It is now clear that transient receptor potential (TRP) channels are sensors of temperature, mechanical force, noxious chemicals, and G protein–coupled receptor–mediated signal transduction. Sequence and topological analyses group TRPs with other tetrameric cation channels (Yu and Catterall, 2004). Each subunit bears extensive N- and C-terminal cytoplasmic domains and an S1-S2-S3-S4-S5-P-S6 motif, where S's are segments of transmembrane α helices and P is the “pore” that houses the ion filter. Although there are no crystal structures of TRPs at this writing, they are expected to be grossly similar to those of K+ channels (Doyle et al., 1998; Long et al., 2007). Indeed, recent electron cryomicroscopy of a TRP channel (TRPV1) at 19-Å resolution is consistent with a tetrameric structure having a compact transmembrane core and a large cytoplasmic domain in the form of a “hanging gondola” (Moiseenkova-Bell et al., 2008). Various aspects of TRP channel research have been reviewed elsewhere (Ramsey et al., 2006; Nilius et al., 2007; Venkatachalam and Montell, 2007). These reviews delve deeply into each of the animal TRP subtypes with regard to their expression, activation, regulation, and medical implications. Here, we intend only to complement these reviews, and in so doing we draw attention to the presence of TRPs beyond animal species, the commonalities among TRP subfamilies, the role of prospective searches in founding these subfamilies, and the continued use of prospective methods in dissecting TRP channels. We find especially striking the repeated identifications of a site at which mutations lead to constitutive channel activity in different TRP subtypes and wish to highlight these forward genetic results.
The Commonalities and Deep Roots of TRPs
Animal TRPs are sorted into seven subfamilies, TRPA (ankyrin), TRPC (canonical), TRPM (melastatin), TRPML (mucolipin), TRPN (NOMPC), TRPP (polycystin), and TRPV (vanilloid), according to sequence features such as ankyrin repeats and “TRP boxes” within the predicted N- or C-terminal cytoplasmic domains. They are grouped into one superfamily based on their 6-S topology and sequence similarities, although confident alignment across the seven subfamilies can only be made from S5 to just beyond S6. In voltage-gated K+ channels, where we do have 3-D structures at atomic resolutions and deep functional knowledge, each subunit in the tetramer consists of two modules: the S1-S2-S3-S4 peripheral module that bears the voltage sensor and the S5-P-S6 core module that forms the ion conduit, the filter, and the gate (Jiang et al., 2003; Long et al., 2007). By analogy, only the ion permeation core shows sequence conservation across the TRP subtypes. The variations in the peripheral modules and the cytoplasmic domains presumably reflect differences in gating stimuli as well as channel-specific modulatory mechanisms.
Although ion channels are mostly studied in multicellular animals, they are in fact also widespread among microbes. Na+ channels, Cl− “channels,” and a large variety of K+ channels (H+-, Ca2+-, glutamate-gated; de- or hyperpolarization-activated) have all been found in prokaryotes and unicellular eukaryotes (Kung and Blount, 2004; Loukin et al., 2005). TRP channels are no exception. Genes predicting a 6-S topology with homologous S5-P-S6 sequences are found with no ambiguity in the genomes of Tetrahymena and Paramecium (ciliates), Dictyostelium (cellular slime mold), Trypanosoma (the agent of African sleeping sickness), and Leishmainia (leishmainasis). Fragments of similarity can also be found in Plasmodium (malaria), Thalassiosira (diatom), and Chlamydomonas (algal flagellate) (Saimi et al., 2007). Homologues of TRPA, TRPC, TRPM, TRPML, and TRPV are found in choanoflagellates, likely the closest known extant relatives of metazoans (Cai, 2008).
Each of the over 30 genomes of fungi (both Ascomycetes and Basidomycetes) also contains one or more TRP channel genes based on the above criteria. TRPY1, the sole TRP sequence in the budding yeast Saccharmoyces cerevisiae, has been examined in detail (Palmer et al., 2001). It is located in the vacuolar membrane and has a 300-pS unitary conductance that rectifies inwardly. Like other TRPs, it passes cations and is polymodal, being activated by cytoplasmic Ca2+, vacuolar H+, and membrane stretch evoked by environmental osmotic changes in vivo (Zhou et al., 2005). Analysis of TRPY1 illustrates that microbial TRPs are not only sequence homologues, but also functional counterparts of animal TRPs. Thus, TRPs apparently have evolved early. Among the tetrameric cation channels, animal K+ channels are devoted to steadying the membrane potential and voltage-gated Ca2+ and Na+ channels to signaling or excitation. TRPs, however, appear to have evolved to sense various environmental stimuli such as temperature, mechanical force, and internal or external ligands (Clapham, 2003).
