Abstract
Epithelial sodium channels (ENaCs) perform diverse physiological roles by mediating Na+ absorption across epithelial surfaces throughout the body. Excessive Na+ absorption in kidney and colon elevates blood pressure and in the airways disrupts mucociliary clearance. Potential therapies for disorders of Na+ absorption require better understanding of ENaC regulation. Recent work has established partial and selective proteolysis of ENaCs as an important means of channel activation. In particular, channel-activating transmembrane serine proteases (CAPs) and cognate inhibitors may be important in tissue-specific regulation of ENaCs. Although CAP2 (TMPRSS4) requires catalytic activity to activate ENaCs, there is not yet evidence of ENaC fragments produced by this serine protease and/or identification of the site(s) where CAP2 cleaves ENaCs. Here, we report that CAP2 cleaves at multiple sites in all three ENaC subunits, including cleavage at a conserved basic residue located in the vicinity of the degenerin site (α-K561, β-R503, and γ-R515). Sites in α-ENaC at K149/R164/K169/R177 and furin-consensus sites in α-ENaC (R205/R231) and γ-ENaC (R138) are responsible for ENaC fragments observed in oocytes coexpressing CAP2. However, the only one of these demonstrated cleavage events that is relevant for the channel activation by CAP2 takes place in γ-ENaC at position R138, the previously identified furin-consensus cleavage site. Replacement of arginine by alanine or glutamine (α,β,γR138A/Q) completely abolished both the Na+ current (INa) and a 75-kD γ-ENaC fragment at the cell surface stimulated by CAP2. Replacement of γ-ENaC R138 with a conserved basic residue, lysine, preserved both the CAP2-induced INa and the 75-kD γ-ENaC fragment. These data strongly support a model where CAP2 activates ENaCs by cleaving at R138 in γ-ENaC.
INTRODUCTION
Abnormal activity of the epithelial sodium channel (ENaC) is implicated in diseases of the cortical collecting ducts of the kidney (Liddle et al., 1963), in the airways (Boucher, 2004; Schild, 2004), and in the middle ear (Guipponi et al., 2002). Novel therapies for these disorders may follow from better understanding of ENaC regulation. Work from several groups identify proteases as important ENaC regulators (Vallet et al., 1997, 2002; Vuagniaux et al., 2000, 2002; Donaldson et al., 2002; Hughey et al., 2003, 2004; Caldwell et al., 2004, 2005; Harris et al., 2007). The results of these studies suggest that ENaC is activated by its partial and selective proteolysis during channel assembly and processing or while channels are resident in the plasma membrane. Despite this progress, detailed knowledge of the molecular mechanism(s) underlying proteolytic activation of ENaC is limited.
Vallet et al. (1997) obtained the first evidence of an epithelial membrane protease–activating ENaC in an autocrine fashion. They cloned a channel-activating protease (CAP), or CAP1 (prostasin), from a Xenopus kidney epithelial cell line (A6) and established that it activates ENaC when coexpressed in Xenopus oocytes (Vallet et al., 1997). Subsequently, two additional serine proteases, mCAP2 (homologue of human transmembrane protease serine 4 [TMPRSS4]) and mCAP3 (MT-SP1/Matriptase or epithin), were identified by homology cloning and found to increase the activity of ENaC coexpressed in Xenopus oocytes (Vuagniaux et al., 2002). These studies found no effects of CAPs on the number of channels at the surface, suggesting that CAPs increase ENaC activity by changing open probability (PO) (Vallet et al., 2002; Vuagniaux et al., 2002; Andreasen et al., 2006). Direct evidence that proteases can increase ENaC PO was first provided by Caldwell et al. (2004, 2005). They found that trypsin or human neutrophil elastase (hNE) added to the outer face of outside-out patches of NIH-3T3 ENaC cells increased PO of “near silent” ENaCs by up to 28-fold (Caldwell et al., 2004, 2005). Subsequently, a 65-kD γ-ENaC fragment generated at the surface by hNE was linked to hNE-stimulated ENaC current (Harris et al., 2007). Adebamiro et al. (2007) identified specific residues in γ-ENaC that were required for elastase to stimulate ENaC current.
ENaC is a multimeric channel consisting of topologically similar α-, β-, and γ-subunits (Canessa et al., 1994). A key link between protease-mediated cleavage of ENaC and channel activity was discovered when Hughey et al. (2003, 2004) detected minimal consensus cleavage sequences for convertases of the furin family in the α- and γ-ENaC subunits. They demonstrated that mutagenesis of these sites eliminated specific fragments of α- and γ-ENaC and caused a reduced basal ENaC current that was recovered by application of exogenous trypsin. Although furin family convertases are known to cleave proteins during trafficking to the cell membrane, they are also active at the cell surface (Thomas, 2002), leaving some doubt as to the subcellular location of furin action on ENaC. In addition, whereas mutagenesis of furin consensus sequences in α-ENaC had the largest effect on basal ENaC current (Hughey et al., 2004), the precise roles of furin-mediated cleavage of the two α- and single γ-ENaC furin sites are not known.
Based on these observations, a simple model for ENaC regulation by proteases emerged (Planes and Caughey, 2007). Cleavage of α- and γ-subunits by furin or related convertases is assumed to occur during channel assembly and delivery to the cell membrane, but to a variable extent. This results in channels arriving at the surface with a range of proteolytic activation. Uncleaved or perhaps mixed multimers of cleaved and uncleaved subunits are thought to be susceptible to further cleavage and stimulation by surface or soluble proteases (Planes and Caughey, 2007).
CAPs are the main candidates identified thus far as surface-bound proteases that may activate ENaC at the cell surface. A polybasic stretch of residues in γ-ENaC was recently identified by mutagenesis as being required for CAP1 stimulation of ENaC (Bruns et al., 2007). However, studies comparing wild-type (WT) and catalytically inactive CAP1-3 mutants have shown that catalytically inactive CAP1 fully stimulates ENaC, suggesting it has an indirect role in activating ENaC (Andreasen et al., 2006). Thus, the mechanism of CAP1 regulation is not entirely clear. In contrast to CAP1, both CAP2 and CAP3 must possess catalytic activity to stimulate ENaC (Andreasen et al., 2006). Here, we have focused on the activation of ENaC by CAP2. Previously, the sites within ENaC where CAP2 cleaves were not known, nor had CAP2-mediated cleavage at a specific ENaC residue been shown to cause an increase in ENaC PO. Here, we report that CAP2 cleaves all three ENaC subunits, both with and without associated stimulation.
MATERIALS AND METHODS
Plasmid Preparation
For biochemical analyses of ENaC subunit proteolysis, cDNAs encoding rat α-, β-, and γ-ENaC with HA–N-terminal (HA-NT) and V5–C-terminal (V5-CT) epitope tags were generated. WT and mutant constructs (α-ENaC: K149A/R164A/K169A/R177A, R205A/R231A, K486A, K501A, R503A, K504A, K512A, R519A, K524A, K544A, R545A, K550A, and K561A; β-ENaC: K411A, R416A, R435A, K443A, K452A, R477A, R487A, K488A, K492A, R503A, and S518C; γ-ENaC: R138A, R138K, R138Q, R153Q, K156Q, K168Q, K170Q, R172Q, R178Q/K179Q/R180Q/K181Q, K185Q, K189Q, K200Q, K201Q, K504A, K510A, and R515A; CAP2: S387A) were generated by PCR and cloned into pCR-BluntII-TOPO (Invitrogen), linearized (HindIII), and in vitro transcribed using T7 RNA polymerase. A PolyA tail was added after transcription (Ambion). Mutations were performed with the quickchange multisite-directed mutagenesis kit (Stratagene). The WT ENaC plasmids were provided by B. Rossier (Université de Lausanne, Lausanne, Switzerland). The sequence of all plasmids was verified at the University of North Carolina sequencing facility.
Functional Studies of Na+ Channels in Xenopus Oocytes
V–VI-stage healthy oocytes were harvested as described previously (Donaldson et al., 2002) and maintained in modified Barth's solution (MBS) at 18°C. Animals were maintained and studied under protocols approved by the University of North Carolina Institutional Animal Care and Use Committee. cRNAs encoding WT of both untagged and HA-NT– and V5-CT–tagged subunits or mutant HA-NT plus V5-CT–tagged subunits rat α-, β-, and γ-ENaC (0.3 ng each) and CAP2 (TMPRSS4) cRNA (1 ng) were coinjected into oocytes. 24 h after injection, two-electrode voltage clamping was performed using a Genclamp amplifier (MDS Analytical Technologies) in a constant perfusion system. Currents were measured in the presence and absence of 10 μM amiloride, with membrane voltage clamped to −100 mV. Currents were digitized and recorded using a Digidata 1200 A/D converter (MDS Analytical Technologies) and Axoscope software. 2 μg/ml trypsin was perfused for 5 min after first measuring the basal amiloride–sensitive current (INa) of each egg. Some eggs were incubated overnight with 50 μg/ml aprotinin, as indicated. All results are expressed as the mean ± SE or as fold stimulation by CAP2 or trypsin. The means of two groups were tested for significant difference using an unpaired Student's t test, and differences between three or more groups were evaluated using ANOVA analysis (GraphPad Software, Inc.). Proteins extracted from control and injected oocytes were analyzed by Western blots to verify expression of ENaC and actin.
