells (Fig. 1 B and Table I). Interestingly, despite the apparently homogeneous pro–B cell phenotype of BaF3 cells transplanted, the leukemias generated from Ahi-1–transduced cells revealed multilineage features that included the production of Gr-1+Mac-1+ (myeloid), Ter119+ (erythroid), B220+ (B-lineage), and CD4+CD8+ (T-lineage; Fig. 1 C), suggesting that overexpression of Ahi-1 induces abnormal differentiation (including lineage switching) in hematopoietic cells. This was also observed in mice injected with Ahi-1 and BCR-ABL cotransduced cells. In addition, YFP+/Ahi-1+ or GFP+/BCR-ABL+ cells purified from BM cells of diseased mice showed increased proliferation and reduced apoptosis compared with control BaF3 cells; these effects were enhanced in Ahi-1– and BCR-ABL–cotransduced cells (Fig. S2 available at http://www.jem.org/cgi/content/full/jem.20072316/DC1). Thus, overexpression of Ahi-1 alone in IL-3–dependent hematopoietic cells has strong transforming activity in vitro and in vivo, and this is additive with the effects of BCR-ABL.
Figure 1.
Overexpression of Ahi-1 induces a lethal leukemia in vivo and these effects can be enhanced by cotransduction of BCR-ABL. (A) Survival curves of NOD/SCID-β2M−/− mice injected with 5 × 106 BaF3 cells transduced with MIY, Ahi-1, BCR-ABL, and Ahi-1 plus BCR-ABL. 8–10 mice were used per each group. (B) Spleen (top) and liver (bottom) weight of mice injected with MIY control cells, Ahi-1–transduced cells, BCR-ABL–transduced cells, and cells cotransduced with Ahi-1 and BCR-ABL. (C) FACS profiles of YFP+ BM cells isolated from a representative moribund mouse with leukemia, after injection of Ahi-1–transduced cells and showing expression by the YFP+ cells of Gr-1/Mac-1, B220, CD4/CD8, Ter119, Sac-1, and c-kit.
Table I.
Pathological parameters of leukemic mice injected with transduced cells in vivo
| Cells injected | Leukemia/mice | Disease latencya | Spleen weight | Liver weight |
|---|---|---|---|---|
| g | g | |||
| MIY control | 0/10 | No disease | 0.035 ± 0.006 | 0.5 ± 0.09 |
| Ahi-1 | 8/8 | 70 ± 9.5 | 0.12 ± 0.04 | 1.1 ± 0.18 |
| BCR-ABL | 8/8 | 41 ± 5.6 | 0.25 ± 0.05 | 1.6 ± 0.25 |
| Ahi-1 + BCR-ABL | 8/8 | 26 ± 4 | 0.41 ± 0.08 | 2.1 ± 0.3 |
Time (in days) to death caused by leukemia.
Overexpression of Ahi-1 alone confers a growth advantage on mouse hematopoietic stem/progenitor cells and enhances the effects of BCR-ABL
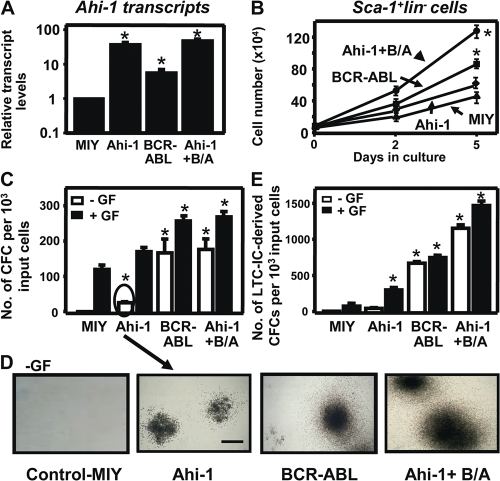
To further investigate the role of Ahi-1 as a potential cooperating oncogene relevant to BCR-ABL–mediated transformation in primitive primary hematopoietic cells, we compared the biological behavior of primitive mouse hematopoietic cells from the BM of 5-FU–treated adult C57BL/6 mice after transduction with MSCV-Ahi-1-IRES-YFP and MSCV-BCR-ABL-IRES-GFP retroviruses, alone or in combination. Q-RT-PCR analysis of RNA from FACS-purified primitive Sca-1+lin−YFP+ (Ahi-1+), Sca-1+lin−GFP+ (BCR-ABL+), and Sca-1+lin−YFP+GFP+ (Ahi-1+BCR-ABL+)–transduced BM cells showed that Ahi-1 transcripts were 40-fold higher in Ahi-1–transduced cells compared with cells transduced with control vector (MIY). Elevated endogenous Ahi-1 expression was also observed in cells transduced with BCR-ABL alone as compared with control cells (8-fold; P < 0.01; Fig. 2 A). 5 d after transduction, the rate of expansion of the Sca-1+lin−YFP+ (Ahi-1+) cells in liquid cultures was approximately twofold higher than in cultures initiated with control cells (Fig. 2 B). Ahi-1–transduced Sca-1+lin− cells produced a slightly greater number of colony-forming cells (CFCs) than MIY-transduced control cells in semisolid cultures in the presence of growth factors (GFs), and some of the Ahi-1+ CFCs (∼15%) were already GF independent (Fig. 2, C and D). After 4 wk in long-term culture-initiating cell (LTC-IC) assays, which are used to measure stem cell activities in vitro, the Sca-1+lin−Ahi-1+ cells produced 3-fold more CFCs than control cells (P < 0.01; Fig. 2 E). All of these endpoints (proliferative activity, GF dependence, and CFC output in LTC-IC assays) are also perturbed by BCR-ABL transduction. In cells cotransduced with Ahi-1 and BCR-ABL, all of these effects were further enhanced compared with cells transduced with either BCR-ABL alone (2–4-fold) or Ahi-1 alone (3–6-fold; P < 0.01; Fig. 2). Thus, overexpression of Ahi-1 alone deregulates proliferative control of primitive mouse hematopoietic cells, and these effects can be enhanced by BCR-ABL.
Figure 2.
Overexpression of Ahi-1 in Sca-1+lin− mouse stem/progenitor BM cells perturbs their in vitro proliferative activity and enhances the effects of BCR-ABL. (A) The levels of Ahi-1 transcripts relative to GAPDH from FACS-purified MIY (Sca-1+lin−YFP+), Ahi-1 (Sca-1+lin−YFP+), BCR-ABL (Sca-1+lin−GFP+), and Ahi-1 plus BCR-ABL (Sca-1+lin−YFP+GFP+) transduced primary mouse BM cells. (B) Growth of each FACS-purified population with GFs. Viable cell numbers were determined by hematocytometer counts of trypan blue–excluding cells. (C) Number of CFC colonies produced in semisolid cultures ± GF from the same FACS-purified mouse BM cells. (D) The appearance of GF-independent CFC colonies is shown. Bar, 250 μm. (E) The numbers of LTC-IC–derived CFCs produced from the same cells ± GF. Values shown are the mean ± SEM of triplicate measurements. * indicates significantly different from MIY control cells.
Suppression or overexpression of AHI-1 mediates transforming activity of K562 CML cells in vitro and in vivo
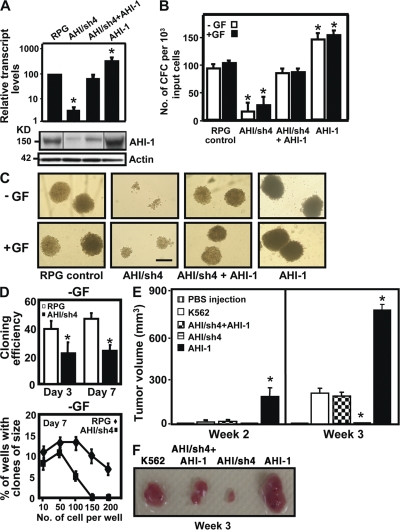
To investigate directly whether deregulated expression of AHI-1 contributes to BCR-ABL–mediated transformation of human CML, knockdown or overexpression of AHI-1 in K562 cells, a cell line derived from a patient with CML and characterized by highly increased expression of AHI-1 (22), was performed, with confirmed down-regulation of AHI-1 in suppressed cells (AHI/sh4) and overexpression of AHI-1 in overexpressed cells (P < 0.01; Fig. 3 A). Interestingly, suppression of AHI-1 expression reduced GF independence in semisolid cultures (up to 5-fold; P < 0.01; Fig. 3 B) compared with cells transduced with control vector (RPG). Colonies formed by AHI-1–suppressed cells were much smaller than those of control cells (Fig. 3 C). Also, the ability to form a clone from a single cell was 2-fold less than that of the controls without GFs in liquid suspension cultures (P < 0.05; Fig. 3 D). In contrast, overexpression of human AHI-1 by transduction of EF1α-AHI-1-IRES-YFP lentivirus in K562 cells resulted in sharply increased colony-forming ability compared with control cells (1.5–2-fold; P < 0.05; Fig. 3, B and C). Importantly, restored expression of AHI-1 both at RNA and protein levels in AHI/sh4 cells reversed growth deficiencies exhibited by suppression of AHI-1 (Fig. 3, A–C).
Figure 3.