Unbiased Searches for Cellular and Environmental Sensors Identified the Founding Members of TRP Subfamilies
Very often, genes and their products become associated with functions for the first time through unbiased searches with no preconceived notions. The classical example of such an approach is “forward genetic” screening, which begins with a mutant phenotype and works toward identifying the underlying gene, without a prior notion of its nature. TRP genes were so identified in the search for genes underlying failures in stimulus sensing as the mutant phenotypes.
A near-blind fly initiated TRP research. In the 70's and 80's, the Pak laboratory generated and examined a large number of blind Drosophila with the explicit purpose of finding the underlying elements in phototransduction (Pak, 1995). Beyond the lack of phototaxis as a behavioral phenotype, these flies were examined by electroretinography, an extracellular recording technique that registers the massive depolarization in the compound eye when light is presented. One type of near-blind fly in question could initiate depolarization like its wild-type counterpart, but it could not sustain this response during the light pulse. The phenotype of the un-sustained transient receptor potential (TRP) became the namesake of these mutant flies and the underlying channel that is responsible for light-evoked depolarization of the photoreceptor (Minke et al., 1975). The corresponding gene was eventually cloned and analyzed (Montell and Rubin, 1989). This founding member of TRP channels, now classified as dTRPC, is in fact one of two similar channels directly responsible for the light-induced membrane depolarization in insects, although the exact molecular mechanism of their activation remains unclear (Minke, 2006) (Leung et al., 2008). The fly also has many behaviors related to mechanosensation. Beginning with mutants defective in balance and touch response of adult flies, Zuker's group (Walker et al., 2000) traced the deficit to mutations in NOMPC, the founding member of the TRPN subfamily. Similarly, mutations blocking the ability of Drosophila larvae to recoil from noxious heat or mechanical stimuli were traced to PAINLESS, now a TRPA (Tracey et al., 2003). Using the nematode Caenorhabditis elegans, the Bargmann laboratory isolated and analyzed mutants defective in their avoidance of hyperosmolar fructose. Positional cloning led to a gene, osm-9, which is distantly related to mammalian TRPVs (Colbert et al., 1997). Similarly, mutations causing defects in the male worm's ability to locate the vulvas of hermaphrodites were traced to LOV-1, a homologue of PKD1 that forms channels by associating with PKD2, now TRPP (Barr and Sternberg, 1999). Of course, cloning genes underlying heritable diseases in humans is the medical equivalent of the phenotype-to-gene forward genetics. In this way, PKD1 and PKD2 were linked to polycystic kidney disease, founding the TRPP subfamily (Hughes et al., 1995; Mochizuki et al., 1996). Likewise, mucolipidosis type IV was traced to MCOLN1, founding the TRPML subfamily (Bargal et al., 2000; Sun et al., 2000).
Expression cloning is another prospective research strategy to find genes without bias toward protein sequence or structure. In the present context, this strategy is based on the ability of cDNAs to confer responsiveness to a given chemical or physical stimulus when introduced into an otherwise nonresponsive cell. In this manner, Caterina et al. (1997) screened a sensory ganglion cDNA library by expression in HEK293 cells for capsaicin-evoked increases in intracellular Ca2+, leading to the identification of the receptor for capsaicin, the founding member of the TRPV subfamily. TRPV1 is also a natural receptor for noxious heat, thereby contributing to mechanisms that underlie thermal pain. Similarly, using menthol as a surrogate of cold, expression cloning revealed TRPM8 as a cold-activated member of the TRP channel family (McKemy et al., 2002).