Surface Labeling
Xenopus oocytes were injected with desired combinations of WT or mutant double epitope–tagged α, β, and γ rat ENaC subunits (0.3 ng each) and with or without CAP2 or CAP1 cRNA (1 ng). After 24 h, 70 oocytes per experimental condition were pre-chilled on ice for 30 min and labeled with 0.7 mg/ml sulfo-NHS-biotin in MBS-Ca2+ (mM), 85 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.41 CaCl, 0.33 Ca(NO3), 16.3 Hepes titrated to pH 8.0 with NaOH, while tumbling gently for 20 min at 4°C. Oocytes were washed twice with chilled MBS-Ca2+ buffer and incubated in MBS-Ca2+ buffer with 100 mM glycine for 10 min at 4°C to quench free biotin. Oocytes were washed again three times with chilled MBS-Ca2+ buffer, and then lysed with lysis buffer (in mM: 20 Tris, 50 NaCl, 50 NaF, 10 β-glycerophosphate, 5 Na4P2O7 pyrophosphate, 1 EDTA, pH 7.5, containing protease inhibitors [complete; Roche], aprotinin [Sigma-Aldrich]). Cell lysates were prepared by passing oocytes through a 27G1/2 needle twice and by centrifugation at 3,600 rpm for 10 min at 4°C. Supernatants were transferred to new tubes, and samples were spun at 14,000 rpm for 20 min at 4°C. Supernatants were discarded and pellets were solubilized in solubilization buffer (in mM: 50 Tris, 100 NaCl, 1% Triton X-100, 1% NP-40, 0.2% SDS, 0.1% Na deoxycholate, 20 NaF, 10 Na4P2O7 pyrophosphate, 10 EDTA plus protease inhibitor cocktail, pH 7.5). Total inputs were taken from whole cell samples representing 4% of total protein. Solubilized proteins were incubated with 100 μl of neutravidin beads (Thermo Fisher Scientific) overnight while tumbling at 4°C. Samples were washed twice with 500 mM NaCl, 50 mM Tris, pH 7.5, buffer and once with 150 mM NaCl, 50 mM Tris, pH 7.5, buffer. Laemmli buffer was added and samples were loaded on a 4–12% gradient Tris-glycine gel after incubation for 10 min at 96°C. Samples were transferred to 0.45 μm polyvinylidene difluoride membranes (Millipore), and Western blot analysis was performed using an anti-V5 (Invitrogen), anti-HA (Covance), and anti-actin (Millipore) monoclonal antibodies. Surface ENaC-biotinylated fragments were quantified using the metamorph imaging 4.5 program (Hooker Microscopy Facility, University of North Carolina). Densitometry of selected bands was performed using uninjected oocyte samples as background signal.
Western Blot Analysis
Proteins were extracted from oocytes as described above. Biotinylated and total proteins were solubilized by boiling in Laemmli sample buffer for 10 min before loading onto 4–12% SDS-PAGE gels. Western blots were performed with anti-V5 (Invitrogen), anti-HA (Covance), anti-express (Invitrogen), and anti-actin (Millipore) antibodies.
RESULTS
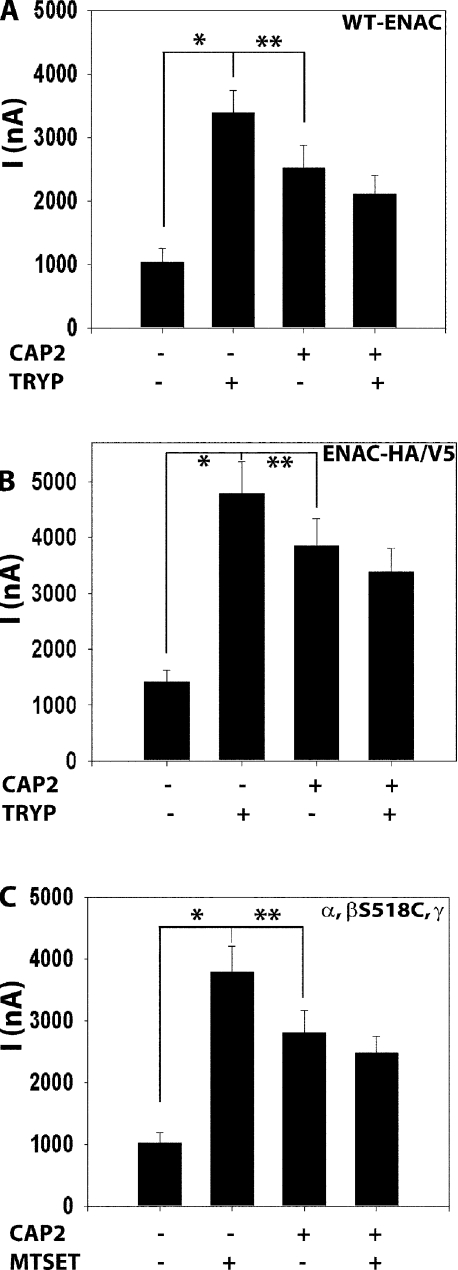
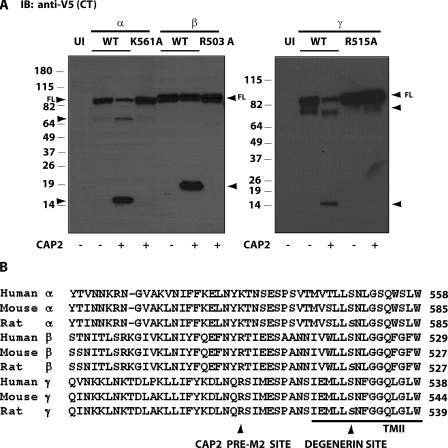
We applied functional and biochemical approaches in parallel to assess the effects of CAP2 on the activity, number, and cleavage of coexpressed ENaC in Xenopus oocytes. To establish that our experimental conditions were informative for these analyses, we first observed that coexpression of CAP2 with WT ENaC elevated basal INa by ∼2.5-fold over WT ENaC alone (Fig. 1 A). Whereas the basal INa of WT ENaC alone was stimulated ∼3.5-fold by the application of trypsin, the already elevated INa of oocytes coexpressing CAP2 and ENaC was not stimulated further by exogenous trypsin. We routinely observed a reduction in INa after 5 min of trypsin exposure of oocytes coexpressing ENaC and CAP2 (Fig. 1, A and B). This apparent inhibitory effect of trypsin is mainly due to rundown of the CAP2-stimulated channels during the interval of trypsin application.
Figure 1.
CAP2 and trypsin effects on amiloride-sensitive INa of WT or HA-NT– and V5-CT–tagged or β-S518C mutant channels in oocytes. WT α-, β-, γ-, or double tagged (HA-NT/V5-CT) and mutant α-, β-S518C, and γ-cRNA (0.3 ng each) ENaC subunits and 1 ng CAP2 cRNA were injected into oocytes. 24 h after injection, two-electrode voltage clamp assays were conducted. Currents measured in the presence and absence of 10 μM amiloride, while clamping the membrane voltage to −100 mV, were digitized and recorded. (A) Stimulation of ENaC WT channels by coexpression of CAP2 and 2 μg/ml of exogenous trypsin (n = 24). (B) CAP2 and trypsin (2 μg/ml) stimulation of ENaC α-, β-, and γ-HA-NT/V5-CT–tagged subunits (n = 18). (C) 1 mM MTSET activation of α-, β-S518C, and γ mutant untagged channels with or without CAP2 (n = 24). (A–C) Batches of oocytes were extracted from four to five different frogs. Results are expressed as the means ± SE. * and **, P < 0.0001, significant difference when CAP2- or trypsin-stimulated INa is compared with control basal INa. Statistical significance was determined using an unpaired Student's t test.
This result indicates that ENaC at the surface of oocytes expressing CAP2 had undergone a proteolytic activation process that was not shared by all channels in the control (ENaC alone) oocytes. To biochemically detect ENaC cleavage concomitant with CAP2 coexpression, we used modified α-, β-, and γ-ENaC subunits with HA-NT– and V5-CT–tagged subunits (Hughey et al., 2003). With this approach, one injected subunit was the double epitope–tagged version of α-, β-, or γ-ENaC, combined with the appropriate WT versions. We first confirmed that CAP2 stimulated ENaC containing the HA-NT–/V5-CT–tagged subunits similarly to WT ENaC (e.g., Fig. 1, B). The effects of CAP2 coexpression and trypsin on INa mediated by these tagged channels were virtually identical to its actions on WT ENaC. Thus, epitope-tagged ENaC subunits can be used to follow the proteolytic actions of CAP2 on ENaC (compare A and B in Fig. 1).
The stimulation of ENaC by active trypsin or elastase is due to an increase in ENaC PO (Caldwell et al., 2004, 2005). To test if CAP2 coexpression increased ENaC PO, we coexpressed CAP2 with ENaC consisting of WT α- and γ-subunits and a mutant β S518C subunit. The cysteine-reactive compound [2-(trimethylammonium)ethyl] methanethiosulfonate bromide (MTSET) locks such channels in the open state (PO = 1.0) and has been used as a means of estimating channel density as well as resting PO (Kellenberger et al., 2002). INa of WT ENaC was not stimulated by 1 mM MTSET (not depicted). However, MTSET typically stimulated INa of eggs coexpressing α, β S518C, and γ-ENaC by three- to sixfold (Fig. 1 C). In eggs injected with α, β S518C, γ-ENaC plus CAP2, basal INa was stimulated ∼2.7-fold as before. The addition of MTSET to these eggs did not change INa (Fig. 1 C). The acute effect of MTSET on the INa of α, β S518C, γ-ENaC was previously attributed to the net of MTSET's actions of simultaneously increasing PO to near 1.0 and reducing single channel conductance by 30% (Kellenberger et al., 2002). Therefore, the absence of change in INa induced by MTSET in eggs coexpressing α, βS518C, γ-ENaC and CAP2 indicates that the PO of ENaC at the surface of oocytes expressing CAP2 is in the range of 0.7, in good agreement with Caldwell's estimation of the PO of trypsin-treated ENaC (Caldwell et al., 2004).