Knockdown or overexpression of AHI-1 in human K562 cells mediates their transforming activity in vitro and in vivo. (A) Q-RT-PCR analysis of the levels of AHI-1 transcripts relative to GAPDH in FACS-purified RPG vector–transduced K562 cells, AHI-1/sh4 cells (with suppression of AHI-1), AHI/sh4 + AHI-1 cells (coexpression of AHI-1 in AHI-1/sh4 cells), and AHI-1 cells (AHI-1 overexpressed cells, top). Western analysis (bottom) of AHI-1 expression in the same transduced cells with an anti–AHI-1 antibody. (B) The numbers of CFC colonies produced in semisolid cultures ± GF (IL-3, GM-CSF, SF) in the same transduced cells. (C) The appearance of CFC colonies produced in semisolid cultures ± GF as shown in B. Bar, 250 μm. (D) Percentage of single control K562 cells or AHI/sh4 cells generating clones (top) and clone size distributions obtained from these cells after being cultured for 7 d (bottom). (E) NOD/SCID-β2M−/− mice were injected subcutaneously with 107 control K562 and transduced cells. Tumor volume is expressed as mean ± SEM areas of each group (n = 4). (F) The appearance of tumors generated by the same cells as shown in E. Values shown are the mean ± SEM of triplicate measurements. * indicates significantly different from K562 or RPG control cells.
To determine if in vitro effects could be replicated in vivo, NOD/SCID-β2m−/− mice were injected subcutaneously with transduced cells to determine their ability to form tumors in vivo. Cells with suppression of AHI-1 failed or had significantly reduced ability to form tumors compared with control K562 cells (Fig. 3, E and F). In contrast, AHI-1 overexpressed cells rapidly generated larger tumors than the control cells within 3 wk (3-fold; P < 0.01). Strikingly, AHI/sh4 cells cotransduced with the AHI-1 construct could reverse in vivo growth deficiencies resulting from knockdown of AHI-1. These results demonstrate that modulation of expression levels of AHI-1 in human CML cells regulates their transforming activity in vitro and in vivo.
Suppression of AHI-1 expression in primitive BCR-ABL–transduced human CB cells and primary CML stem/progenitor cells reduces their growth autonomy
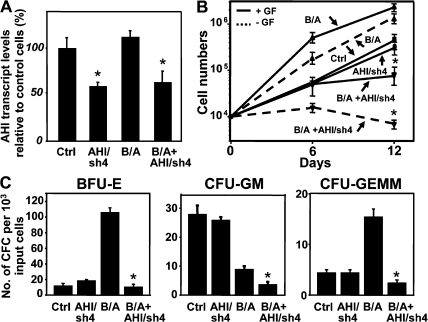
We next evaluated the effects of down-regulation of AHI-1 in BCR-ABL–transduced primitive human CB stem/progenitor cells (Lin−CD34+) using lentiviral-mediated RNA interference. Q-RT-PCR analysis indicated that endogenous AHI-1 expression was suppressed by 40–50% when the shRNA construct (AHI/sh4) was introduced (Fig. 4 A). Interestingly, BCR-ABL–transduced CB cells expanded rapidly in the presence or absence of GFs as expected, but the growth of the BCR-ABL–transduced cells with suppression of AHI-1 was significantly suppressed (2.2 × 106 versus 4.8 × 105 cells with GF; 106 versus 5 × 103 cells without GF; P < 0.01; n = 3; Fig. 4 B). Similarly, BCR-ABL–transduced CB cells with suppression of AHI-1 had up to 10-fold less CFC output compared with BCR-ABL–transduced CB cells (P < 0.01; Fig. 4 C). A similar reduction of CFC output was observed in the absence of GFs (data not shown). Notably, burst-forming-units-erythroids (BFU-Es) were especially increased in BCR-ABL–transduced CB cells compared with control cells, which remained predominantly myeloid as previously reported (110 ± 8 versus 18 ± 3; P < 0.01; Fig. 4 C) (25). Suppression of AHI-1 expression in BCR-ABL-transduced CB cells significantly reduced myeloid CFC (BFU-E and colony-forming units-granulocyte-macrophage [CFU-GM]) output, as well as primitive myeloid progenitors (colony-forming units-granulocyte-erythrocite-monocyte-megakaryocyte [CFU-GEMM]), suggesting that AHI-1 may play a role in regulation of overproduction of myeloid cells in CML.
Figure 4.
Lentiviral-mediated suppression of AHI-1 expression causes reduced proliferative activity and GF independence of BCR-ABL–transduced human CB cells. (A) Q-RT-PCR analysis of the levels of human AHI-1 transcripts relative to GAPDH in transduced lin−CD34+ CB cells, including cells transduced with a control vector (Ctrl), AHI/sh4 cells (with suppression of AHI-1), BCR-ABL (B/A), and AHI/sh4 cotransduced cells (B/A + AHI/sh4). (B) Growth of each transduced population in suspension cultures ± GF for 12 d. Viable cell numbers were determined by hematocytometer counts of trypan blue-excluding cells. (C) Number of BFU-E, CFU-GM, and CFU-GEMM colonies produced in semisolid cultures from the same cells as shown in B. Values shown are the mean ± SEM of triplicate measurements. * indicates significant difference between BCR-ABL–transduced cells alone and cells cotransduced with BCR-ABL and AHI/sh4.
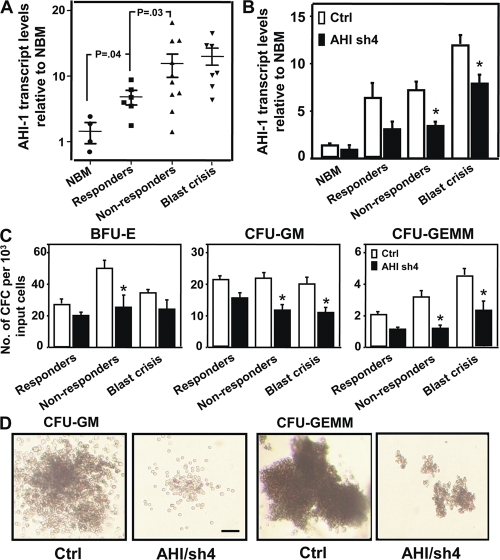
To further elucidate the role of AHI-1 in primary CML stem/progenitor cells, AHI-1 transcript levels were evaluated in pretreatment lin−CD34+ cells from 16 chronic phase patients with subsequent clinical responses to IM therapy (6 responders and 10 nonresponders) and from 7 patients in blast crisis. Increased levels of AHI-1 expression were observed in lin−CD34+ CML stem/progenitor cells from all patient samples studies (2.5–18-fold; P < 0.04; Fig. 5 A), compared with lin−CD34+ normal BM cells. Interestingly, lin−CD34+ cells from IM nonresponders expressed higher levels of AHI-1 transcripts compared with the same cells isolated from IM responders (P = 0.03; Fig. 5 A). Lin−CD34+ cells from blast crisis patients also expressed relatively higher levels of AHI-1 transcripts (P < 0.03, compared with IM responders). A lentiviral-mediated AHI-1 shRNA construct was directly transduced into lin−CD34+ stem/progenitor cells isolated from three IM responders, three IM nonresponders, and three blast crisis patients. Lentiviral-mediated suppression of AHI-1 expression resulted in a reduction in CFC output by ∼28–60% in lin−CD34+ CML cells from all patient samples studied (with AHI-1 expression partially suppressed (∼40–55%; Fig. 5, B and C). CFC colonies formed by CML cells with suppression of AHI-1 were found to be smaller than those resulting from cells transduced with a control vector (Fig. 5 D). It was interesting that more significant reduction in CFC numbers was observed in transduced primary CML cells from IM nonresponders and blast crisis compared with IM responders (Fig. 5 C).
Figure 5.
Elevated AHI-1 transcript levels in CML stem/progenitor cells and reduced CFC production in the same cells when AHI-1 expression is suppressed. (A) Q-RT-PCR analysis of the levels of human AHI-1 transcripts relative to GAPDH in lin−CD34+ normal bone marrow (NBM) and lin−CD34+ CML cells from IM responders, nonresponders, and blast crisis patients. (B) Q-RT-PCR analysis of the levels of human AHI-1 transcripts relative to GAPDH in transduced lin−CD34+ normal BM and CML cells (three IM responders, three nonresponders, and three blast crisis patients) ± suppression of AHI-1 expression, including cells transduced with a control vector (Ctrl) or a AHI/sh4 vector. (C) The yield of BFU-E, CFU-GM, and CFU-GEMM colonies from primary CD34+lin− CML cells transduced with a control vector or AHI/sh4 vector. (D) The appearance of CFC colonies produced in semisolid cultures from control and AHI/sh4-transduced cells from an IM nonresponder patient. Bar, 100 μm. Values shown are the mean ± SEM of triplicate measurements. * indicates significant difference between transduced control cells and cells transduced with AHI/sh4 vector.