Thus, members of TRP subfamilies (TRPA, TRPC, TRPV, TRPN, TRPP, TRPM, or TRPML) were first independently discovered or functionally defined by virtue of unbiased, function-based searches, rather than through sequence or homology-based screening methods. Starting with the organism's ability to sense light, osmolarity, touch, balance, temperature extremes, or their chemical surrogates, the convergence of these multiple function-based studies onto members of the same superfamily of ion channels is remarkable. Indeed, this convergence of discoveries from unbiased independent screens strongly endorses the central role of TRPs in sensory biology as essential components of cellular signaling pathways that sense the environment.
A Deeper Forward Genetics Dissection
Once a TRP channel has been identified from the above searches or by homology with founding members, it is possible to perform site-directed mutageneses to examine the effects of channel mutations, usually by expressing the mutant TRP in oocytes or cultured mammalian cells. An often-overlooked opportunity is to take the forward genetics approach a step further to dissect TRPs. Much like randomly mutating the entire genome and following a phenotype to a single gene, one should be able to randomly mutate an entire gene and follow the phenotype to a single amino acid. The logic is the same: even though a protein has hundreds of amino acid residues and each can be mutated into 19 others, we can, without a preconceived bias, simply ask which single amino acid change(s) among a very large number of induced mutations produces significant changes in molecular activities. This method, however, is currently unavailable with animal (the worm, the fly, zebrafish, and the mouse) or plant (the cress) models because it is, at the moment, impractical to generate the thousands of single-gene transformants en masse that would be required for such a forward genetic approach. The ease in transformation and the large populations of microbes, on the other hand, make the procedure facile therein. After a random mutagenesis of a single plasmid-borne gene in a microbe, it is relatively simple to find mutations that alter its molecular activities, corresponding to so-called “gain-” or “loss-of-function” mutations and “reduced-” or “altered-function” mutations. Indeed, the search and study of “gain-of-function” alleles of the TRPY1 channel native to yeast have been performed (Su et al., 2007; Zhou et al., 2007). This method can also be extended to the analysis of animal or plant channels by mutating their genes on plasmids, expressing these plasmids in a microbe or cultured cell, and leveraging the power of a genetic selection or function-based screen to uncover mutants that specifically alter a given property of the channel. Such strategies have been described for the identification of functionally interesting residues or domains in TRPV1 (Myers et al., 2008), TRPV3 (Grandl et al., 2008), and TRPM8 (Bandell et al., 2006).
An Uncanny Convergence
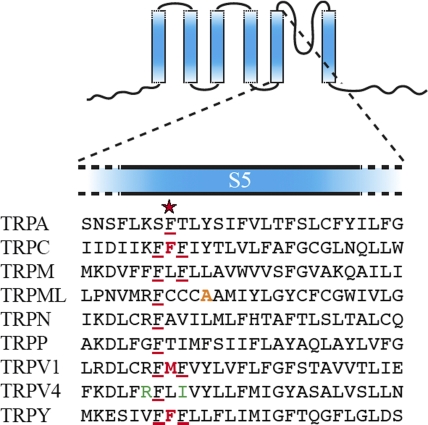
The founding mutations of TRPs that blind the fly are loss-of-function alleles. As noted above, the inability to sustain the light-induced receptor potential results from loss of function for the Drosophila TRPC1 channel, originally called trp (Cosens and Manning, 1969; Minke et al., 1975; Montell and Rubin, 1989). A lone gain-of-function dTRPC1 allele was later identified, which results not only in blindness, but also rapid and severe retinal degeneration (Yoon et al., 2000). In this fly, receptor potential is sustained not only during the light presentation, but long after the light is turned off. The retinal degeneration is presumably caused by the cytotoxicity of Ca2+ entering through the constitutively active channels. The causative mutation was identified as F550I (Fig. 1, red), located near the beginning of the predicted S5 (Hong et al., 2002), within a cluster of three phenylalanines. In fact, one or more phenylalanines can be found in this region of members of fungal and animal TRP subfamilies (Fig. 1, underlined).
Figure 1.