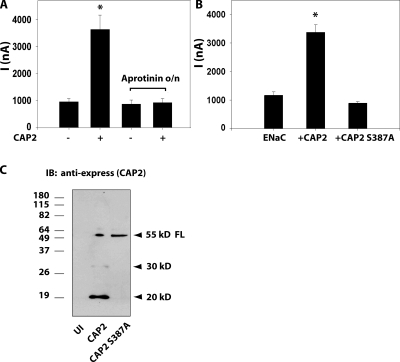
To establish cleavage of ENaC in the presence of CAP2 as a potential mechanism for stimulation of INa, we next asked if CAP2 catalytic activity was required for stimulation of coexpressed ENaC. We addressed this question in two ways. Because CAP2-mediated proteolysis is inhibited by aprotinin, we asked if aprotinin blocked CAP2 regulation of ENaC. Overnight incubation of oocytes with 50 μg/ml aprotinin prevented the stimulation of currents typically seen from coexpressing ENaC and CAP2 (Fig. 2 A). This result does not rule out indirect action of CAP2 on ENaC through another aprotinin-sensitive protease, as has been proposed for CAP1 (Andreasen et al., 2006). Therefore, we constructed an inactive CAP2 control by mutating serine 387 in its catalytic triad (CAP2s387a) and tested its effect on ENaC. This mutation alters the HDS triad at the catalytic site of CAP2 resulting in inactivation (Andreasen et al., 2006). As shown in Fig. 2 B, mutation of the catalytic triad of CAP2 abolished the protease's ability to stimulate basal INa.
Figure 2.
Catalytic activity of CAP2 is required to stimulate ENaC. (A) INa stimulated by CAP2 is sensitive to aprotinin. Eggs coexpressing ENaC plus CAP2 were preincubated or not with 50 μg/ml aprotinin overnight (n = 18). ENaC WT plus CAP2 versus ENaC WT plus CAP2 plus aprotinin. *, P < 0.0001. (B) Inactive CAP2 s387a (CAP2s387a) does not activate ENaC. Amiloride-sensitive INa stimulated by WT but not by inactive CAP2 (n = 30). (A and B) Batches of oocytes were extracted from four to five different frogs. Results are expressed as the means ± SE. *, P < 0.0001. Statistical significance was determined using an unpaired Student's t test. (C) Inactive CAP2 is expressed as a full-length precursor. Western blots of WT and inactive express-tagged CAP2. Representative experiment is shown.
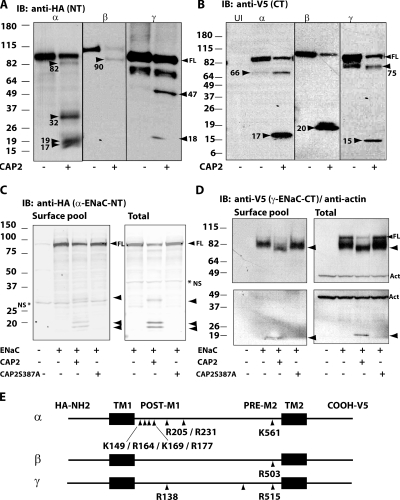
The observation that trypsin and MTSET do not stimulate the INa of ENaC coexpressed with active CAP2 indicates that ENaC at the surface of these oocytes are maximally proteolytically stimulated to a high PO. To identify ENaC cleavage(s) in the presence of CAP2 that could increase PO, we examined the fragmentation pattern of ENaC when coexpressed with CAP2 using HA-NT– and V5-CT–tagged subunits (Hughey et al., 2003) (Fig. 3). With ENaC alone, we typically observed a pattern of fragments similar to that reported (Hughey et al., 2003). Western blots stained for epitope-tagged α-ENaC or γ-ENaC each variably demonstrated fragments (α-ENaC HA-NT, ∼32 kD; γ-ENaC HA-NT, ∼18 kD; α-ENaC V5-CT, ∼66 kD; γ-ENaC V5-CT, ∼75 kD), consistent with cleavage at identified consensus cleavage sites for members of the protein convertase family, whereas Western blots of β-ENaC, as reported previously, did not indicate cleavage under basal conditions (Hughey et al., 2003).
Figure 3.
Cleavage of α-, β-, and γ-ENaC–tagged subunits by WT but not inactive CAP2 S387A in oocytes. WT α-, β-, and γ-ENaC–double tagged (HA-NT/V5-CT) and α-, β-, or γ-ENaC–untagged subunits (0.3 ng each) and 1 ng CAP2 cRNA were injected into oocytes. Total protein lysates were prepared from oocytes, and Western blots analysis were conducted using anti-HA (A) and anti-V5 (B) monoclonal antibodies. Western blots of surface biotinylated and total pools of α-ENaC HA-NT (C) and γ-ENaC V5-CT (D) fragments caused by WT but not inactive CAP2 (CAP2 S387A). Actin expression was detected with an anti-actin monoclonal antibody as control (D). A β-ENaC 90-kD HA-NT fragment was detected at the surface pool (not depicted). NS, nonspecific. Batches of oocytes were extracted from three different frogs. Representative experiments are shown (n = 3). (E) Linear diagram of ENaC residues cleaved by CAP2. NH2, amino terminus; COOH, carboxy terminus; TM1, transmembrane domain 1; TM2, transmembrane domain 2.
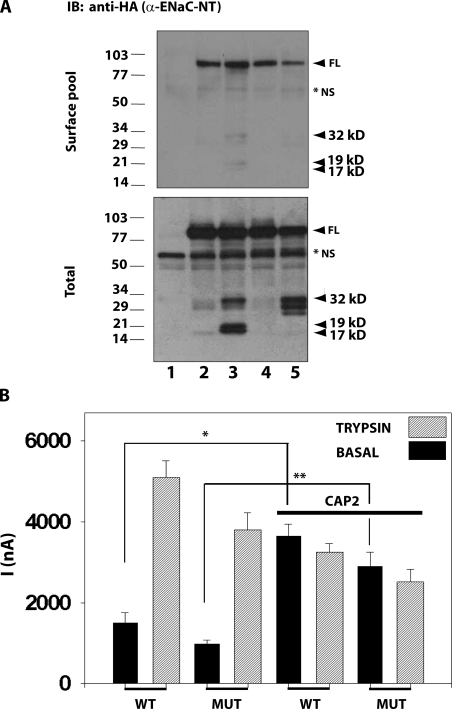
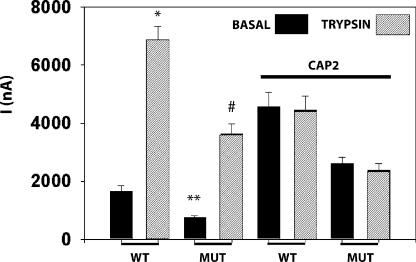
CAP2 coexpression decreased the amount of full-length protein corresponding to all three ENaC subunits in whole cell lysates (Fig. 3). The loss of full-length ENaC coincided with the somewhat variable appearance of smaller fragments. CAP2 coexpression consistently enhanced ∼32-kD HA-NT and complementary ∼66-kD V5-CT staining bands of α-ENaC, suggesting increased cleavage at the furin-consensus sites (Fig. 3, A and B). In some experiments, CAP2 coexpression with α-ENaC generated an HA-NT band of ∼82 kD (Fig. 3 A), and more consistently yielded an ∼17-kD V5-CT band (Fig. 3 B). These complementary α-ENaC fragments predict cleavage close to the second transmembrane, or pre-M2, region. The relationship between CAP2-stimulated cleavage at the α-ENaC furin and pre-M2 regions to CAP2-stimulated INa is addressed in detail below. In addition, unique HA-NT fragments of α-ENaC of ∼17 and ∼19 kD were induced by CAP2, consistent with cleavage just distal of the first transmembrane segment (Fig. 3 A). Mutagenesis of four conserved basic residues in this region to alanine (K149A/R164A/K169A/R177A) prevented ∼17 and ∼19 kD N-terminal fragments with CAP2 coexpression, but it did not affect stimulation of INa (Fig. 4). Thus, we conclude that CAP2-mediated cleavage in the proximal region of the extracellular loop of the α-ENaC subunit is not required for CAP2 to stimulate ENaC.
Figure 4.
Effect of α-ENaC K149A/R164A/K169A/R177A mutant on CAP2-induced INa and α-ENaC N-terminal fragments. (A) Western blots of α-ENaC N-terminal 17- and 19-kD novel fragments at the surface (top) and total pools (bottom). Lane 1, uninjected eggs; lane 2, ENaC alone; lane 3, ENaC plus CAP2; lane 4, α-ENaC mutant alone; lane 5, α-ENaC mutant plus CAP2. FL, full-length. A representative experiment is shown (n = 3). (B) INa of WT and mutant α-ENaC stimulated by CAP2. Batches of oocytes were extracted from three different frogs (n = 18). * and **, P < 0.0001. Amiloride-sensitive currents were measured as described in Fig. 1.
Previous work with other proteases revealed no cleavage of β-ENaC (Hughey et al., 2003; Harris et al., 2007). To our surprise, CAP2 induced ∼90-kD HA-NT and ∼20-kD V5-CT fragments (Fig. 3, A and B) of double epitope–tagged β-ENaC, which complement each other, as the expected size of full-length β-ENaC protein is ∼110 kD. These β-ENaC fragments indicate that, similar to its action on α-ENaC, CAP2 promotes cleavage of β-ENaC at the pre-M2 region.
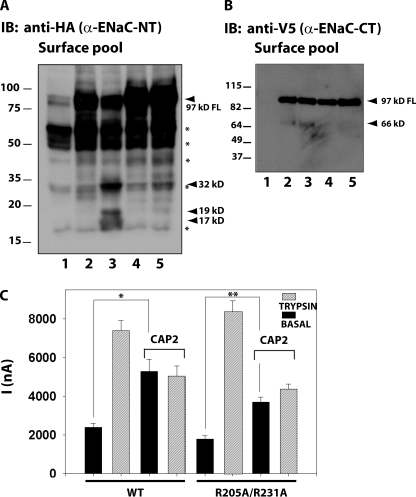
CAP2 coexpression induced the formation of HA-NT γ-ENaC fragments of ∼47 and ∼18 kD (Fig. 3 A). We did not pursue the ∼47-kD fragment due to a lack of complementarity with a corresponding V5-CT fragment of the expected size. The HA-NT band at 18 kD matches a fragment reported to be generated by a convertase family member, furin (Hughey et al., 2003). CAP2 induced a complementary V5-CT fragment of γ-ENaC at ∼75 kD, also consistent with cleavage at the γ-ENaC furin site. There was also an ∼15-kD V5-CT γ-ENaC band in lysates of oocytes expressing active CAP2 (Fig. 3 B). This band suggests cleavage at a site near the pre-M2 region of γ-ENaC (Fig. 3 B).