Coexpression of Ahi-1 in BCR-ABL–inducible cells regulates transforming activities of BCR-ABL+ cells
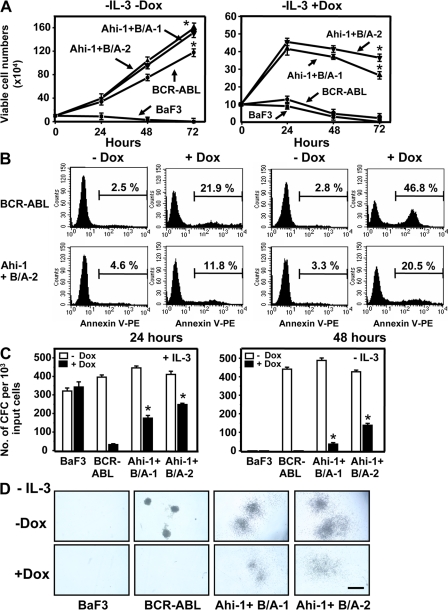
To further investigate potential regulatory roles of Ahi-1 in mediating BCR-ABL–transforming activities, we evaluated cooperative effects of Ahi-1 in a BCR-ABL–transduced BaF3 cell line in which the level of expression of p210BCR-ABL can be variably down-regulated by exposure to doxycycline (Dox) (26). Reduction in BCR-ABL protein expression in the presence of Dox results in a corresponding decrease in GF independence of BaF3 cells both in liquid suspension cultures and in semisolid cultures in vitro (Fig. 6). As shown in Fig. 6 (A and B), cell growth and survival was dramatically reduced when Dox was added to culture at 48 h (>90% inhibition of proliferation, Fig. 6 A, right; ∼50% detectable Annexin V+ apoptotic cells, Fig. 6 B, top right), while the same cells showed a marked increase in cell expansion in the absence of both IL-3 and Dox (no down-regulation of BCR-ABL, Fig. 6 A, left). Similarly, down-regulation of BCR-ABL expression completely inhibited CFC generation in semisolid cultures in the absence of IL-3 (Fig. 6, C and D). Interestingly, introduction of Ahi-1 into cells with inhibited BCR-ABL expression under these stringent conditions enabled them to grow continuously in liquid suspension culture, to have less Annexin V+ apoptotic cells, and to produce more factor-independent CFCs than cells transduced with BCR-ABL alone (∼4 × 105 versus 103 cells after 48 h in liquid culture and ∼100 CFCs versus 0 in the CFC assay; P < 0.01; Fig. 6). These results were consistently observed in two individual clonal cell lines. In the presence of IL-3 and Dox, BCR-ABL–transduced BaF3 cells showed a significant reduction in CFC production; addition of IL-3 cannot rescue inhibitory effects caused by suppression of BCR-ABL expression. This can be enhanced significantly by cotransduction of Ahi-1 (10–15-fold; P < 0.01; Fig. 6 C). Collectively, these results demonstrate the ability of Ahi-1 to immediately reverse in vitro growth deficiencies resulting from down-regulation of BCR-ABL in vitro, and provide direct evidence of the regulatory role of Ahi-1 in BCR-ABL–mediated transformation.
Figure 6.
Overexpression of Ahi-1 rescues growth suppression induced by the inhibition of BCR-ABL expression in BCR-ABL–inducible BaF3 cells. (A) Growth of control BaF3, BCR-ABL–inducible cells, and two Ahi-1–transduced, BCR-ABL–inducible clonal lines without IL-3 ± Dox (+Dox = suppression of BCR-ABL expression). Viable cell numbers were determined by hematocytometer counts of trypan blue–excluding cells. (B) Annexin V-PE/7-AAD staining of BCR-ABL–inducible cells and Ahi-1–transduced BCR-ABL–inducible cells (Ahi-1 + B/A-2) after culture without IL-3 ± Dox for 24 and 48 h. Percentages of Annexin V+ cells are indicated. (C) CFC colonies produced in semisolid media ± IL-3 and Dox from the same cells as shown in A. (D) The appearance of GF-independent CFC colonies in Ahi-1 and BCR-ABL–cotransduced cells without IL-3 ± Dox. Bar, 250 μm. Values shown are the mean ± SEM of triplicate measurements. * indicates significant difference between BCR-ABL inducible cells alone and inducible cells cotransduced with Ahi-1.
Coexpression of Ahi-1 in BCR-ABL–inducible cells sustains tyrosine phosphorylation of BCR-ABL and enhances activation of JAK2 and STAT5
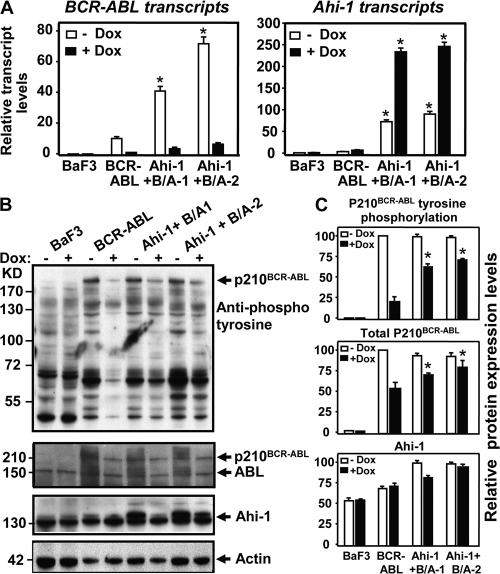
To determine whether coexpression of Ahi-1 in BCR-ABL–inducible BaF3 cells influences protein expression and tyrosine kinase activity of BCR-ABL, and candidate downstream signaling that reflects enhanced transforming phenotypes observed, we performed Q-RT-PCR and Western blot analyses in these cells in the presence and absence of Dox. As expected, BCR-ABL transcripts were highly expressed without Dox and down-regulated in the presence of Dox in the BCR-ABL–inducible cells after 24–48 h in culture (Fig. 7 A). Elevated BCR-ABL transcript levels were again observed in BCR-ABL– and Ahi-1–cotransduced cells generated from two individual cells lines (3–6-fold; P < 0.01). As expected, Ahi-1 transcripts were highly elevated in cotransduced cells when compared with control BaF3 cells (>50-fold; P < 0.01; Fig. 7 A). Strikingly, tyrosine phosphorylation of p210BCR-ABL could not sufficiently be suppressed in Ahi-1- and BCR-ABL–cotransduced cells, as compared with BCR-ABL–transduced cells alone in the presence of the same amount of Dox (30–40% inhibition versus ∼80% inhibition; P < 0.01; Fig. 7, B and C). In all cases, BCR-ABL phosphorylation was not completely suppressed after 24 h in culture with addition of Dox. These results were consistently observed in two individual cotransduced cell lines. Similarly, BCR-ABL protein expression was also suppressed to a lesser degree in cotransduced cells than in cells transduced with BCR-ABL only, and Ahi-1 protein expression was found to be higher in cotransduced cells than in cells transduced with BCR-ABL only.
Figure 7.
Sustained tyrosine phosphorylation and protein expression of P210BCR-ABL in BCR-ABL–inducible BaF3 cells cotransduced with Ahi-1. (A) Q-RT-PCR analysis of the levels of BCR-ABL (left) and Ahi-1 (right) transcripts relative to GAPDH in control BaF3, BCR-ABL inducible cells and two Ahi-1-transduced BCR-ABL inducible clonal lines cultured without IL-3 ± Dox for 24 h. (B) Western analyses of cell lysates from the same cells for 24 h. Antibodies used are indicated. (C) Tyrosine phosphorylation and protein expression of p210BCR-ABL and Ahi-1 relative to actin, as compared with BCR-ABL–transduced cells alone. Values shown are the mean ± SEM of triplicate measurements. * indicates significant difference between BCR-ABL–inducible cells alone and the inducible cells cotransduced with Ahi-1.
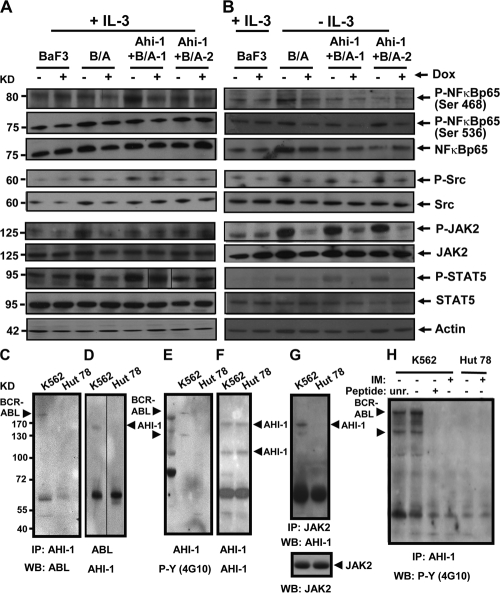
We next evaluated the effect of increased BCR-ABL expression and tyrosine phosphorylation of cotransduced cells on the activity of candidate signaling mechanisms. We observed increased levels of phosphorylation of JAK2, STAT5, NF-κB p65 (at Ser 563 and Ser 468), and Src (at Tyr 416) in BCR-ABL–inducible cells and Ahi-1–cotransduced BCR-ABL–inducible cells compared with control BaF3 cells (Fig. 8, A and B). Interestingly, phosphorylation of most downstream proteins was down-regulated when BCR-ABL expression was inhibited by Dox, but sustained phosphorylation of JAK2 and STAT5 was consistently observed in cotransduced cells in the presence of IL-3 and Dox (Fig. 8 A, lanes 6 and 8 compared with lane 4). In the absence of IL-3, phosphorylation of JAK2 and STAT5 was reduced in cotransduced cells when BCR-ABL expression was suppressed. A similar, albeit less pronounced, finding of sustained phosphorylation of Src was also observed, particularly in Ahi-1 + BCR-ABL-1 cells in the presence of IL-3. These results suggest that Ahi-1 may play a regulatory role in mediation of BCR-ABL activity associated with enhanced activation of JAK2 and STAT5 through the IL-3 signaling pathway.