An alignment of the S5's (the predicted fifth transmembrane α helices) of different TRP subtypes. In three independent unbiased searches, the same site (red star) was discovered at which mutations cause constitutive channel activities in TRPC1, TRPV1, and TRPY (bold red letters). The site is in a small cluster of phenylalanines, members of which are found in all TRP subtypes (underlined red). The mutations in TRPV4 that cause brachyolmia in human (green) and the mutation in TRPML that causes the varitint-waddler mouse phenotype are nearby (orange). Shown are subfamily representatives: TRPA (painless of Drosophila), TRPC (the canonical TRPC of Drosophila), TRPM2 (human), TRPML3 (mouse), TRPN (zebra fish), TRPP2 (mouse), TRPV1 (rat), TRPV4 (rat), and TRPY1 (budding yeast). Analyses began with a large-scale alignment of the full-length sequences of all current members of the TRP superfamily, along with several other 6-S cation channels, using the CLUSTAL W algorithm (Gonnet 250 matrix) by way of the CLUSTAL X interface (1.81) (Thompson et al., 1997). Comparisons with pfam00520 (Finn et al., 2006) and applications of various transmembrane and secondary-structure prediction algorithms (e.g., PROFphd; Thompson et al., 1997; Rost et al., 2004; Finn et al., 2006) predict the sequences shown here to be the transmembrane α helices preceding the pore helix.
As explained in the previous section, it is possible to systematically search with microbes for significant channel mutations, rather than relying on happenstance, as was the case for identification of the Drosophila TRP mutant described above. In a search for gain-of-function mutations, the cloned TRPY1 was mutagenized and transformed into trpy1Δ yeast cells, and transformants were screened for mutations. It is not practical to use the labor-intensive patch clamp as a screening device. Instead, a more convenient phenotyping procedure was used. Osmotic upshock can activate the mechanosensitive TRPY1 in the vacuolar membrane to release vacuolar Ca2+ into the cytoplasm. Using transgenic aequorin as a reporter, this Ca2+ signal can be followed in vivo, and the unusually large signal appearing upon mild osmotic shock was used as the criterion for identifying overly active mutant channels (Zhou et al., 2007). Upon patch clamp scrutiny, the mutant channels have higher open probabilities than the wild-type channel under various conditions, as expected. 7 of 10 such gain-of-function mutations in the first harvest were found to be located in predicted transmembrane helices. Surprisingly, one (F380L) aligns perfectly with the gain-of-function mutation of the Drosophila TRPC1 described above (Su et al., 2007) (Fig. 1, red).
The budding yeast has also been used to study a mammalian TRP channel. Expression of rat TRPV1 is toxic to yeast when its ligand, capsaicin, is present in the growth medium. It thus became possible, after random mutagenesis, to search for gain-of-function mutants that hamper growth even in the absence of ligand (but which can be rescued by the addition of the channel blocker, ruthenium red) (Myers et al., 2008). The mutants identified in this manner were examined electrophysiologically in Xenopus oocytes. Consistent with the cellular phenotype in yeast, some of the gain-of-function mutants were found to potentiate the effect of acidity, a known gating modality of TRPV1. Others caused constitutive activation, i.e., passing substantial current even at room temperature and in the absence of chemical stimuli. Amazingly, one of these constitutive alleles, M581T, sandwiched between two phenylalanines, aligned exactly with F550 of dTRPC1 and F380 of the yeast TRPY1 (Myers et al., 2008) (Fig. 1, red)!
Because the experimental systems described above are so very different, the electrophysiological methods are also necessarily different: The dTRPC1 channel activities were deduced from the fly's electroretinographs; the rTRPV1 activities were from macroscopic currents when expressed in oocytes; and the yeast TRPY1 single-channel activities were directly examined by patch clamp. Despite the differences in methods, the gain-of-function mutant channels were all found to be overly active. Pinpointing same-site mutations (Fig. 1, red star) that lead to constitutive channel activities from three independent unbiased searches is not likely to be a coincidence.