To identify those ENaC fragments at the cell surface that could have derived from the catalytic activity of CAP2, we compared fragments detected when ENaC was coexpressed with CAP2 or a catalytically inactive CAP2 mutant. With coexpression of inactive CAP2 (CAP2 S387A), we observed a migration pattern of ENaC on gels, corresponding to full-length subunits and ENaC “furin fragments” indicative of endogenous furin-like activity, indistinguishable from WT ENaC alone (Fig. 3, C and D). In addition, INa due to ENaC coexpressed with CAP2 S387A was similar to that of ENaC alone (Fig. 2 B). To be sure the lack of any effect was not due to poor expression of the mutant CAP2, we performed Western blot analyses of WT and mutant CAP2 expression. These results indicated that the mutant was efficiently expressed (Fig. 2 C). As previously observed (Andreasen et al., 2006), WT CAP2 appears as the glycosylated precursor (∼55 kD) and cleaved N-terminus glycosylated forms (∼30 and ∼20 kD), whereas mutant CAP2 S387A appears as a single precursor band of ∼55 kD (Fig. 2 C). In paired experiments with active CAP2, the surface pool contained the fragments of α- and γ-ENaC described above, which indicated cleavage in the pre-M2 region. In addition, we routinely observed increased intensity of the bands believed to represent furin fragments: In α-ENaC, an HA-NT band of ∼32 kD, complemented by an ∼66-kD V5-CT band; for γ-ENaC, an HA-NT band of ∼18 kD complemented by a V5-CT band of ∼75 kD (Fig. 3). The surface pool also contained bands (HA-NT of ∼90 kD and V5-CT of ∼20 kD) of β-ENaC consistent with cleavage in the pre-M2 region (not depicted).
Thus, active CAP2 induces fragments in all three subunits at the oocyte surface. Because these fragments represent novel cleavage events at the pre-M2 region in α-, β-, and γ-ENaC, and apparent increased cleavage at the convertase consensus site in α- and γ-ENaC, we directed additional studies to these regions. We used site-directed mutagenesis to locate the specific residues cleaved and to link specific cleavages to stimulation of INa.
We first sought to identify specific residues necessary for CAP2-stimulated fragments in the pre-M2 regions of α-, β-, and γ-ENaC. We analyzed α-, β-, and γ-ENaC sequences corresponding to the extracellular loop at the pre-M2 region with a cleavage prediction model for CAP3/matriptase available at http://pops.csse.monash.edu.au/pops.html. Although there is no established model to predict cleavage sites for CAP2, we have observed that CAP2 and CAP3/matriptase generate an identical C terminal–fragmentation pattern in the α-ENaC subunit (not depicted). From this observation, we reasoned that these CAPs could share some substrate affinity. Based on the apparent molecular mass of the ∼15–17-kD C terminal fragments generated by CAP2 in all three subunits, we surveyed a stretch of 40 amino acids in the pre-M2 region of α-, β-, and γ-ENaC. Whereas multiple basic residues in this region had high predictive scores for matriptase cleavage, we noted an aligned basic residue at position K561 in α-ENaC, R503 in β-ENaC, and R515 in γ-ENaC that was conserved across all three subunits of human, mouse, and rat ENaCs (Fig. 5 B). When we replaced basic residues at this residue with alanines by site-directed mutagenesis, the appearance of the pre-M2 fragments with CAP2 coexpression was eliminated (Fig. 5 A). Interestingly, in the mutants there was a trend toward lighter intensity of bands associated with cleavage at the furin sites in α- and γ-ENaC. Mutagenesis of other basic residues (α-ENaC: K486A, K501A, R503A, K504A, K512A, R519A, K524A, K544A, R545A, K550A, and K556A; β-ENaC: K411A, R416A, R435A, K443A, K452A, R477A, R487A, K488A, and K492A; γ-ENaC: K504A and K510A) in the 80-residue stretch had no effects on CAP2-mediated fragments (not depicted).
Figure 5.
CAP2 cleaves α-, β-, and γ-ENaC at a conserved basic residue in the pre-M2 region. WT α-, β-, and γ-ENaC–untagged or –double tagged (HA-NT/V5-CT) or α-K561A, β-R503A, and γ-R515A ENaC–tagged subunits (0.3 ng each) and 1 ng CAP2 cRNA were injected into oocytes. Total protein lysates were prepared from oocytes and Western blot analysis was conducted using anti-V5 monoclonal antibodies (A). Black arrows indicate full-length (FL) and cleaved ENaC subunits. Batches of oocytes were extracted from three different frogs. Representative experiments are shown (n = 3). (B) α-, β-, and γ-ENaC human, mouse, and rat pre-M2 region sequences alignment. TMII, transmembrane 2.
The highly conserved residue we found to be required for pre-M2 CAP2 fragments is 15 residues upstream of the degenerin site present in each subunit (Fig. 5 B). To assess the functional effect of this cleavage event in the pre-M2 region of the channel, we coexpressed a triple mutant channel (α-K561A, β-R503A, and γ-R515A) in oocytes with or without CAP2 and measured INa. We used trypsin to evaluate the proteolytic state of the WT or mutant channels, given previous reports by Caldwell et al. (2004) that trypsin activates near-silent channels in excised patches. Although the basal INa of the triple mutant channel was significantly diminished (751 ± 56 nA) compared with the basal INa of WT channels (1,652 ± 184 nA) (Fig. 6), subsequent trypsin exposure activated the INa of mutant channels (3.9-fold) and WT channels (3.9-fold) similarly. CAP2 coexpression increased basal INa of triple mutant channels by 3.43-fold, and trypsin had no further effect. In comparison, WT channels were activated by CAP2 (2.43-fold), and no further trypsin response was observed (Fig. 6). Thus, we identified a specific and highly conserved residue in the pre-M2 region of α-, β-, and γ-ENaC that is cleaved when the channels are expressed with CAP2. However, this cleavage event is not required for the stimulation of ENaC by coexpressed CAP2.
Figure 6.
Amiloride-sensitive INa of triple mutant α-K561A, β-R503A, and γ-R515A channels activated by CAP2. WT α-, β-, γ-, or mutant α-K561A, β-R503A, and γ-R515A ENaC cRNA (0.3 ng each) and 1 ng CAP2 cRNA were injected into oocytes. 24 h after injection, two-electrode voltage clamp assays were conducted. Currents measured in the presence and absence of 10 μM amiloride, while clamping the membrane voltage to −100 mV, were digitized and recorded. Batches of oocytes were extracted from four different frogs (n = 24). Results are expressed as the means ± SE. Statistical significance was determined using an unpaired Student's t test. Basal WT INa versus WT trypsin-stimulated INa: *, P < 0.0001; basal WT INa versus basal triple mutant INa: **, P < 0.0011; basal triple mutant INa versus triple mutant trypsin-stimulated INa: #, significant P < 0.0001.
Previous findings by Hughey et al. (2003) ascribed HA-NT α- and γ-ENaC fragments (α: 30 kD; γ: 18 kD) and complementary V5-CT α- and γ-ENaC fragments (α: 65 kD; γ: 75 kD) to cleavage by furin family proprotein convertases (Hughey et al., 2003). Prevention of these fragments by mutagenesis of the furin consensus cleavage sites diminished basal proteolytic activation of ENaC (Hughey et al., 2003, 2004). Because CAP2 consistently increased the intensity bands corresponding to α- and γ-ENaC furin fragments, we asked if this apparent enhanced cleavage contributed to CAP2 stimulation of basal INa. First, we compared the effects of CAP2 expression on WT and ENaC channels mutagenized to prevent cleavage by convertases. Although furin fragments were not prominent in every control (WT ENaC alone) experiment, coexpression with CAP2 reliably increased intensity of the HA-NT 32-kD and V5-CT 66-kD bands (furin fragments), and induced the novel HA-NT bands of 17, 19 (Fig. 7, A and B, compare lanes 2 and 3), and 82 kD (Fig. 3 A). For example, in the gel shown, the 32-kD HA-NT band (furin fragment) was hardly noticeable above background in the surface pool of control oocytes (WT ENaC alone) (Fig. 7 A, lane 2). This band is similarly indistinct in α-R205A/R231A, β-, and γ-ENaC–expressing oocytes (Fig. 7 A, lane 4). CAP2 coexpression with WT ENaC robustly increased the intensity of the 32-kD band, as well as generating the novel 17- and 19-kD bands (Fig. 7 A, lane 3). However, this effect of CAP2 coexpression was not seen in oocytes expressing α-R205A/R231A, β-, and γ-ENaC (Fig. 7 A, lane 5). Although Western blot sensitivity does not permit the presence of fragments to be ruled out, it is clear even from overexposed gels that CAP2 has minimal effects on the 32-kD band (taking into account background staining in the overexposed gel and slightly unequal loading of the lanes). The 66-kD V5-CT fragment was visible at the surface pool of WT ENaC eggs and enhanced when CAP2 was coexpressed; however, this fragment was not detectable in oocytes expressing α-R205A/R231A, β-, and γ-ENaC channels with/without CAP2 (Fig. 7 B). Not only did coexpression of CAP2 with α-R205A/R231A, β-, and γ-ENaC channels not increase the furin fragments significantly, but it also did not generate the smaller novel ∼17- and ∼19-kD α-ENaC N-terminal fragments seen in WT ENaC plus CAP2 experiments. (Fig. 7 A, lane 5 vs. lane 3). Nonetheless, the fold increase ratio of INa due CAP2 coexpression with the mutant channels was very similar (∼2.2-fold) to that of WT channels (∼2.0-fold) (Fig. 7 C) despite the absence of CAP2-induced fragments of α-ENaC (Fig. 7, A and B). Importantly, the elevated basal INa of oocytes coexpressing α-R205A/R231A, β, and γ-ENaC with CAP2 was unresponsive to exogenous trypsin, suggesting that CAP2 achieved full proteolytic activation of these mutant channels (Fig. 7 C). Basal INa of oocytes expressing α-R205A/R231A, β, and γ-ENaC was significantly smaller than INa of oocytes expressing WT channels, but responded briskly to trypsin, as reported by Hughey et al. (2004). These results do not support a strong correlation between apparent increased generation of α-ENaC furin fragments by CAP2 and stimulated INa. The elimination of the other CAP2-induced fragments by mutating the furin sites in α-ENaC suggests that they may have been generated secondarily to cleavage at the furin sites.