Figure 8.
Enhanced activation of JAK2 and STAT5 in BCR-ABL inducible BaF3 cells cotransduced with Ahi-1 and detection of a AHI-1–BCR-ABL–JAK2 interaction complex in human K562 cells. (A and B) Western blot analyses of cell lysates from control BaF3, BCR-ABL–inducible cells, and two Ahi-1–transduced BCR-ABL–inducible clonal lines cultured in the presence (A) and absence (B) of IL-3 ± Dox for 24 h. Antibodies used are indicated. (C–H) Protein lysates of K562 cells (BCR-ABL+) and Hut 78 cells (BCR-ABL−) were used to immunoprecipitate AHI-1 (C, E-F, and H), ABL (D), and JAK2 (G), and proteins were detected by Western blotting with antibodies to ABL (C), AHI-1 (D, F, and G), JAK-2 (G), and phosphotyrosine (E and H) as indicated. In H, before immunoprecipitation, lysates were incubated for 1 h with the indicated AHI-1 peptide; unrelated peptide (unr.), no peptide (-), and antigenic peptide (+) or cells were incubated ± IM (5 μM) for 6 h before preparation of protein lysates.
AHI-1 physically interacts with BCR-ABL
To detect a physical interaction between AHI-1 and BCR-ABL in CML cells, coimmunoprecipitation (co-IP) experiments were performed. We demonstrated a direct interaction between AHI-1 and BCR-ABL at endogenous levels by detection of BCR-ABL in human CML cells (K562) after IP with a human AHI-1 antibody (Fig. 8 C). This interaction was not found in a BCR-ABL− T cell line (Hut 78; Fig. 8 C, lane 2) or in control antibody coimmunoprecipitated K562 cells (not depicted). The result was confirmed by detection of AHI-1 in the same cells after IP with a specific antibody to ABL (Fig. 8 D). Importantly, tyrosine phosphorylated p210BCR-ABL could be detected in K562 cells using an antiphosphotyrosine antibody (4G10) after IP with the AHI-1 antibody (Fig. 8 E). Interestingly, this protein interaction complex is also associated with a 120-kD tyrosine-phosphorylated protein (Fig. 8 E). To determine whether this 120-kD protein could be an AHI-1 isoform, the same membrane was washed and reprobed with an AHI-1 antibody. The full-length (150 kD) and a 100-kD isoform of AHI-1 protein (but not a 120-kD protein) were identified by Western blot analyses (Fig. 8 F). Using co-IP, we examined several candidate 120-kD tyrosine-phosphorylated proteins known to interact with BCR-ABL, including CBL and JAK2. We determined that JAK2 was associated with this protein interaction complex, as AHI-1 could be detected by an anti–AHI-1 antibody in K562 cells after IP with a specific antibody to JAK2 (Fig. 8 G); this interaction was further confirmed by reverse detection of JAK2 after IP with the anti-AHI-1 antibody (not depicted). In addition, the antigenic peptide derived from the sequence of AHI-1 specifically blocked the ability of the AHI-1 antibody to precipitate both tyrosine-phosphorylated BCR-ABL and the 120-kD protein, whereas an unrelated peptide had no effect (Fig. 8 H, lane 3). Interestingly, this interaction complex was found to be modulated by tyrosine kinase activity of BCR-ABL, as IM treatment (5 μM) of K562 cells for 6 h resulted in inability to detect both tyrosine-phosphorylated BCR-ABL and JAK2 (Fig. 8 H, lane 4). These results indicate that AHI-1 and BCR-ABL can interact and form a complex involving tyrosine-phosphorylated JAK2.
AHI-1 regulates response of BCR-ABL+ primitive CML cells to TKIs
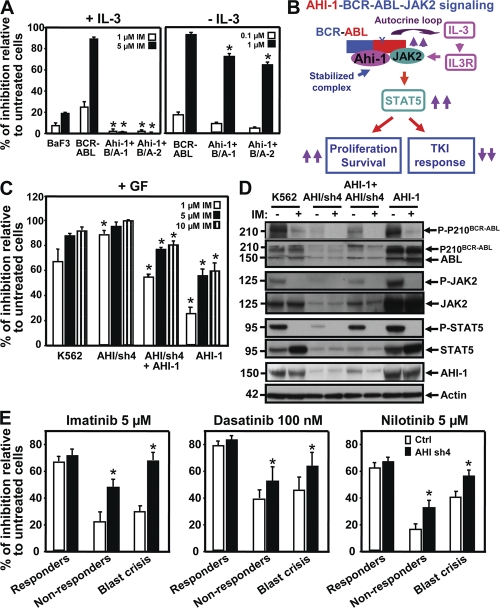
To determine whether AHI-1–BCR-ABL–JAK2 interaction complex may mediate IM sensitivity/resistance of BCR-ABL+ cells, BCR-ABL–transduced inducible BaF3 cells and cells cotransduced with Ahi-1 were treated with various doses of IM (0–10 μm). As expected, BCR-ABL–transduced cells showed a significant reduction in CFC output in response to IM treatment in the presence and absence of IL-3 (∼90% inhibition with 5 μM IM + IL-3 and 1 μM IM − IL-3; Fig. 9 A). Strikingly, BaF3 cells cotransduced with Ahi-1 and BCR-ABL showed no response to IM and produced as many CFCs in the presence of IL-3 as were produced by the same cells without IM treatment (Fig. 9 A, left). Moreover, cotransduced cells also displayed greater resistance to IM in CFC output in the absence of IL-3, although these cells were more sensitive to IM treatment than those in the presence of IL-3 (Fig. 9 A, right). These results indicate that Ahi-1 is capable of overcoming IM-induced growth suppression in BCR-ABL+ cells when IL-3 signaling is activated in these cells.
Figure 9.
AHI-1 mediates response/resistance of TKIs as assessed by overexpression or suppression of AHI-1 in BCR-ABL+ primitive CML cells. (A) Percentage of inhibition of CFC colonies generated in semisolid media ± IL-3 and IM (0–5 μM) from control BaF3, BCR-ABL–inducible cells, and two Ahi-1–transduced BCR-ABL–inducible clonal cell lines. (B) Model of AHI-1–BCR-ABL–JAK2 complex regulation of constitutive activation of BCR-ABL and JAK2–STAT5 pathway, resulting in increased proliferation and reduced TKI response of CML stem/progenitor cells. (C) Percentage of inhibition of CFC generation in semisolid media with GF and IM (0–10 μM) from control K562, AHI/sh4–transduced cells (suppression of AHI-1), overexpression of AHI-1 in AHI/sh4 cells, and AHI-1–transduced K562 cells. * indicates significantly different from K562 control cells. (D) Western analyses of cell lysates from the same cells ± IM (5 μM) for 6 h. Antibodies used are indicated. (E) Inhibition of CFCs in semisolid media with IM (5 μM), DS (100 nM), and NL (5 μM) in lin−CD34+ CML cells from IM responders (n = 3), nonresponders (n = 3), and blast crisis (n = 3) patient samples transduced with either a control vector or AHI-1/sh4 vector. Values shown are the mean ± SEM of triplicate measurements. * indicates significantly different between BCR-ABL–inducible cells and inducible cells cotransduced with Ahi-1, and between lin−CD34+ cells transduced with a control vector or transduced with the AHI-1/sh4 vector.
Similarly, overexpression of human AHI-1 in K562 cells resulted in greater resistance to IM treatment, as assessed by the CFC assay, (∼2-fold; P < 0.01) in comparison to K562 control cells (Fig. 9 C). Conversely, suppression of AHI-1 (AHI/sh4) resulted in increased sensitivity to IM, particularly in the presence of a low concentration of IM (1 μM). Strikingly, restored expression of AHI-1 by overexpression of an AHI-1 construct in AHI/sh4 cells (suppression of AHI-1) restored IM resistance to AHI/sh4 cells (Fig. 9 C). Western analysis revealed increased tyrosine-phosphorylated BCR-ABL, JAK2, and STAT5 in K562 cells with AHI-1 overexpression and reduced levels of these phosphorylated proteins when AHI-1 expression is suppressed (Fig. 9 D). Interestingly, phosphorylated BCR-ABL, JAK2, and STAT5 levels could be restored in the AHI-1/sh4 cells when AHI-1 construct was reintroduced into the same cells. Importantly, expression of AHI-1 not only modulates phosphorylation of BCR-ABL, JAK2, and STAT5 in BCR-ABL+ K562 cells, but also regulates protein expression of these genes, as demonstrated by significantly enhanced expression of these proteins when AHI-1 is overexpressed, reduced expression when AHI-1 is suppressed, and restored expression in AHI-1–suppressed cells where AHI-1 expression has been rescued by introduction of an AHI-1 construct (Fig. 9 D). These results further support our findings in the BCR-ABL–inducible system that AHI-1 plays a critical role in mediation of BCR-ABL and JAK2–STAT5 activities.