Recent genetic studies lend further support to the idea that the hydrophobic region at the base of S5 forms a conserved part of the gating apparatus in multiple TRP channels. A set of gain-of-function alleles was uncovered in TRPV4 through genetic mapping of autosomal dominant brachyolmia, a class of human inherited skeletal dysplasias (Rock et al., 2008). By examining the pedigree of a large family with autosomal dominant brachyolmia, the authors traced the occurrence of the disease to mutations in the TRPV4 locus. When engineered into the cloned channel, such mutations cause increased basal activity, providing a correlation between hyperactive TRPV4 alleles and skeletal disease. Interestingly, both of the disease-causing TRPV4 alleles map near to the cytoplasmic end of S5 (R616Q, V620I; Fig. 1, green), in close proximity to the gain-of-function alleles described above for rat TRPV1, yeast TRPY1, and the Drosophila TRPC1. Furthermore, a gain-of-function allele of TRPML3 is known to underlie the varitint-waddler phenotype of the mouse (Di Palma et al., 2002). Expression of the mutant TRPML3 allele in cultured cells (Grimm et al., 2007; Kim et al., 2007; Xu et al., 2007; Nagata et al., 2008) leads to a constitutively active current. The causative mutation, A419P, likely introduces a kink in the α-helical turn downstream from the S5 site identified by the studies described above (Fig. 1, orange).
A Common Gating Mechanism among Different TRP Subfamilies?
Much research has been devoted to understand how individual TRP channels, belonging to a certain subtype, detect their cognate stimuli or relevant modulatory factors. Less is known about the possible structural or mechanistic commonalities among subtypes. Note that the mammalian TRPV1 is a heat/inflammation/pain sensor; fungal TRPY1 is a mechanosensitive channel that monitors osmolarity in vivo, whereas Drosophila TRPC1 is a “receptor-operated” channel that functions as a component of the insect phototransduction pathway. That mutations at the same residue near the beginning of S5 were found to have the same molecular effects in three TRPs that play disparate biological roles strongly indicates a common molecular mechanism in the gating of these channels. This insight could hardly have been derived from sequence comparison alone because homology among the different TRPs is limited, even within the S5-P-S6 region (Fig. 1).
In K+ channels, the convergence of the cytoplasmic ends of the four S6's closes the gate, i.e., the hydrophobic occlusion of the ion pathway. The gate can be opened in several ways. In the case of some Ca2+-activated K+ channels, the conformational changes of Ca2+ binding in the cytoplasmic domain mechanically pull open the gate through the peptide linking this domain to the end of the S6's (Jiang et al., 2002). In the cases of voltage-gated channels, the charged S4 moves with the change in transmembrane voltage. Here, each of the S4-S5 linkers binds to a site in S6 near the occlusion, in a “hand-and-handle” manner, to transmit the mechanical gating force (Jiang et al., 2003). The residue identified in the three forward genetic studies is predicted to reside near the inner end of S5 in TRPs. Even though the region linking S4 and S5 of TRPs bears little resemblance to that of K+ channels, a different set of hand-and-handle interactions may be at work here.
The phenylalanine pinpointed by the TRPY1 and TRPC1 mutations above often has phenylalanine neighbors in other TRP channels, forming a cluster (Fig. 1, underlined). In the TRPY1 study, many of the gain-of-function mutations involve aromatic amino acids. A residue near the predicted hydrophobic occlusion and may be a part of the “handle,” Y458, was studied thoroughly. All functional channels among the 19 possible substitutions, except Y458F and Y458W, have severe abnormal gating kinetics, indicating that aromaticity, rather than size or shape, is the crucial parameter for normal gating (Zhou et al., 2007). Why aromatics? The cluster of phenylalanines (Fig. 1, underlined) in the “hand” may fulfill a similar requirement for aromaticity of the handle, represented by Y458 in the TRPY1 experiment, assuming a “hand-handle” mechanism indeed operates here. Alternatively or additionally, aromatics may form networks to stabilize proteins in the lipid bilayer. Most crystallized membrane proteins are found to have aromatic belts in their interior. For example, KcsA, the first K+ channel crystallized, shows two such belts (Doyle et al., 1998). These belts are aligned approximately along the two interfaces of the bilayer where surface tension is maximal (Nyholm et al., 2007). It appears that KcsA has evolved to use the natural tendency of the aromatic rings to dwell at the interface (Yau et al., 1998) to maintain its structures. The belt near the outside, for instance, is apparently used to keep the K+ filter from collapsing (Doyle et al., 1998). In TRPY1, the aromatic mutations identified by the gain-of-function screens all show severe gating abnormalities. One possibility is that these aromatics are parts of networks that maintain the various open and closed conformations of the gate. If so, this may be a feature of TRP channels in general, as suggested by the convergence of mutations described above.