Figure 7.
Effect of α-R205A/R231A, β-, and γ-ENaC mutant on INa and fragments stimulated by CAP2. (A and B) Surface biotinylated α-ENaC N-terminal and C-terminal fragments were visualized by Western blot analysis using anti-HA or anti-V5 monoclonal antibodies. (A) Many nonspecific (*, NS) bands evident in film overexposed to reveal the fate of the furin (32 kD) and novel fragments (19 and 17 kD). FL, full-length. Lane 1, uninjected eggs; lane 2, WT ENaC alone; lane 3, WT ENaC plus CAP2; lane 4, α-R205A/R231A, β-, and γ-ENaC alone; lane 5, α-R205A/R231A, β-, and γ-ENaC plus CAP2. (C) CAP2-mediated INa of WT or mutant channels were measured as described above. Batches of oocytes were extracted from five different frogs (n = 31). Results are expressed as the means ± SE. * and **, P < 0.0001. Statistical significance was determined using an unpaired Student's t test.
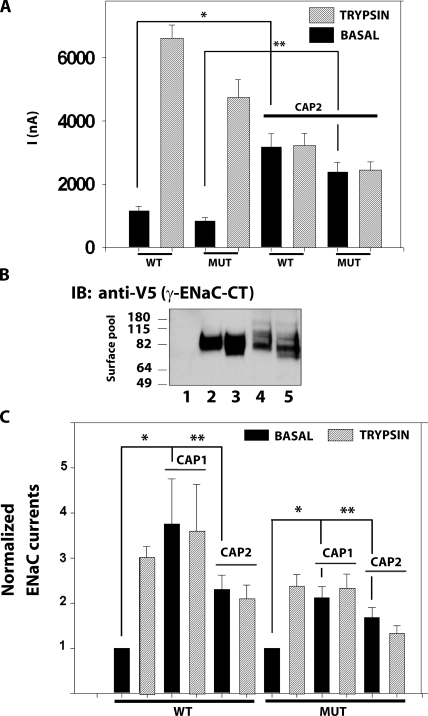
We next tested the role of γ-ENaC's single furin consensus site (135–138 RKRR) in CAP2 stimulation by substituting alanine for the arginine at residue 138 in γ-ENaC (135–138 RKRA). This change in the critical P1 site in the furin consensus cleavage site is expected to eliminate cleavage by furin-like convertases, and indeed, γ-R138A in the surface pool appeared on Western blot predominantly as the full-length channel, with the ∼75-kD C-terminal band that has been termed a furin fragment nearly eliminated (Fig. 8 A, compare lanes 2 [WT γ] and 4 [γ-R138A]). For WT γ-ENaC at the surface of CAP2-expressing oocytes, a broad, ∼75-kD band was markedly increased in intensity (Figs. 8 A and 3 D). In ENaC containing γ-R138A, most staining represented the full-length subunit, and CAP2 coexpression produced no marked increase in the ∼75-kD band (Fig. 8 A, lanes 4 and 5). An ∼73-kD band was also visible in lysates of γ-furin mutants and enhanced by CAP2 coexpression. Densitometric quantification of the 75-kD fragment of WT γ-ENaC showed an ∼2.4-fold increase induced by CAP2 coexpression (Fig. 8 C), which correlates with a routinely two- to threefold increase in INa observed with CAP2 coexpression (Fig. 8 B). Basal INa and fold trypsin stimulation for ENaC containing γ-R138A was not much different from WT ENaC. However, CAP2 coexpression did not increase basal current of oocytes expressing α-, β-, and γ-R138A. Notably, subsequent exposure of the α-, β-, and γ-R138A plus CAP2 oocytes to trypsin stimulated INa by more than twofold. These results identify R138 in γ-ENaC as a residue required for γ-ENaC cleavage and stimulation of INa in the presence of CAP2.
Figure 8.
Effect of α-, β-, and γ-R138A-ENaC mutant on INa and fragments stimulated by CAP2. (A) (Top) Surface biotinylated γ-ENaC CT 75-kD fragment induced by CAP2 was visualized by Western blot analysis using an anti-V5 monoclonal antibody. (Bottom) Total whole cell lysates (input 4%) were probed for actin as loading control. FL, full-length. Lane 1, uninjected eggs; lane 2, WT ENaC alone; lane 3, WT ENaC plus CAP2; lane 4, α-, β-, and γ-R138A ENaC alone; lane 5, α-, β-, and γ-R138A ENaC plus CAP2. A representative experiment is shown (n = 3). (B) CAP2-mediated INa of WT and α-, β-, and γ-R138A mutant channels were measured as described in Results. Batches of oocytes were extracted from six different frogs (n = 36). Results are expressed as the means ± SE. * and **, P < 0.0001. Statistical significance was determined using an unpaired Student's t test. (C) Semi-quantification of the 75-kD γ-ENaC surface fragment induced by CAP2 (IOD, integrated optic density; n = 4).
In light of previous work on the activation of ENaC by prostasin, we noted with interest that CAP2 coexpression with WT ENaC promoted the contribution of a slightly faster migrating band of γ-ENaC than the 75-kD band associated with furin cleavage, and under different exposure conditions, this staining resolved into distinct bands of ∼75 and ∼73 kD (Figs. 3 D and 8 A). Thus, we suspected that CAP2 was cleaving γ-ENaC downstream from the furin site. Bruns et al. (2007) had observed a similar fragment due to prostasin coexpression, which they concluded arose from prostasin cleavage of mouse γ-ENaC after the polybasic tract 183–186 RKRK (178–181 RKRK in rat γ-ENaC). In the previous study, both generation of the 73-kD fragment and the stimulation of INa by prostasin were eliminated by replacing RKRK with QQQQ (Bruns et al., 2007). This, and other results, led the authors to conclude that prostasin and furin worked in concert to fully activate ENaC by excising the residues between the furin site and the end of the polybasic tract (Bruns et al., 2007).
To determine if the polybasic tract 178–181 RKRK in rat γ-ENaC was important for CAP2 stimulation of rat ENaC, we replaced 178–181 RKRK with 178–181 QQQQ and determined the effect of CAP2 coexpression on INa and rat γ-ENaC fragments. We found that CAP2-stimulated INa in oocytes expressing this quadruple γ-ENaC mutant were indistinguishable from INa activated by CAP2 coexpressed with WT ENaC (Fig. 9 A). In addition, the fragment pattern induced by CAP2 in either WT or mutant γ-ENaC was identical (Fig. 9 B). These data demonstrate that CAP2 does not require the cluster of arginine and lysines (178–181) to activate rat ENaC, in contrast to the effect of CAP1 on mouse ENaC (Bruns et al., 2007). Interestingly, in our hands, CAP1/prostasin coexpression stimulated the QQQQ mutant as well as WT ENaC (threefold for WT and 2.8-fold for γ-ENaC 178–181 QQQQ; not depicted). Like Bruns et al. (2007), we conjectured that CAP2 was able to cleave the QQQQ mutant at nearby basic residues, and we therefore studied the effect of additional mutations (178–181 QQQQ plus K185Q, K189Q, K200Q, and K201Q). Both CAP1 and CAP2 stimulated basal currents of these mutant channels by 2.2- and 1.8-fold, respectively, and the stimulated basal INa was not further increased by trypsin (Fig. 9 C). Thus, other than the γ-ENaC furin site, we did not locate a second site containing basic residues that was required for stimulation of ENaC by CAP1 or CAP2 coexpression.
Figure 9.
Effect of α-, β-, and γ-178-181 QQQQ or α-, β-, and γ-178-181 QQQQ, K185Q, K189Q, K200Q, and K201Q ENaC mutants on INa stimulated by CAP2 and CAP1. (A) CAP2 induced INa of WT and 178–181 QQQQ ENaC mutant channels. Amiloride-sensitive currents were measured as described in Fig. 1. Batches of oocytes were extracted from three different frogs (n = 12). Results are expressed as the means ± SE. * and **, P < 0.0001. Statistical significance was determined using an unpaired Student's t test. (B) Western blot of surface pool of WT and 178–181 QQQQ γ-ENaC mutant channels. Lane 1, uninjected eggs; lane 2, WT ENaC alone; lane 3, WT ENaC plus CAP2; lane 4, α-, β-, and γ-178-181 QQQQ ENaC alone; lane 5, α-, β-, and γ-178-181 QQQQ ENaC plus CAP2. A representative experiment is shown (n = 3). (C) CAP2 and CAP1 induced INa of WT and 178–181 QQQQ, K185Q, K189Q, K200Q, and K201Q ENaC mutant channels. Amiloride-sensitive currents were measured as described in Fig. 1. Batches of oocytes were extracted from three different frogs (n = 12). Results are expressed as the means ± SE. * and **, difference P < 0.05. Statistical significance was determined using ANOVA.