We next assessed sensitivity of lin−CD34+ CML stem/progenitor cells, with and without suppression of AHI-1 expression, to the TKIs IM, DS, and NL. Cells were obtained from three IM responders, three IM nonresponders, and three blast crisis patients with partial suppression of AHI-1 expression in transduced CML cells (∼40–50%) as shown in Fig. 5 B. Interestingly, in all cases, lin−CD34+ CML cells were more sensitive to DS treatment (100 nM) than to IM (5 μM) or NL (5 μM), as assessed by their ability to generate CFCs (Fig. 9 E), whereas lin−CD34+ cells with suppression of AHI-1 expression, particularly cells from the clinically IM-resistant and blast crisis patients, were more sensitive to all three inhibitors (∼2-fold; P < 0.05). Collectively, these data suggest that AHI-1 plays an important role in modulating sensitivity to IM and other selective BCR-ABL TKIs in BCR-ABL+ CML cells.
DISCUSSION
In this study, we demonstrate for the first time that Ahi-1/AHI-1 is a new oncogene that cooperates in transforming activities with BCR-ABL both in vitro and in vivo through a direct physical interaction. First, in a mouse system, overexpression of mouse Ahi-1 confers a proliferative advantage in vitro to IL-3–dependent BaF3 cells and a stem cell–enriched Sca-1+lin− population from 5-FU–treated mouse BM cells, and induces a lethal leukemia in vivo. This deregulated proliferative activity, GF independence, and leukemogenic potential is enhanced by introduction of BCR-ABL. Thus, there is a direct biological correlation between Ahi-1 and BCR-ABL in regulating transforming activity of these cells. Second, in a human system, AHI-1 expression appears to regulate transforming activities of BCR-ABL–transduced human CB stem/progenitor cells (lin−CD34+), as indicated by their significantly reduced autonomous growth when endogenous AHI-1 expression is stably inhibited. These effects were further demonstrated in CML patient samples; reduced autonomous growth was observed in primary CML stem/progenitor cells in all patient samples studied with knockdown of AHI-1. The effects were more significant in CML stem/progenitor cells from IM-resistant patients and blast crisis patients who expressed relatively higher levels of AHI-1 (Fig. 5). Knockdown of AHI-1 expression in BCR-ABL–transduced human CB cells not only inhibited all differentiated myeloid cells (CFU-GM and CFU-GEMM) but also significantly inhibited differentiating erythroid cells (BFU-E; Fig. 4 C) that are produced at a high frequency from BCR-ABL–transduced CD34+ stem/progenitor cells independent of their apparent prior lineage commitment status caused by modulation of P210BCR-ABL activity (25). Interestingly, we also observed that overexpression of Ahi-1 in pro–B BaF3 cells altered their differentiation pattern in vivo (Fig. 1 C), suggesting that modulation of Ahi-1/AHI-1 expression alters progenitor cell differentiation, including lineage switching, as previous reports have suggested for other oncogenes (25, 27–30). Third, a regulatory role of Ahi-1 in BCR-ABL–mediated transformation is evident as coexpression of Ahi-1 in BCR-ABL–inducible cells can reverse growth deficiencies induced by down-regulation of BCR-ABL. This biological effect is associated with sustained phosphorylation of BCR-ABL and enhanced activities of JAK2–STAT5. Finally, an AHI-1–BCR-ABL–-JAK2 interaction complex has been identified in human CML cells. This complex mediates sensitivity/resistance of primitive CML stem/progenitor cells to TKIs, as demonstrated by either coexpression or inhibition of Ahi-1/AHI-1 expression in BCR-ABL+ CML cells with a resulting modulation of CML cell sensitivity to these inhibitors. Collectively, these findings provide strong evidence of the importance of persistent deregulated expression of Ahi-1/AHI-1 to the sustained in vitro and in vivo transforming activities associated with CML and in the modulation of BCR-ABL-mediated malignant transformation and response of CML stem/progenitor cells to TKIs. This research has further identified Ahi-1/AHI-1 as a novel modulator of BCR-ABL that is specifically involved in the JAK2–STAT5 pathway.
Identification of an AHI-1–BCR-ABL–JAK2 interaction complex raises many interesting questions about the nature of the interaction, how it is regulated, and how it contributes to malignant transformation, altered BCR-ABL signaling, and IM response/resistance of CML stem/progenitor cells. Particularly notable is the observation that coexpression of Ahi-1 in BCR-ABL–inducible cells can rescue GF-independent cell growth that is inhibited by down-regulation of BCR-ABL. Interestingly, this renewed GF independence with introduction of Ahi-1 appears to be regulated by sustained phosphorylation of BCR-ABL, rather than its continual expression, as these effects were observed in cotransduced cells where BCR-ABL expression was inducibly suppressed in vitro (Fig. 7 B). These results suggest that physical interaction between Ahi-1 and BCR-ABL may stabilize a protein–protein interaction complex that enables constitutively active BCR-ABL tyrosine kinase activity and further alters specific downstream BCR-ABL signaling pathways that deregulate cellular proliferation and apoptosis control (Fig. 9 B). This is further supported by identification of JAK2 as an associated protein in this interaction complex and observation of enhanced activity of the JAK2–STAT5 pathway in BCR-ABL–inducible cells with cotransduction of Ahi-1 in the presence of GF stimulation with IL-3 (Fig. 8). Interestingly, all these findings can be replicated significantly in a human CML cell line system in which changes in AHI-1 expression are found to modulate tyrosine phosphorylation and protein expression of BCR-ABL and JAK2–STAT5 (Fig. 9 D). It is known that BCR-ABL signaling closely mimics signaling pathways of cytokine receptors and that both IL-3/GM-CSF receptor activation and the BCR-ABL oncoprotein can induce a tyrosine phosphorylation cascade of which numerous proteins, including JAK2 and STAT5, are common substrates (5, 6, 31). Interestingly, BCR-ABL–expressing cells have many similarities to cells induced by IL-3 stimulation (32) or cells with forced IL-3 overexpression (33, 34). Early studies have shown that BCR-ABL can interact with the common β chain of IL-3/GM-CSF receptor and constitutively activate JAK2 (35). In particular, increased phosphorylation of STAT5, which was previously thought to be an immediate function of the BCR-ABL oncoprotein, has now been shown in primary CML CD34+ progenitor cells to occur largely as a consequence of BCR-ABL–induced activation of IL-3 autostimulation, leading to activation of STAT5 (36). It has been reported that targeted disruption of the STAT5A and STAT5B genes reduces myeloid progenitor numbers, suggesting a nonredundant role for STAT5 in primitive normal hematopoiesis (37). Recent studies further suggest that autocrine production of GM-CSF may contribute to IM and NL-resistance in BCR-ABL+ progenitors through activation of JAK2 and STAT5 pathway (38), and inhibition of STAT5 expression by a shRNA approach significantly reduced colony formation of CD34+ CML progenitor cells in vitro (39). In addition, JAK2 is known to interact with the C-terminal region of BCR-ABL, and recent studies further show that mouse hematopoietic cells transformed by the T315I mutant of BCR-ABL can be induced to undergo apoptosis by a JAK2 inhibitor (AG490) (40–42). Collectively, these results indicate that activation of the JAK2–STAT5 pathway in CML stem/progenitors is likely to be an important mechanism contributing to responses to BCR-ABL–targeted therapies and identification of Ahi-1/AHI-1 as a novel mediator involved in this pathway suggests AHI-1 alone or in combination with JAK2 and STAT5, as potential additional therapeutic targets.
The most revealing result presented here is the finding that expression of BCR-ABL was consistently associated with an up-regulation of endogenous Ahi-1/AHI-1 transcripts and an increase in Ahi-1/AHI-1 protein expression in several overexpression and inducible experimental systems. Up-regulated AHI-1 expression has been noted in K562 cells, a cell line derived from a patient with CML in blast crisis (22). In particular, modulation of AHI-1 expression in K562 cells regulates BCR-ABL and JAK2–STAT5 activities, as described above. Importantly, AHI-1 is highly deregulated in primary CML stem/progenitor cells where levels of BCR-ABL transcripts are also highly elevated, suggesting that deregulated expression of BCR-ABL and AHI-1 may be clinically relevant to their cooperative activities that result in a permanently expanding clone of deregulated stem cells during leukemia development. Indeed, our study further demonstrates that highly elevated levels of AHI-1 in CML stem/progenitor cells from IM nonresponders and blast crisis patients correlate with their relative resistance to TKIs. Similarly, a potential mechanism for a physical interaction between Ahi-1/AHI-1 and BCR-ABL is revealed based on their molecular structures (20, 43), which are compatible with specific protein–protein interactions. Ahi-1 may interact with BCR-ABL through its SH3 domain or its SH3 binding sites (i.e., the SH3 domain of one protein interacting with the SH3 binding sites of the other) or through the SH2 domain of BCR-ABL if Ahi-1 is tyrosine phosphorylated (Ahi-1 contains two potential tyrosine phosphorylation sites). In addition, Ahi-1 may bind to a SH2-containing protein that is a substrate of BCR-ABL, thus forming a complex, as it is known that BCR-ABL is extensively tyrosine-phosphorylated, providing numerous, potential docking sites for SH2 domain-containing proteins (44). Moreover, Ahi-1 may interact with multiple domains of BCR-ABL, as demonstrated by other BCR-ABL–interacting proteins (45). Although more detailed structural mapping will be required to define specific protein–protein interactions between Ahi-1 and BCR-ABL, in association with JAK2, our findings that coexpression of Ahi-1 in BCR-ABL–transduced cells can completely rescue IM-induced suppression of cell growth in the presence of IL-3 suggests that Ahi-1 may not be a direct substrate of the BCR-ABL tyrosine kinase, but rather a modular protein that forms a stable protein interaction complex with other tyrosine phosphorylated proteins to mediate IL-3–dependent BCR-ABL and JAK2–STAT5 activities. In addition, this protein interaction complex seems to be disrupted by suppression of tyrosine phosphorylation of BCR-ABL by IM. Studies are underway to define how AHI-1 specifically interacts with BCR-ABL and JAK2. It was interesting to note that enhanced phosphorylation of Src was observed in Ahi-1 coexpressed BCR-ABL–inducible cells in the presence of IL-3 (Fig. 8), suggesting that other kinases are also activated with stimulation of IL-3 when BCR-ABL and AHI-1 are coexpressed. This finding also explains the observation that CML progenitor cells with suppression of AHI-1 experienced greater inhibition of CFC generation in response to dasatinib, a more potent TKI that also inhibits Src activity (14).