The Continued Role of Forward Genetics in TRP Research
Both the yeast TRPY1 and rat TRPV1 screens, though not saturated, yielded multiple gain-of-function mutations, besides the noted coincidental ones. For example, the F640L mutation of TRPV1 in the predicted pore-helix domain (Myers et al., 2008) and the F458H mutation of TRPY1 at a point near the predicted hydrophobic occlusion (Zhou et al., 2007) can be expected to have profound effects on channel conformations. These mutations, in turn, experimentally validate the domain assignments based only on sequence analyses. Interestingly, the constitutive pore helix mutations recovered from the TRPV1 yeast screen were found to produce similar effects when introduced into TRPV2 and TRPV3, supporting the notion that this region of TRPV channels forms a conserved part of the gating machinery (Myers et al., 2008). Along these lines, it might be informative to engineer the mutations recovered from various unbiased screens into other TRP subtypes to assess the generality of their effects, particularly in the S5-P-S6 region, where sequences can be aligned with greatest confidence. Function-altering mutations are most often destructive, leading to failures not only in channel function per se, but also in expression, trafficking, etc. Therefore, a gain-of-function screen is a reasonable first exploration. Nevertheless, mutations causing specific loss of function can also be very informative. For example, upon expressing TRPV3 in cultured cells, Grandl et al. (2008) used high throughput calcium imaging to screen for TRPV3 mutants that abolish heat activation but retain activation by camphor or 2-aminoethoxydiphenyl borate. These mutations therefore located residues of the channel required for heat activation. Such studies, whether based on gain- or loss-of-function phenotypes, will continue to be exceedingly useful for dissecting the structure and function of polymodal ion channels, such as TRPs.
In summary, assumption-free unbiased searches have pioneered many areas of biological research, a strategy that has now been extended to the analysis of TRP channel physiology. We expect that this forward genetics approach will continue to complement hypothesis-driven targeted mutational analyses, as well as future insights from crystallographic and other biophysical studies.
Acknowledgments
We thank Dr. W. John Haynes and Ms. Leanne Olds for preparing Fig. 1.
This work is supported by the American Heart Association (to B.R. Myers), the National Institutes of Health (NIH; National Institute of Neurological Disorders and Stroke; to D. Julius), NIH GM054867 (to Y. Saimi), NIH GM047856 to (C. Kung), and the Vilas Trust of the University of Wisconsin.
Edward N. Pugh Jr. served as editor.
Abbreviation used in this paper: TRP, transient receptor potential.
References
- Bandell, M., A.E. Dubin, M.J. Petrus, A. Orth, J. Mathur, S.W. Hwang, and A. Patapoutian. 2006. High-throughput random mutagenesis screen reveals TRPM8 residues specifically required for activation by menthol. Nat. Neurosci. 9:493–500. [DOI] [PubMed] [Google Scholar]
- Bargal, R., N. Avidan, E. Ben-Asher, Z. Olender, M. Zeigler, A. Frumkin, A. Raas-Rothschild, G. Glusman, D. Lancet, and G. Bach. 2000. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 26:118–123. [DOI] [PubMed] [Google Scholar]
- Barr, M.M., and P.W. Sternberg. 1999. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 401:386–389. [DOI] [PubMed] [Google Scholar]
- Cai, X. 2008. Unicellular Ca2+ signaling ‘toolkit’ at the origin of metazoa. Mol. Biol. Evol. 25:1357–1361. [DOI] [PubMed] [Google Scholar]
- Caterina, M.J., M.A. Schumacher, M. Tominaga, T.A. Rosen, J.D. Levine, and D. Julius. 1997. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 389:816–824. [DOI] [PubMed] [Google Scholar]
- Clapham, D.E. 2003. TRP channels as cellular sensors. Nature. 426:517–524. [DOI] [PubMed] [Google Scholar]
- Colbert, H.A., T.L. Smith, and C.I. Bargmann. 1997. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J. Neurosci. 17:8259–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosens, D.J., and A. Manning. 1969. Abnormal electroretinogram from a Drosophila mutant. Nature. 224:285–287. [DOI] [PubMed] [Google Scholar]