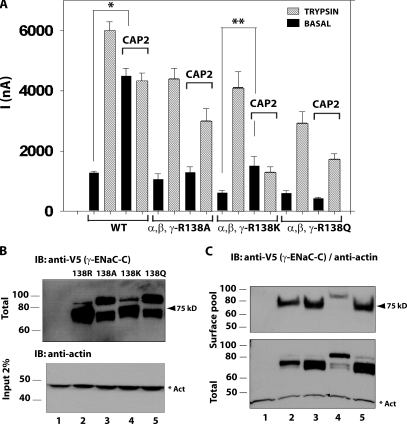
Our results with previously validated furin mutants indicate that cleavage at R138 in γ-ENaC is largely responsible for the stimulation of INa by coexpressed CAP2. Because this residue occupies the critical P1 site for furin and similar convertases, we next asked if CAP2 was actually cleaving after R138 or if CAP2 stimulated furin or another convertase to cleave there. The minimal P4-P1 recognition sequence for furin is R/K-X-X-R, and the requirement for arginine at P1 is stringent (Thomas, 2002; Wheatley and Holyoak, 2007). Interestingly, recent crystallization and other work reveal a similar, but not identical, optimal sequence for matriptase binding and cleavage, R-X-X-K/R (Friedrich et al., 2002; Netzel-Arnett et al., 2003; Desilets et al., 2006). Furthermore, a similar P4-P1 preferred sequence for CAP1/prostasin, R/K-H/R/K-X-R/K, was identified by screening peptide libraries (Shipway et al., 2004). These published observations indicate that CAP1 and CAP3 prefer an arginine, but tolerate a lysine in the P1 position. We speculated that if CAP2 shared this relaxed stringency at P1, CAP2 coexpression would stimulate the furin-resistant ENaC mutant α-, β-, and γ-R138K. We also tested the effect of CAP2 coexpression on α-, β-, and γ-R138Q ENaC, in which γ residue 138 should not be a substrate for either furin or CAP2. We found that basal INa of ENaC containing γ-R138A or γ-R138Q was not increased by coexpressed CAP2, but was robustly stimulated by exogenous trypsin (Fig. 10 A). In contrast, when ENaC contained γ-R138K, CAP2 coexpression significantly increased basal INa. Importantly, trypsin did not further stimulate INa of oocytes expressing α-, β-, and γ-R138K plus CAP2, indicating that ENaC in these oocytes had already been proteolytically activated. Thus, CAP2 stimulated ENaC when the critical 138 (P1) residue in the γ-subunit was a lysine. These results strongly suggest that cleavage induced by CAP2 at R138 in γ-ENaC is responsible for CAP2 stimulation of INa.
Figure 10.
Effect of replacement of γ-ENaC R138 (P1) by alanine, lysine, or glutamine on INa and fragments stimulated by CAP2. (A) CAP2 effect on INa of WT and α-, β-, and γ-R138A or α-, β-, and γ-R138K or α-, β-, and γ-R138Q ENaC mutant channels. Amiloride-sensitive currents were measured as described in Fig. 1. Batches of oocytes were extracted from three different frogs (n = 18). Results are expressed as the means ± SE. * and **, P < 0.0001. (B) Western blots of total whole cell extracts. CAP2-induced fragment pattern of WT and γ-ENaC mutant channels (top) and actin as loading control (bottom). Lane 1, uninjected eggs; lane 2, WT ENaC plus CAP2; lane 3, α-, β-, and γ-R138A ENaC plus CAP2; lane 4, α-, β-, and γ-R138K ENaC plus CAP2; lane 5, α-, β-, and γ-R138Q ENaC plus CAP2. A representative experiment is shown (n = 3). (C) Western blots of surface biotinylated (top), total γ-ENaC pools (bottom), and actin. Lane 1, uninjected eggs; lane 2, WT ENaC alone; lane 3, WT ENaC plus CAP2; lane 4, α-, β-, and γ-R138K ENaC alone; lane 5, α-, β-, and γ-R138K ENaC plus CAP2. A representative experiment is shown (n = 3).
In agreement with the functional results, we found that CAP2 coexpression generated a 75-kD V5-CT γ-ENaC fragment when coexpressed with α-, β-, and γ-R138 or α-, β-, and γ-R138K, but not when coexpressed with α-, β-, and γ-R138A or α-, β-, and γ-R138Q (Fig. 10, B and C). This is again consistent with cleavage at position 138 in γ-ENaC–mediating CAP2 stimulation of INa. The appearance or disappearance of the 75-kD fragment consistently correlates with ENaC activation by CAP2. We routinely detected a faster migrating fragment (∼73 kD) with variable intensity, more evident in the R138A/Q mutants (Figs. 8–10). The fact that this faster migrating form appears in these mutants strongly suggests that this cleavage event by itself is not linked to CAP2-stimulated INa.
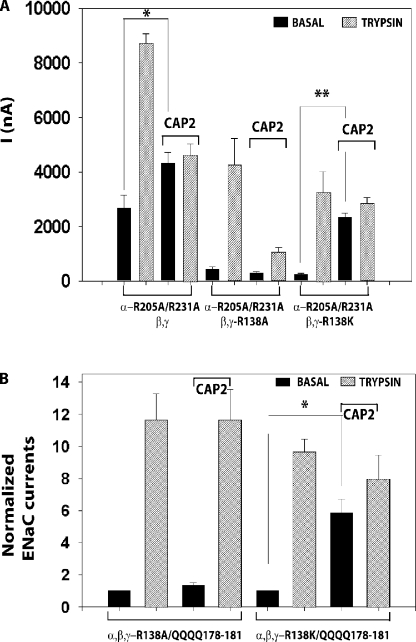
To further test the importance of γ-ENaC residue 138 in CAP2 stimulation of ENaC, we coexpressed either WT γ or γ-R138A or γ-R138K with α-R205A/R231A and WT β-ENaC. INa was stimulated by CAP2 coexpression only when an arginine (WT) or a lysine was present at position 138 in γ-ENaC (Fig. 11 A). We further isolated the action of CAP2 by coexpressing CAP2 with WT α- and β-ENaC combined with γ-178-181 QQQQ mutants that contained either alanine or lysine at γ-ENaC residue 138. These mutant γ-subunits are not expected to be cleaved by convertases acting at the γ-ENaC furin site, and they lack the polybasic tract implicated in CAP1 stimulation of mouse ENaC. CAP2 did not stimulate basal INa of ENaC with the double mutant γ-subunit containing alanine at residue 138; however, INa was markedly stimulated by subsequent trypsin (Fig. 11 B). In contrast, CAP2 coexpression clearly enhanced basal INa of ENaC that contained the mutant γ-ENaC with lysine at residue 138; moreover, subsequent trypsin exposure did not significantly add to the elevated basal INa (Fig. 11 B). These results indicate that ability of CAP2 coexpression to stimulate ENaC INa is exquisitely dependent on the residue at position 138 in γ-ENaC.
Figure 11.
CAP2 stimulated α-R205A/R231A, β-, and γ-138K or α-, β-, and γ-138K plus 178–181 QQQQ but not α-R205A/R231A, β-, and γ-138A or α-, β-, and γ-138A plus 178–181 QQQQ mutant channels. (A) CAP2 induced INa of WT and α-R205A/R231A, β-, γ-, or α-R205A/R231A, β-, and γ-R138A or α-R205A/R231A, β-, and γ-R138K ENaC mutant channels (n = 12). (B) CAP2 induced INa of α-, β-, and γ-R138A, R178Q, K179Q, R180Q, and K181Q or α-, β-, and γ-R138K, R178Q, K179Q, R180Q, and K181Q ENaC mutant channels. Amiloride-sensitive currents were measured as described in Fig. 1. Batches of oocytes were extracted from three different frogs (n = 12). Results are expressed as the means ± SE. * and **, P < 0.0001.
DISCUSSION
CAP2/TMPRSS4 is a member of a family of S1 serine proteases that are anchored directly to cell surfaces, with their catalytic domains located extracellularly (Netzel-Arnett et al., 2003). The activity of such proteases on the cell surface is controlled by an activation step and by interaction with native inhibitors, potentially permitting regulation of events on the cell surface. Because CAP2, as well as related serine proteases CAP1/prostasin and CAP3/matriptase, regulate ENaC (Andreasen et al., 2006), it seems reasonable to assume that proteolytic activation of ENaC is among those events. The notion of near-silent ENaC being delivered to the cell surface and activated in proportion to a balance between surface proteases and protease inhibitors fits well with emerging views of proteolytic regulation ENaC (Rossier, 2004; Tarran et al., 2006). However, we do not yet possess a detailed understanding of how CAPs regulate ENaC. For example, CAP1/prostasin stimulates ENaC and appears from knockdown studies to play the dominant role in proteolytic regulation of ENaC (Tong et al., 2004). Yet, Andreasen et al. (2006) reported that prostasin regulation of ENaC does not require prostasin itself to be catalytically active, suggesting that its mode of regulation may involve other surface proteases. Because CAP2 is highly expressed in some epithelia, and regulates ENaC through a mechanism that requires it to be proteolytically active, we thought it was important to examine how CAP2 regulates ENaC coexpressed in Xenopus oocytes. In particular, we sought to identify the origin of CAP2-induced ENaC fragments at the cell surface and to determine the functional consequence of each identified cleavage event.
We found that CAP2 induced cleavage of all three ENaC subunits, and that resulting fragments appear at the cell surface. Fragments were used to map cleavage of ENaC subunits: (1) in α-ENaC, novel post-M1 fragments required the basic conserved residues K149, R164, K169, and R177; (2) in all three subunits in the pre-M2 region at α-K561, β-R503, and γ-R515, a highly conserved basic residue just before the important degenerin site; and (3) at the conserved convertase consensus sites in α-ENaC (R205/R231) and γ-ENaC (R138). Somewhat surprisingly, of these demonstrated sites of proteolysis, only cleavage at R138 of γ-ENaC could be linked to CAP2-mediated activation of ENaC. Our results add to a growing body of work that identifies γ-ENaC as a critical target of proteases that increase ENaC PO (Adebamiro et al., 2007; Bruns et al., 2007; Harris et al., 2007).