In conclusion, our studies demonstrate that Ahi-1/AHI-1 is a novel BCR-ABL interacting protein that is also physically associated with JAK2. This AHI-1–BCR-ABL–AK2 complex seems to modulate BCR-ABL–transforming activity and TKI response/resistance of CML stem/progenitor cells through the IL-3–dependent BCR-ABL and JAK2–STAT5 pathway. These results suggest that a more promising potential therapeutic approach would be the combined suppression of BCR-ABL tyrosine kinase activity, Ahi-1/AHI-1 expression, and JAK2–STAT5 signaling.
MATERIALS AND METHODS
Retroviral and lentiviral vectors and virus production.
An MSCV-Ahi-1-IRES-YFP retroviral vector was constructed by ligating a 3.5-kb fragment containing full-length Ahi-1 cDNA (20) to the upstream IRES of the MIY vector. The MSCV-BCR-ABL-IRES-GFP (BCR-ABL) retroviral vector was constructed as previously described (46). To generate a BCR-ABL lentiviral vector, the full-length BCR-ABL cDNA (J. Griffin, Dana Farber Cancer Institute, Boston, MA) was cloned into the pMNDU3-pGK-eGFP lentiviral vector (D. Kohn, University of Southern California, Los Angeles, CA) through an EcoRI site. To generate human AHI-1 lentiviral vector, plasmid containing full-length AHI-1 cDNA (AL136797, obtained from Resource Center Primary Database, Berlin, Germany) was subcloned into the EF1α-IRES-YFP lentiviral vector (with substitution of eGFP with YFP from an EF1α-IRES-eGFP lentiviral vector; P. Leboulch, Massachusetts Institute of Technology, Boston, MA) between AscI and PacI sites. To generate the AHI sh4 lentiviral construct, the H1 promoter-driven small hairpin against human AHI-1 was released from a modified pSUPER AHI sh4 construct (23) and inserted into an EF1α-IRES-YFP lentiviral vector. A similar subcloning procedure was performed to insert only the H1 promoter into the BamHI site of the EF1α-IRES-YFP lentiviral vector as control. All constructs were verified by restriction enzyme digestion analysis and DNA sequencing.
Lentiviral production was performed after a previously described procedure (47). The titers of these concentrated viruses were measured with HeLa cells as ∼1 × 108 infectious U/ml for the BCR-ABL virus and 2 × 108 infectious U/ml for control, AHI sh4, and AHI-1 viruses. Helper-free retrovirus was obtained by transfecting ecotropic Phoenix packaging cells (48) cultured in DMEM plus 10% fetal calf serum using the calcium-phosphate precipitation method, as previously described (46).
Human primary cells and FACS sorting.
Peripheral blood was obtained from 23 CML patients who had elevated white blood cell counts (105 × 109 cells/liter to 420 × 109 cells/liter) and had not been treated with any inhibitors, 10 of whom subsequently developed IM resistance. CB cells were obtained from mothers undergoing cesarean delivery of healthy, full-term infants. In all cases, informed consent was obtained, and the procedures used, including the Declaration of Helsinki protocol, were approved by the Research Ethics Board of the University of British Columbia (Vancouver).
Lin−CD34+ CB cells from six individuals were isolated using the EasySep cell separation kit (Stem Cell Technologies) according to the manufacturer's instruction. The purity of the enriched cells was measured by FACS as >95%. FACS isolation of BaF3 cells, Sca-1+lin− mouse BM cells, and K562 cells transduced with GFP, YFP, or GFP and YFP using a FACStar Plus or FACSVantage (BD Biosciences) was previously described (46).
Transduction of murine and human hematopoetic cells.
BM cells from 6–10-wk-old C57BL/6 mice were harvested 4 d after injection of 150 mg/kg 5-FU. Low-density cells were isolated and cultured for 48 h in serum-free medium (SFM) supplemented with BIT (StemCell Technologies), to which 100 ng/ml mouse Steel Factor (SF; STEMCELL Technologies), 10 ng/ml mouse IL-3 (StemCell Technologies), and 10 ng/ml human IL-6 (Cangene) were also added. The BM cells, as well as IL-3–dependent BaF3 cells, were then collected and resuspended at 5 × 105 cells/ml in VCS diluted 1:3 in DMEM containing 15% FCS, protamine sulfate (5 μg/ml), and SF, IL-3, and IL-6 at the same concentrations as described above. BaF3 cells were cultured with IL-3 only. The cells were then transferred to Petri dishes that had been precoated with fibronectin (Sigma-Aldrich) and preloaded with virus as previously described (46).
CD34+ CB cells were prestimulated with a cocktail of cytokines and transduced with virus as previously described (25). BCR-ABL–transduced cells were sorted for GFP+ cells after 3 d of culture and sorted GFP+BCR-ABL+ cells were retransduced with either control or AHI/sh4 virus. Cells transduced with only control or AHI/sh4 were cultured for 6 d before FACS sorting. The GFP+YFP+ cells (BCR-ABL+AHI-1/sh4+) were FACS-sorted after 3 d of expansion. Lin−CD34+ CML cells were prestimulated and transduced with either control or AHI/sh4 virus in a similar manner to that described for CB cells. After 3–4 d of expansion, YFP+CD34+ cells were FACS sorted.
Cell culture.
FACS-sorted cells were cultured in SFM with or without GFs. At various time points, viable cell numbers were determined by hemocytometer counts of trypan blue-excluding cells. For single cells sorting, YFP+–transduced K562 cells were sorted into the wells of a 96 well plate by FACS. Subsequent clone formation (presence of ≥2 refractile cells) and clone size (total number of refractile cells) in wells seeded with single viable (PI−) cells was assessed by direct visual inspection using an inverted microscope.
Proliferation assays.
Aliquots of 2 × 104 cells were cultured in triplicate in round bottom wells of 96-well Falcon plates in 100 μl media per well. The cells were incubated for 24 and 48 h at 37°C. The amount of tritiated [3H]thymidine incorporated during a 4-h pulse of the culture was measured. In brief, 1 μCi of tritiated [3H]thymidine was added to each well. 4 h later, the cells were harvested onto a membrane using a Skatron instruments combi Harvester (LKB Wallace-PerkinElmer). The amount of tritiated [3H]thymidine was measured with a LKB Betaplate scintillation counter.
Apoptosis analysis.
The apoptosis analysis was performed using an Apoptosis Detection kit (BD Biosciences) following the manufacturer's instruction. In brief, aliquots of 1 × 105 cells were washed twice with PBS, resuspended in 100 μl binding buffer, and incubated with 5 μl of 7-amino-actinomycin D and 5 μl of PE-conjugated Annexin V antibody at room temperature in the dark for 15 min. 400 μl binding buffer was added for FACS analysis.
Progenitor assays.
CFCs and LTC-ICs for mouse BM cells were assayed using culture media from Stem Cell Technologies. Colony counts were performed, using standard scoring criteria, on the basis of the ability to produce colonies containing a minimum of 20 cells after 12 to 14 d of incubation (49). LTCs were set up and maintained using preestablished irradiated mouse marrow LTC feeder layers and weekly half medium changes of LTC medium as previously described (46). CFC assays for transduced K562 cells, human CB cells and CML cells were performed in FCS-containing methylcellulose medium (StemCell Technologies) in the presence or absence of GFs ± imatinib (0.1–10 μM), dasatinib (10–150 nM), and NL (0.1–10 μM).
Q-RT-PCR.
Total RNA was extracted from aliquots of cells using the Absolutely RNA Microprep kit (Stratagene) or TRIzol (Invitrogen), and RT reactions were performed as previously described (46). Real-time PCR was performed using 25 μl of 2X SYBR Green PCR Master Mix (Applied Biosystems), 1 μl of 20 pM of specific primers, 1–2 μl cDNA, and water to a final volume of 50 μl, as previously described (19). Specific forward and reverse primers to detect total expression levels were as follows: AHI-1, 5′-GCCGAGATAGCCCGGTTTATC-3′ and 5′-TCAGTTCGGTGAATGTAAACTCC-3′; BCR-ABL, 5′-CATTCCGCTGACCATCAATAAG-3′ and 5′-GATGCTACTGGCCGCTGAAG-3′; GAPDH, 5′-CCCATCACCATCTTCCAGGAG-3′ and 5′-CTTCTCCATGGTGGTGAAGACG-3′. Optimal conditions for the amplification of these gene products and for normalization of the Ct values of the gene of interest to that of GAPDH were previously described (19).