- Di Palma, F., I.A. Belyantseva, H.J. Kim, T.F. Vogt, B. Kachar, and K. Noben-Trauth. 2002. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc. Natl. Acad. Sci. USA. 99:14994–14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, D.A., J. Morais Cabral, R.A. Pfuetzner, A. Kuo, J.M. Gulbis, S.L. Cohen, B.T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Finn, R.D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, et al. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandl, J., H. Hu, M. Bandell, B. Bursulaya, M. Schmidt, M. Petrus, and A. Patapoutian. 2008. Pore region of TRPV3 ion channel is specifically required for heat activation. Nat. Neurosci. 11:1007–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, C., M.P. Cuajungco, A.F. van Aken, M. Schnee, S. Jors, C.J. Kros, A.J. Ricci, and S. Heller. 2007. A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc. Natl. Acad. Sci. USA. 104:19583–19588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y.S., S. Park, C. Geng, K. Baek, J.D. Bowman, J. Yoon, and W.L. Pak. 2002. Single amino acid change in the fifth transmembrane segment of the TRP Ca2+ channel causes massive degeneration of photoreceptors. J. Biol. Chem. 277:33884–33889. [DOI] [PubMed] [Google Scholar]
- Hughes, J., C.J. Ward, B. Peral, R. Aspinwall, K. Clark, J.L. San Millan, V. Gamble, and P.C. Harris. 1995. The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat. Genet. 10:151–160. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, M. Cadene, B.T. Chait, and R. MacKinnon. 2002. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B.T. Chait, and R. MacKinnon. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., Q. Li, S. Tjon-Kon-Sang, I. So, K. Kiselyov, and S. Muallem. 2007. Gain-of-function mutation in TRPML3 causes the mouse Varitint-Waddler phenotype. J. Biol. Chem. 282:36138–36142. [DOI] [PubMed] [Google Scholar]
- Kung, C., and P. Blount. 2004. Channels in microbes: so many holes to fill. Mol. Microbiol. 53:373–380. [DOI] [PubMed] [Google Scholar]
- Leung, H.T., J. Tseng-Crank, E. Kim, C. Mahapatra, S. Shino, Y. Zhou, L. An, R.W. Doerge, and W.L. Pak. 2008. DAG lipase activity is necessary for TRP channel regulation in Drosophila photoreceptors. Neuron. 58:884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, S.B., X. Tao, E.B. Campbell, and R. MacKinnon. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382. [DOI] [PubMed] [Google Scholar]
- Loukin, S.H., M.M. Kuo, X.L. Zhou, W.J. Haynes, C. Kung, and Y. Saimi. 2005. Microbial K+ channels. J. Gen. Physiol. 125:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKemy, D.D., W.M. Neuhausser, and D. Julius. 2002. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature. 416:52–58. [DOI] [PubMed] [Google Scholar]
- Minke, B., and M. Parnas. 2006. Insights on TRP channels from in vivo studies in Drosophila. Annu. Rev. Physiol. 68:649–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke, B., C.-F. Wu, and W.L. Pak. 1975. Induction of photoreceptor voltage noise in the dark in Drosophila mutant. Nature. 258:84–87. [DOI] [PubMed] [Google Scholar]
- Mochizuki, T., G. Wu, T. Hayashi, S.L. Xenophontos, B. Veldhuisen, J.J. Saris, D.M. Reynolds, Y. Cai, P.A. Gabow, A. Pierides, et al. 1996. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 272:1339–1342. [DOI] [PubMed] [Google Scholar]
- Moiseenkova-Bell, V.Y., L.A. Stanciu, I.I. Serysheva, B.J. Tobe, and T.G. Wensel. 2008. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA. 105:7451–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell, C., and G.M. Rubin. 1989. Molecular characterization of the Drosophila trp locus: a putative integral membrane protein required for phototransduction. Neuron. 2:1313–1323. [DOI] [PubMed] [Google Scholar]