As our work unfolded, it became clear that some CAP2-mediated cleavages of ENaC do not contribute to proteolytic activation of ENaC. Using site-directed mutagenesis, we modified four basic residues found in the post-M1 region of α-ENaC, which prevented the appearance of the short N-terminal fragments at the cell surface but had no effect on the CAP2 regulation of ENaC current. Due to lack of a role of this cleavage in CAP2 stimulation of ENaC, we did not identify the precise residues involved. Although the appearance of these fragments at the oocyte surface was dependent on catalytically active CAP2, any role for cleavage in the early extracellular loop in regulating ENaC remains obscure. We did note a tendency for trypsin-stimulated INa of coexpressing ENaC and CAP2 to be smaller than typsin-stimulated WT control currents (Figs. 1, 4, and 6–8). Therefore, we considered the possibility that these small N-terminal fragments might arise from CAP2-mediated degradation of active channels at the cell surface. However, our functional study of the post-M1 mutants offers no hint that preventing these α-ENaC fragments counters a decrease of INa that is part of mixed stimulatory and inhibitory effects of CAP2. Nonetheless, if these fragments are shown in the future to be directly generated by CAP2, it may help in mapping the physical association between ENaC and serine surface proteases.
We next turned our attention to CAP2-mediated cleavage of the pre-M2 region of ENaC. An ∼82-kD N-terminally tagged fragment predicted to arise from cleavage at the pre-M2 region of α-ENaC has also been seen by others (Planes et al., 2002; Ergonul et al., 2006; Myerburg et al., 2006), but its role in proteolytic regulation of ENaC has not been studied. Our data expand upon these previous observations. We sporadically detected a similar sized N-terminal α-ENaC fragment at the surface and total cellular pools when CAP2 was coexpressed with ENaC (Figs. 3 and 7). We speculate that the 82-kD HA-NT band is missing in some experiments due to a variable extent of additional cleavage. However, we consistently detected a CAP2-induced ∼17-kD C-terminal fragment in α-ENaC that fully complemented the ∼82-kD N-terminal band. We unequivocally found this fragment at the cell surface. Interestingly, we detected similar sized C-terminal–tagged fragments of β- (∼19 kD) and γ-ENaC (∼15 kD) at the surface of oocytes coexpressing catalytically active CAP2 and ENaC. To the best of our knowledge, this is the first documented cleavage of β-ENaC by a coexpressed protease. Specific basic residues in each ENaC subunit were required for CAP2 to stimulate the appearance of these pre-M2 fragments. These basic residues were conserved across all ENaC subunits and species, aligned 15 amino acids upstream of the degenerin site in each subunit, and embedded in a region, the palm domain (β sheet 12), homologous to that described for the acid-sensing ion channel 1 structure (Jasti et al., 2007). Due to the critical role of the degenerin site in ENaC gating, we were surprised when triple mutant channels engineered with alanines in place of the conserved basic residues were normally stimulated by CAP2 coexpression. Again, we considered the possibility that the lower maximum current of CAP2 coexpressing oocytes, compared with ENaC-alone controls, could result from cleavage/degradation of active (surface) ENaC by CAP2. However, the triple mutant channels that were not cleaved in the pre-M2 region exhibit lower rather than greater stimulated current. Unexpectedly, we noticed that the mutant α-K561A, β-R503A, and γ-R515A not only completely abolished the small V5-CT pre-M2 fragments, but also notably reduced the furin α- and γ-ENaC fragments (Fig. 5).
CAP2 coexpression with WT ENaC consistently increased staining intensity of bands that Hughey et al. (2003) attributed to cleavage at the furin sites. Logically, increased cleavage at furin sites should increase basal current and decrease trypsin responsiveness, which adequately describes the effects of CAP2 on ENaC-mediated currents. Therefore, we designed studies to determine if cleavage at recognized furin sites was indeed required for CAP2 stimulation of ENaC. Using α-ENaC in which the furin sites had been mutated through substitution of alanines for the critical P1 arginines, we found that CAP2-enhanced cleavage at the α-furin sites was effectively suppressed, without affecting CAP2 stimulation of INa. Interestingly, the 17–19-kD post-M1 and the ∼82-kD pre-M2 α-ENaC fragments produced by CAP2 coexpression with WT α-ENaC were also not generated when the α-ENaC double furin mutant was used (Fig. 7 A). Thus, the numerous cleavages of α-ENaC observed with CAP2 coexpression may occur in a sequence that begins with cleavage at the furin sites, but also appear to be unrelated to the ability of CAP2 to stimulate ENaC basal current by two- to threefold.
In contrast to the lack of consequence of α-ENaC furin site mutations, the R138A or R138Q mutations of the P1 residue in the γ-ENaC furin site completely abrogated CAP2 stimulation of ENaC, and dramatically suppressed the appearance of fragments associated with cleavage at the γ-ENaC furin site. These mutants clearly produced channels that trafficked to the surface and could be stimulated by trypsin, but they were not cleaved at the furin site or activated when coexpressed with CAP2. Cleavage of γ-ENaC downstream of the documented furin site (rat residues 135–138) has been linked to activation of ENaC by hNE (rat residues 180–200) and by prostasin (rat residues 178–181) (Adebamiro et al., 2007; Bruns et al., 2007). These observations focus attention on this region of γ-ENaC as critical to stimulation of ENaC by proteases.
Accordingly, we performed extensive mutagenesis of the region beginning with the γ-furin site and extending downstream ∼70 residues to determine if CAP2 was directly cleaving ENaC in this region. Published work suggests that cleavage at V182 and V193 in human γ-ENaC accounts for activation of ENaC by neutrophil elastase, whereas porcine pancreatic elastase cleaves at A190 (Adebamiro et al., 2007). Consistent with this observation, neutrophil elastase generates a slightly faster migrating fragment than the furin fragment on SDS gels (Harris et al., 2007). Moreover, the polybasic tract 183–186 RKRK in mouse ENaC was required for CAP1/prostasin to both activate mouse ENaC and to generate a faster migrating fragment consistent with cleavage at that site (Bruns et al., 2007). CAP2 increased the intensity of bands at the size of the furin fragment and at a slightly smaller molecular mass, suggesting that CAP2 cleaved in this elastase and CAP1-sensitive region of γ-ENaC. Initially, we were drawn to the polybasic tract 183–186 RKRK because of the CAP1/prostasin results (Bruns et al., 2007). Surprisingly, mutation of the homologous site (178–181 RKRK) in rat γ-ENaC to QQQQ did not abolish CAP2 regulation of ENaC, nor did it eliminate a specific band that migrated ahead of the furin fragment. Based on the persistence of fragments slightly smaller than the furin fragment with the RKRK tract converted to QQQQ, we explored the contribution of flanking basic amino acids. Despite converting multiple basic residues (R153Q, K156Q, K168Q, K170Q, R172Q, R178Q, K179Q, R180Q, K181Q, K185Q, K189Q, K200Q, and K201Q) to glutamines in this region and generating different mutant combinations (Fig. 9 C), we never generated a mutant γ-subunit that was not activated by CAP2, and we never completely eliminated the fragment indicative of some cleavage downstream from the furin site. Although our study focuses on CAP2 stimulation of rat ENaC, these results differ significantly from the finding that conversion of the RKRK tract to QQQQ eliminated both a smaller fragment and activation of mouse ENaC by CAP1/prostasin (Bruns et al., 2007). We cannot account for this discrepancy. However, we found that CAP1/prostasin continued to stimulate the RKRK to QQQQ mutant of rat γ-ENaC. Thus, in our hands, CAP1/prostasin and CAP2 behave similarly, i.e., they activate ENaC independently of the polybasic tract 178–181 RKRK in γ-ENaC (Fig. 9).
Faced with these observations, the apparent increase in cleavage at the furin site of γ-ENaC assumed greater significance. We noted that furin or other convertases of the constitutive secretory pathway have a stringent requirement for arginine in the P1 position of their cleavage site (Thomas, 2002; Wheatley and Holyoak, 2007). In the context of the P4-P1 residues present in the γ-furin site (135–138 RKRR), converting 138R to 138K effectively eliminated the furin fragment of γ-ENaC in eggs expressing ENaC alone (Fig. 10 C). In contrast to its effect on the furin fragment, conversion of this key residue to lysine had no effect on generation of a 75-kD fragment and activation of ENaC by CAP2. CAP1 and CAP3/matriptase are reported to accept a lysine in the P1 position (Friedrich et al., 2002; Netzel-Arnett et al., 2003; Shipway et al., 2004; Desilets et al., 2006). Our data indicate that CAP2 may share this flexibility for basic residues at P1, and importantly, strongly suggest that CAP2 cleaves γ-ENaC directly.
Our results raise an important question: Is a single cleavage event in the γ subunit at R138 sufficient for CAP2 to activate ENaC? Whereas cleavage at γ-R138 is clearly essential for CAP2 to stimulate ENaC, our data do not rule out a requirement for additional cleavage downstream of the furin site. Although en masse substitution of glutamines for the 178–181 RKRK polybasic sequence downstream of the furin site and other nearby basic residues did not prevent activation of rat ENaC by CAP2, the maneuver also did not eliminate the occurrence of the ∼73-kD fragment. It is a formal possibility that a different protease with conceivably a nonbasic amino acid requirement, endogenously expressed in oocytes, cleaves in this region of γ-ENaC and that such cleavage contributes to CAP2 stimulation of ENaC.
The results of our study add to a body of evidence identifying γ-ENaC as the ENaC subunit that is targeted by proteases that increase ENaC PO by cleavage after specific residues. Thus, it appears that CAP2, human neutrophil, and pancreatic elastases are now demonstrated to cleave either at the furin site or in a region ∼55 residues downstream of the furin site of γ-ENaC. In addition, thrombin activates ENaC with a thrombin cleavage sequence (LVPRG) engineered into γ-hENaC at position 186 (Adebamiro et al., 2007). All of these proteases can be inferred to act at the extracellular surface of the cell; elastases and thrombin because they act acutely after addition to the cell surface, and CAP2 because CAPs are thought to be activated at the cell surface. This suggests that in the ENaC heteromer, the furin site of γ-ENaC and a region downstream of the furin site at ∼55 residues from it are exposed to proteases at the cell surface. Future structural studies to reveal the molecular basis of this access, and to provide insight as to how cleavage in this region increases ENaC PO, await more definitive reconciliation of ENaC primary sequence with the proposed ASIC1 crystal structure (Stockand et al., 2008).
Acknowledgments
We thank Dr. Ray Caldwell, Dr. Scott Donaldson, and Dr. Martina Gentzsch for critical reading of this manuscript.