Immunoprecipitation and Western blot analyses.
Cells were lysed in PBS as previously described (19). For immunoprecipitation, protein extracts were diluted to ∼1 μg total protein/μl with PBS containing proteinase inhibitors, and specific antibodies were added and incubated at 4°C overnight. The immunocomplexes were then captured by protein G bead slurry, rocked, and separated from the beads by washing. The samples were then heated at 70°C for 10 min before Western blotting. Protein lysates were then separated on NuPAGE Novex bis-tris gels (Invitrogen), and the filters were probed with specific antibodies as previously described (19). Primary antibodies used in these studies included a monoclonal anti-ABL antibody (8E9; BD Biosciences), an antiphosphotyrosine antibody (4G10; Millipore), anti–phospho-JAK2 (Y1007/Y1008) and anti-JAK2 antibodies (Millipore) in transduced BaF3 cells, anti–phospho-JAK2 (Y1007/Y1008) and anti-JAK2 antibodies (Cell Signaling Technology) in transduced K562 cells, anti–phospho-Src (Tyr 416) and anti-Src antibodies (Cell Signaling Technology), anti–phospho-Akt and anti-Akt antibodies (Cell Signaling Technology), anti–phospho-STAT5 (Tyr694) and anti-STAT5 antibodies (Cell Signaling Technology), anti–phospho-NF-κB p65 and anti- NF-κB p65 antibodies (Cell Signaling Technology), polyclonal anti–Ahi-1 antibody (C-terminal) (23), C-terminal monoclonal anti–Ahi-1 antibody (C-mAhi-1 M5) generated by a standard procedure as previously described (50), and polyclonal N-terminal AHI-1 antibody, IMX-3395 (IMGENEX). Relative expression levels were normalized to expression levels of human actin (Sigma-Aldrich) after quantification with ImageQuant software. The peptide competition assay was completed as per an online protocol (http://www.rockland-inc.com/commerce/misc/methods) by Rockland Immunochemicals, Inc.
Animals.
NOD/SCID-β2m−/− mice were bred and maintained in microisolator cages and provided with autoclaved food and water. Mice were irradiated at 8–10 wk of age with 350 cGy 137Cs γ-rays. Cells were injected into mice intravenously a few hours after they had been irradiated. Mice were monitored daily for signs of weight loss or lethargy. Peripheral blood was collected every 1–2 wk, and leukocytes were analyzed by FACS as previously described (46). For tumor formation studies, 10 million cells were injected subcutaneously into the left and right flank of mice, and the mice were monitored daily for the tumor growth. The tumor volume was calculated from the greatest transverse (width) and longitudinal (length) diameter of the tumor using the formula: tumor volume = length × width2/2. All animal experiments were performed in the Animal Resource Centre of BC Cancer Research Centre, and the procedures used were approved by the Animal Care Committee of the University of British Columbia (Vancouver).
Statistical analysis.
Results are shown as the mean ± SEM of values obtained in independent experiments. Differences between groups were assessed using the Student's t test for paired samples.
Online supplemental material.
Fig. S1 shows that overexpression of Ahi-1 confers growth advantage on BaF3 cells and enhances effects of BCR-ABL with increased protein expression and tyrosine phosphorylation of p210BCR-ABL. Fig. S2 shows that Ahi-1 and BCR-ABL–transduced cells purified from BM cells of diseased mice have increased proliferative and antiapoptotic activities and these effects can be enhanced by BCR-ABL. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20072316/DC1.
Acknowledgments
The authors thank K. Saw and K. Lambie for excellent technical assistance; Dr. D. Forrest and other members of the Leukemia/Bone Marrow Transplant Program of British Columbia for providing patient samples; the Terry Fox Laboratory FACS Facility for assistance in cell sorting; the Stem Cell Assay Laboratory for cell processing and cryopreservation of normal and CML samples; Novartis for imatinib and NL; Bristol-Myers Squibb for dasatinib; and Dr. C. Eaves for useful discussion.
This work was supported by grants from the Cancer Research Society and in part by the Leukemia and Lymphoma Society of Canada (X. Jiang). Y. Zhao was a recipient of a Stem Cell Network Fellowship and a Leukemia and Lymphoma Society of Canada Fellowship and A. Ringrose was a recipient of a BC Cancer Studentship. D. DeGeer received a Medical Genetics Graduate Entrance Studentship from University of British Columbia and a BC Cancer Agency Recruitment Training Incentive Studentship. E. Kennah was a recipient of a Medical Genetics Graduate Entrance Studentship from University of British Columbia. X. Jiang is a Michael Smith Foundation for Health Research Scholar.
The authors have no conflicting financial interests.
Abbreviations used: Ahi-1, Abelson helper integration site 1; BFU-E, burst-forming-units-erythroid; CB, cord blood; CFC, colony-forming cell; CFU-GEMM, colony-forming units-granulocyte-erythrocyte-monocyte-megakaryocyte; CFU-GM, colony-forming units-granulocyte-macrophage; CML, chronic myeloid leukemia; co-IP, coimmunoprecipitation; Dox, doxycycline; DS, dasatinib; GF, growth factor; IM, imatinib mesylate; IRES, internal ribosomal entry site; LTC-IC, long-term culture-initiating cell; NL, nilotinib; Q-RT-PCR, quantitative real-time-PCR; PI3K, phosphatidylinositol 3-kinase; SFM, serum-free medium; TKI, tyrosine kinase inhibitor.
References
- 1.Goldman, J.M., and J.V. Melo. 2003. Chronic myeloid leukemia–advances in biology and new approaches to treatment. N. Engl. J. Med. 349:1451–1464. [DOI] [PubMed] [Google Scholar]
- 2.Jiang, X., C. Smith, A. Eaves, and C. Eaves. 2007. The challenges of targeting chronic myeloid leukemia stem cells. Clin. Lymph. Myeloma. 7:S71–S80. [DOI] [PubMed] [Google Scholar]
- 3.Lugo, T.G., A.M. Pendergast, A.J. Muller, and O.N. Witte. 1990. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science. 247:1079–1082. [DOI] [PubMed] [Google Scholar]
- 4.Druker, B.J., S. Tamura, E. Buchdunger, S. Ohno, G.M. Segal, S. Fanning, J. Zimmermann, and N.B. Lydon. 1996. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 2:561–566. [DOI] [PubMed] [Google Scholar]
- 5.Sattler, M., and J.D. Griffin. 2003. Molecular mechanisms of transformation by the BCR-ABL oncogene. Semin. Hematol. 40:4–10. [DOI] [PubMed] [Google Scholar]
- 6.Jiang, X. 2007. Molecular and cellular mechanisms of deregulated hematopoietic stem cell functions in chronic myeloid leukemia. In Cancer Nanotechnology. American Scientific Publishers. pp. 135–157.