- Myers, B.R., C.J. Bohlen, and D. Julius. 2008. A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron. 58:362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, K., L. Zheng, T. Madathany, A.J. Castiglioni, J.R. Bartles, and J. Garcia-Anoveros. 2008. The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc. Natl. Acad. Sci. USA. 105:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius, B., G. Owsianik, T. Voets, and J.A. Peters. 2007. Transient receptor potential cation channels in disease. Physiol. Rev. 87:165–217. [DOI] [PubMed] [Google Scholar]
- Nyholm, T.K., S. Ozdirekcan, and J.A. Killian. 2007. How protein transmembrane segments sense the lipid environment. Biochemistry. 46:1457–1465. [DOI] [PubMed] [Google Scholar]
- Pak, W.L. 1995. Drosophila in vision research. The Friedenwald Lecture. Invest. Ophthalmol. Vis. Sci. 36:2340–2357. [PubMed] [Google Scholar]
- Palmer, C.P., X.L. Zhou, J. Lin, S.H. Loukin, C. Kung, and Y. Saimi. 2001. A TRP homolog in Saccharomyces cerevisiae forms an intracellular Ca(2+)-permeable channel in the yeast vacuolar membrane. Proc. Natl. Acad. Sci. USA. 98:7801–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, I.S., M. Delling, and D.E. Clapham. 2006. An introduction to TRP channels. Annu. Rev. Physiol. 68:619–647. [DOI] [PubMed] [Google Scholar]
- Rock, M.J., J. Prenen, V.A. Funari, T.L. Funari, B. Merriman, S.F. Nelson, R.S. Lachman, W.R. Wilcox, S. Reyno, R. Quadrelli, et al. 2008. Gain-of-function mutations in TRPV4 cause autosomal dominant brachyolmia. Nat. Genet. 40:999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saimi, Y., X.-L. Zhou, S.H. Loukin, W.J. Haynes, and C. Kung. 2007. Microbial TRP channels and their mechanosensitivity. In Current Topics in Membranes. O.P. Hamill, editor. Elsevier, Boston. 311–327.
- Su, Z., X. Zhou, W.J. Haynes, S.H. Loukin, A. Anishkin, Y. Saimi, and C. Kung. 2007. Yeast gain-of-function mutations reveal structure function relationships conserved among different subfamilies of transient receptor potential channels. Proc. Natl. Acad. Sci. USA. 49:19607–19612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, M., E. Goldin, S. Stahl, J.L. Falardeau, J.C. Kennedy, J.S. Acierno Jr., C. Bove, C.R. Kaneski, J. Nagle, M.C. Bromley, et al. 2000. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 9:2471–2478. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin, and D.G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey, W.D. Jr., R.I. Wilson, G. Laurent, and S. Benzer. 2003. painless, a Drosophila gene essential for nociception. Cell. 113:261–273. [DOI] [PubMed] [Google Scholar]
- Venkatachalam, K., and C. Montell. 2007. TRP channels. Annu. Rev. Biochem. 76:387–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, R.G., A.T. Willingham, and C.S. Zuker. 2000. A Drosophila mechanosensory transduction channel. Science. 287:2229–2234. [DOI] [PubMed] [Google Scholar]
- Xu, H., M. Delling, L. Li, X. Dong, and D.E. Clapham. 2007. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc. Natl. Acad. Sci. USA. 104:18321–18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau, W.M., W.C. Wimley, K. Gawrisch, and S.H. White. 1998. The preference of tryptophan for membrane interfaces. Biochemistry. 37:14713–14718. [DOI] [PubMed] [Google Scholar]
- Yoon, J., H.C. Ben-Ami, Y.S. Hong, S. Park, L.L. Strong, J. Bowman, C. Geng, K. Baek, B. Minke, and W.L. Pak. 2000. Novel mechanism of massive photoreceptor degeneration caused by mutations in the trp gene of Drosophila. J. Neurosci. 20:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.H., and W.A. Catterall. 2004. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci. STKE. 2004:re15. [DOI] [PubMed] [Google Scholar]
- Zhou, X.-L., S.H. Loukin, R. Coria, C. Kung, and Y. Saimi. 2005. Heterologously expressed fungal transient receptor potential channels retain mechanosensitivity in vitro and osmotic response in vivo. Eur. Biophys. J. 34:413–422. [DOI] [PubMed] [Google Scholar]
- Zhou, X., Z. Su, A. Anishkin, W.J. Haynes, E.M. Friske, S.H. Loukin, C. Kung, and Y. Saimi. 2007. Yeast screens show aromatic residues at the end of the sixth helix anchor transient receptor potential channel gate. Proc. Natl. Acad. Sci. USA. 104:15555–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]