This work is supported by grant P01-HL034322-II.
Lawrence G. Palmer served as editor.
Abbreviations used in this paper: CAP, channel-activating protease; ENaC, epithelial sodium channel; HA-NT, HA–N-terminal; hNE, human neutrophil elastase; MBS, modified Barth's solution; MTSET, [2-(trimethylammonium)ethyl] methanethiosulfonate bromide; PO, open probability; V5-CT, V5–C-terminal; WT, wild-type.
References
- Adebamiro, A., Y. Cheng, U.S. Rao, H. Danahay, and R.J. Bridges. 2007. A segment of γ ENaC mediates elastase activation of Na+ transport. J. Gen. Physiol. 130:611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen, D., G. Vuagniaux, N. Fowler-Jaeger, E. Hummler, and B.C. Rossier. 2006. Activation of epithelial sodium channels by mouse channel activating proteases (mCAP) expressed in Xenopus oocytes requires catalytic activity of mCAP3 and mCAP2 but not mCAP1. J. Am. Soc. Nephrol. 17:968–976. [DOI] [PubMed] [Google Scholar]
- Boucher, R.C. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23:146–158. [DOI] [PubMed] [Google Scholar]
- Bruns, J.B., M.D. Carattino, S. Sheng, A.B. Maarouf, O.A. Weisz, J.M. Pilewski, R.P. Hughey, and T.R. Kleyman. 2007. Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the gamma-subunit. J. Biol. Chem. 282:6153–6160. [DOI] [PubMed] [Google Scholar]
- Caldwell, R.A., R.C. Boucher, and M.J. Stutts. 2004. Serine protease activation of near-silent epithelial Na+ channels. Am. J. Physiol. Cell Physiol. 286:C190–C194. [DOI] [PubMed] [Google Scholar]
- Caldwell, R.A., R.C. Boucher, and M.J. Stutts. 2005. Neutrophil elastase activates near-silent epithelial Na+ channels and increases airway epithelial Na+ transport. Am. J. Physiol. Lung Cell. Mol. Physiol. 288:L813–L819. [DOI] [PubMed] [Google Scholar]
- Canessa, C.M., L. Schild, G. Buell, B. Thorens, I. Gautschi, J.D. Horisberger, and B.C. Rossier. 1994. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 367:463–467. [DOI] [PubMed] [Google Scholar]
- Desilets, A., J.M. Longpre, M.E. Beaulieu, and R. Leduc. 2006. Inhibition of human matriptase by eglin c variants. FEBS Lett. 580:2227–2232. [DOI] [PubMed] [Google Scholar]
- Donaldson, S.H., A. Hirsh, D.C. Li, G. Holloway, J. Chao, R.C. Boucher, and S.E. Gabriel. 2002. Regulation of the epithelial sodium channel by serine proteases in human airways. J. Biol. Chem. 277:8338–8345. [DOI] [PubMed] [Google Scholar]
- Ergonul, Z., G. Frindt, and L.G. Palmer. 2006. Regulation of maturation and processing of ENaC subunits in the rat kidney. Am. J. Physiol. Renal Physiol. 291:F683–F693. [DOI] [PubMed] [Google Scholar]
- Friedrich, R., P. Fuentes-Prior, E. Ong, G. Coombs, M. Hunter, R. Oehler, D. Pierson, R. Gonzalez, R. Huber, W. Bode, and E.L. Madison. 2002. Catalytic domain structures of MT-SP1/matriptase, a matrix-degrading transmembrane serine proteinase. J. Biol. Chem. 277:2160–2168. [DOI] [PubMed] [Google Scholar]
- Guipponi, M., G. Vuagniaux, M. Wattenhofer, K. Shibuya, M. Vazquez, L. Dougherty, N. Scamuffa, E. Guida, M. Okui, C. Rossier, et al. 2002. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum. Mol. Genet. 11:2829–2836. [DOI] [PubMed] [Google Scholar]
- Harris, M., D. Firsov, G. Vuagniaux, M.J. Stutts, and B.C. Rossier. 2007. A novel neutrophil elastase inhibitor prevents elastase activation and surface cleavage of the epithelial sodium channel expressed in Xenopus laevis oocytes. J. Biol. Chem. 282:58–64. [DOI] [PubMed] [Google Scholar]
- Hughey, R.P., G.M. Mueller, J.B. Bruns, C.L. Kinlough, P.A. Poland, K.L. Harkleroad, M.D. Carattino, and T.R. Kleyman. 2003. Maturation of the epithelial Na+ channel involves proteolytic processing of the alpha- and gamma-subunits. J. Biol. Chem. 278:37073–37082. [DOI] [PubMed] [Google Scholar]
- Hughey, R.P., J.B. Bruns, C.L. Kinlough, K.L. Harkleroad, Q. Tong, M.D. Carattino, J.P. Johnson, J.D. Stockand, and T.R. Kleyman. 2004. Epithelial sodium channels are activated by furin-dependent proteolysis. J. Biol. Chem. 279:18111–18114. [DOI] [PubMed] [Google Scholar]
- Jasti, J., H. Furukawa, E.B. Gonzales, and E. Gouaux. 2007. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 449:316–323. [DOI] [PubMed] [Google Scholar]
- Kellenberger, S., I. Gautschi, and L. Schild. 2002. An external site controls closing of the epithelial Na+ channel ENaC. J. Physiol. 543:413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddle, G.W., T. Bledsoe, and W.S. Coppage Jr. 1963. A familial renal disorder simulating primary aldosteronism but with negligible aldosterone secretion. Trans. Assoc. Am. Physicians. 76:199–213. [Google Scholar]
- Myerburg, M.M., M.B. Butterworth, E.E. McKenna, K.W. Peters, R.A. Frizzell, T.R. Kleyman, and J.M. Pilewski. 2006. Airway surface liquid volume regulates ENaC by altering the serine protease-protease inhibitor balance: a mechanism for sodium hyperabsorption in cystic fibrosis. J. Biol. Chem. 281:27942–27949. [DOI] [PubMed] [Google Scholar]
- Netzel-Arnett, S., J.D. Hooper, R. Szabo, E.L. Madison, J.P. Quigley, T.H. Bugge, and T.M. Antalis. 2003. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 22:237–258. [DOI] [PubMed] [Google Scholar]
- Planes, C., and G.H. Caughey. 2007. Regulation of the epithelial Na+ channel by peptidases. Curr. Top. Dev. Biol. 78:23–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planes, C., M. Blot-Chabaud, M.A. Matthay, S. Couette, T. Uchida, and C. Clerici. 2002. Hypoxia and beta 2-agonists regulate cell surface expression of the epithelial sodium channel in native alveolar epithelial cells. J. Biol. Chem. 277:47318–47324. [DOI] [PubMed] [Google Scholar]
- Rossier, B.C. 2004. The epithelial sodium channel: activation by membrane-bound serine proteases. Proc. Am. Thorac. Soc. 1:4–9. [DOI] [PubMed] [Google Scholar]
- Schild, L. 2004. The epithelial sodium channel: from molecule to disease. Rev. Physiol. Biochem. Pharmacol. 151:93–107. [DOI] [PubMed] [Google Scholar]
- Shipway, A., H. Danahay, J.A. Williams, D.C. Tully, B.J. Backes, and J.L. Harris. 2004. Biochemical characterization of prostasin, a channel activating protease. Biochem. Biophys. Res. Commun. 324:953–963. [DOI] [PubMed] [Google Scholar]
- Stockand, J.D., A. Staruschenko, O. Pochynyuk, R.E. Booth, and D.U. Silverthorn. 2008. Insight toward epithelial Na(+) channel mechanism revealed by the acid-sensing ion channel 1 structure. IUBMB Life. 60:620–628. [DOI] [PubMed] [Google Scholar]
- Tarran, R., L. Trout, S.H. Donaldson, and R.C. Boucher. 2006. Soluble mediators, not cilia, determine airway surface liquid volume in normal and cystic fibrosis superficial airway epithelia. J. Gen. Physiol. 127:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3:753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Z., B. Illek, V.J. Bhagwandin, G.M. Verghese, and G.H. Caughey. 2004. Prostasin, a membrane-anchored serine peptidase, regulates sodium currents in JME/CF15 cells, a cystic fibrosis airway epithelial cell line. Am. J. Physiol. Lung Cell. Mol. Physiol. 287:L928–L935. [DOI] [PubMed] [Google Scholar]
- Vallet, V., A. Chraibi, H.P. Gaeggeler, J.D. Horisberger, and B.C. Rossier. 1997. An epithelial serine protease activates the amiloride-sensitive sodium channel. Nature. 389:607–610. [DOI] [PubMed] [Google Scholar]
- Vallet, V., C. Pfister, J. Loffing, and B.C. Rossier. 2002. Cell-surface expression of the channel activating protease xCAP-1 is required for activation of ENaC in the Xenopus oocyte. J. Am. Soc. Nephrol. 13:588–594. [DOI] [PubMed] [Google Scholar]
- Vuagniaux, G., V. Vallet, N.F. Jaeger, C. Pfister, M. Bens, N. Farman, N. Courtois-Coutry, A. Vandewalle, B.C. Rossier, and E. Hummler. 2000. Activation of the amiloride-sensitive epithelial sodium channel by the serine protease mCAP1 expressed in a mouse cortical collecting duct cell line. J. Am. Soc. Nephrol. 11:828–834. [DOI] [PubMed] [Google Scholar]
- Vuagniaux, G., V. Vallet, N.F. Jaeger, E. Hummler, and B.C. Rossier. 2002. Synergistic activation of ENaC by three membrane-bound channel-activating serine proteases (mCAP1, mCAP2, and mCAP3) and serum- and glucocorticoid-regulated kinase (Sgk1) in Xenopus oocytes. J. Gen. Physiol. 120:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley, J.L., and T. Holyoak. 2007. Differential P1 arginine and lysine recognition in the prototypical proprotein convertase Kex2. Proc. Natl. Acad. Sci. USA. 104:6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]