- 7.Druker, B.J., M. Talpaz, D.J. Resta, B. Peng, E. Buchdunger, J.M. Ford, N.B. Lydon, H. Kantarjian, R. Capdeville, S. Ohno-Jones, and C.L. Sawyers. 2001. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 344:1031–1037. [DOI] [PubMed] [Google Scholar]
- 8.Kantarjian, H., C. Sawyers, A. Hochhaus, F. Guilhot, C. Schiffer, C. Gambacorti-Passerini, D. Niederwieser, D. Resta, R. Capdeville, U. Zoellner, et al. 2002. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N. Engl. J. Med. 346:645–652. [DOI] [PubMed] [Google Scholar]
- 9.O'Brien, S.G., F. Guilhot, R.A. Larson, I. Gathmann, M. Baccarani, F. Cervantes, J.J. Cornelissen, T. Fischer, A. Hochhaus, T. Hughes, et al. 2003. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. New. Eng. J. Med. 348:994–1004. [DOI] [PubMed] [Google Scholar]
- 10.Deininger, M., E. Buchdunger, and B.J. Druker. 2005. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 105:2640–2653. [DOI] [PubMed] [Google Scholar]
- 11.O'Hare, T., A.S. Corbin, and B.J. Druker. 2006. Targeted CML therapy: controlling drug resistance, seeking cure. Curr. Opin. Genet. Dev. 16:92–99. [DOI] [PubMed] [Google Scholar]
- 12.Azam, M., R.R. Latek, and G.Q. Daley. 2003. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 112:831–843. [DOI] [PubMed] [Google Scholar]
- 13.Shah, N.P., J.M. Nicoll, B. Nagar, M.E. Gorre, R.L. Paquette, J. Kuriyan, and C.L. Sawyers. 2002. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2:117–125. [DOI] [PubMed] [Google Scholar]
- 14.Shah, N.P., C. Tran, F.Y. Lee, P. Chen, D. Norris, and C.L. Sawyers. 2004. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 305:399–401. [DOI] [PubMed] [Google Scholar]
- 15.Weisberg, E., P.W. Manley, W. Breitenstein, J. Bruggen, S.W. Cowan-Jacob, A. Ray, B. Huntly, D. Fabbro, G. Fendrich, E. Hall-Meyers, et al. 2005. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 7:129–141. [DOI] [PubMed] [Google Scholar]
- 16.Graham, S.M., H.G. Jorgensen, E. Allan, C. Pearson, M.J. Alcorn, L. Richmond, and T.L. Holyoake. 2002. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 99:319–325. [DOI] [PubMed] [Google Scholar]
- 17.Copland, M., A. Hamilton, L.J. Elrick, J.W. Baird, E.K. Allan, N. Jordanides, M. Barow, J.C. Mountford, and T.L. Holyoake. 2006. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML, but does not eliminate the quiescent fraction. Blood. 107:4532–4539. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, X., Y. Zhao, C. Smith, M. Gasparetto, A. Turhan, A. Eaves, and C. Eaves. 2007. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 21:926–935. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, X., K.M. Saw, A. Eaves, and C. Eaves. 2007. Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J. Natl. Cancer Inst. 99:680–693. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, X., Z. Hanna, M. Kaouass, L. Girard, and P. Jolicoeur. 2002. Ahi-1, a novel gene encoding a modular protein with WD40-repeat and SH3 domains, is targeted by the Ahi-1 and Mis-2 provirus integrations. J. Virol. 76:9046–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard, L., Z. Hanna, N. Beaulieu, C.D. Hoemann, C. Simard, C.A. Kozak, and P. Jolicoeur. 1996. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 10:1930–1944. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, X., Y. Zhao, W.Y. Chan, S. Vercauteren, E. Pang, S. Kennedy, F. Nicolini, A. Eaves, and C. Eaves. 2004. Deregulated expression in Ph+ human leukemias of AHI-1, a gene activated by insertional mutagenesis in mouse models of leukemia. Blood. 103:3897–3904. [DOI] [PubMed] [Google Scholar]
- 23.Ringrose, A., Y. Zhou, E. Pang, L. Zhou, A.E. Lin, G. Sheng, X.J. Li, A. Weng, M.W. Su, M.R. Pittelkow, and X. Jiang. 2006. Evidence for an oncogenic role of AHI-1 in Sezary Syndrome, a leukemic variant of human cutaneous T-cell lymphomas. Leukemia. 20:1593–1601. [DOI] [PubMed] [Google Scholar]
- 24.Jamieson, C.H., L.E. Ailles, S.J. Dylla, M. Muijtjens, C. Jones, J.L. Zehnder, J. Gotlib, K. Li, M.G. Manz, A. Keating, et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351:657–667. [DOI] [PubMed] [Google Scholar]
- 25.Chalandon, Y., X. Jiang, G. Hazelwood, S. Loutet, E. Conneally, A. Eaves, and C. Eaves. 2002. Modulation of p210BCR-ABL activity in transduced primary human hematopoietic cells controls lineage reprogramming. Blood. 99:3197–3204. [DOI] [PubMed] [Google Scholar]
- 26.Dugray, A., J.F. Geay, A. Foudi, M.L. Bonnet, W. Vainchenker, F. Wendling, F. Louache, and A.G. Turhan. 2001. Rapid generation of a tetracycline-inducible BCR-ABL defective retrovirus using a single autoregulatory retroviral cassette. Leukemia. 15:1658–1662. [DOI] [PubMed] [Google Scholar]
- 27.Era, T., and O.N. Witte. 2000. Regulated expression of P210 Bcr-Abl during embryonic stem cell differentiation stimulates multipotential progenitor expansion and myeloid cell fate. Proc. Natl. Acad. Sci. USA. 97:1737–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elefanty, A.G., and S. Cory. 1992. bcr-abl-Induced cell lines can switch from mast cell to erythroid or myeloid differentiation in vitro. Blood. 79:1271–1281. [PubMed] [Google Scholar]
- 29.Borzillo, G.V., R.A. Ashmun, and C.J. Sherr. 1990. Macrophage lineage switching of murine early pre-B lymphoid cells expressing transduced fms genes. Mol. Cell. Biol. 10:2703–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chalandon, Y., X. Jiang, S. Loutet, A.C. Eaves, and C.J. Eaves. 2004. Growth autonomy and lineage switching in BCR-ABL-transduced human cord blood cells depend on different functional domains of BCR-ABL. Leukemia. 18:1006–1012. [DOI] [PubMed] [Google Scholar]
- 31.Baker, S.J., S.G. Rane, and E.P. Reddy. 2007. Hematopoietic cytokine receptor signaling. Oncogene. 26:6724–6737. [DOI] [PubMed] [Google Scholar]
- 32.Steelman, L.S., S.C. Pohnert, J.G. Shelton, R.A. Franklin, F.E. Bertrand, and J.A. McCubrey. 2004. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 18:189–218. [DOI] [PubMed] [Google Scholar]
- 33.Chang, J.M., D. Metcalf, R.A. Lang, T.J. Gonda, and G.R. Johnson. 1989. Nonneoplastic hematopoietic myeloproliferative syndrome induced by dysregulated multi-CSF (IL-3) expression. Blood. 73:1487–1497. [PubMed] [Google Scholar]
- 34.Just, U., M. Katsuno, C. Stocking, E. Spooncer, and M. Dexter. 1993. Targeted in vivo infection with a retroviral vector carrying the interleukin-3 (multi-CSF) gene leads to immortalization and leukemic transformation of primitive hematopoietic progenitor cells. Growth Factors. 9:41–55. [DOI] [PubMed] [Google Scholar]
- 35.Wilson-Rawls, J., J. Liu, P. Laneuville, and R.B. Arlinghaus. 1997. P210 Bcr-Abl interacts with the interleukin-3 beta c subunit and constitutively activates Jak2. Leukemia. 11:428–431. [PubMed] [Google Scholar]
- 36.Jiang, X., A. Lopez, T. Holyoake, A. Eaves, and C. Eaves. 1999. Autocrine production and action of IL-3 and granulocyte colony-stimulating factor in chronic myeloid leukemia. Proc. Natl. Acad. Sci. USA. 96:12804–12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teglund, S., C. McKay, E. Schuetz, J.M. van Deursen, D. Stravopodis, D. Wang, M. Brown, S. Bodner, G. Grosveld, and J.N. Ihle. 1998. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 93:841–850. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Y., D. Cai, C. Brendel, C. Barett, P. Erben, P.W. Manley, A. Hochhaus, A. Neubauer, and A. Burchert. 2007. Adaptive secretion of granulocyte-macrophage colony-stimulating factor (GM-CSF) mediates imatinib and nilotinib resistance in BCR/ABL+ progenitors via JAK-2/STAT-5 pathway activation. Blood. 109:2147–2155. [DOI] [PubMed] [Google Scholar]
- 39.Scherr, M., A. Chaturvedi, K. Battmer, I. Dallmann, B. Schultheis, A. Ganser, and M. Eder. 2006. Enhanced sensitivity to inhibition of SHP2, STAT5, and Gab2 expression in chronic myeloid leukemia (CML). Blood. 107:3279–3287. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto, N., K. Sugita, K. Goi, T. Inukai, K. Lijima, T. Tezuka, S. Kojika, M. Nakamura, K. Kagami, and S. Nakazawa. 2001. The JAK2 inhibitor AG490 predominantly abrogates the growth of human B-precursor leukemic cells with 11q23 translocation or Philadelphia chromosome. Leukemia. 15:1758–1768. [DOI] [PubMed] [Google Scholar]
- 41.Xie, S., Y. Wang, J. Liu, T. Sun, M.B. Wilson, T.E. Smithgall, and R.B. Arlinghaus. 2001. Involvement of Jak2 tyrosine phosphorylation in Bcr-Abl transformation. Oncogene. 20:6188–6195. [DOI] [PubMed] [Google Scholar]
- 42.Samanta, A.K., H. Lin, T. Sun, H. Kantarjian, and R.B. Arlinghaus. 2006. Janus kinase 2: a critical target in chronic myelogenous leukemia. Cancer Res. 66:6468–6472. [DOI] [PubMed] [Google Scholar]
- 43.Van Etten, R.A. 2002. Studying the pathogenesis of BCR-ABL+ leukemia in mice. Oncogene. 21:8643–8651. [DOI] [PubMed] [Google Scholar]
- 44.Seet, B.T., I. Dikic, M.M. Zhou, and T. Pawson. 2006. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7:473–483. [DOI] [PubMed] [Google Scholar]
- 45.Dai, Z., and A.-M. Pendergast. 1995. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 9:2569–2582. [DOI] [PubMed] [Google Scholar]
- 46.Jiang, X., E. Ng, C. Yip, W. Eisterer, Y. Chalandon, M. Stuible, A. Eaves, and C.J. Eaves. 2002. Primitive interleukin 3 null hematopoietic cells transduced with BCR-ABL show accelerated loss after culture of factor-independence in vitro and leukemogenic activity in vivo. Blood. 100:3731–3740. [DOI] [PubMed] [Google Scholar]
- 47.Dull, T., R. Zufferey, M. Kelly, R.J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinsella, T.M., and G.P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405–1413. [DOI] [PubMed] [Google Scholar]
- 49.Eaves, C.J. 1995. Assays of hemopoietic progenitor cells. In Williams Hematology. E. Beutler, M.A. Lichtman, B.S. Coller, and T.J.Kipps, editors. McGraw-Hill, Inc. L22–L26.
- 50.Harlow, E., and D. Lane. 1988. Antibodies A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.