Abstract
Genomic hypomethylation is a consistent finding in both human and animal tumors and mounting experimental evidence suggests a key role for epigenetic events in tumorigenesis. Furthermore, it has been suggested that early changes in DNA methylation and histone modifications may serve as sensitive predictive markers in animal testing for carcinogenic potency of environmental agents. Alterations in metabolism of methyl donors, disturbances in activity and/or expression of DNA methyltransferases, and presence of DNA single-strand breaks could contribute to the loss of cytosine methylation during carcinogenesis; however, the precise mechanisms of genomic hypomethylation induced by chemical carcinogens remain largely unknown. This study examined the mechanism of DNA hypomethylation during hepatocarcinogenesis induced by peroxisome proliferators WY-14,643 (4-chloro-6-(2,3-xylidino)-pyrimidynylthioacetic acid) and DEHP (di-(2-ethylhexyl)phthalate), agents acting through non-genotoxic mode of action. In the liver of male Fisher 344 rats exposed to WY-14,643 (0.1% (w/w), 5 months), the level of genomic hypomethylation increased by ~2-fold, as compared to age-matched controls, while in the DEHP group (1.2% (w/w), 5 months) DNA methylation did not change. Global DNA hypomethylation in livers from WY-14,643 group was accompanied by the accumulation of DNA single-strand breaks, increased cell proliferation, and diminished expression of DNA methyltransferase 1, while the metabolism of methyl donors was not affected. In contrast, none of these parameters changed significantly in rats fed DEHP. Since WY-14,643 is much more potent carcinogen than DEHP, we conclude that the extent of loss of DNA methylation may be related to the carcinogenic potential of the chemical agent, and that accumulation of DNA single-strand breaks coupled to the increase in cell proliferation and altered DNA methyltransferase expression may explain genomic hypomethylation during peroxisome proliferator-induced carcinogenesis.
Keywords: DNA hypomethylation, DNA damage, Cell proliferation, Peroxisome proliferators
1. Introduction
Recent developments in clinical and experimental cancer research show that cancer development is affected not only by changes to DNA sequence [1], but also by the alterations of the cellular epigenome [2]. Genomic hypomethylation is a consistent finding in human tumors [3], including liver cancer [4,5]. Aberrant patterns of DNA methylation and/or modification of histones have been also observed with both genotoxic and non-genotoxic environmental agents [6–8] and mounting evidence suggests that epigenetic events may be crucial in the mode of action of various carcinogenic chemicals [9]. It was also suggested that epigenetic changes should be used as sensitive predictive indicators in the studies on the mechanism and prognosis of human tumors [10,11], as well as in assessment of the carcinogenic potential of environmental chemicals since changes in DNA methylation and histone modifications usually precede the development of pre-carcinogenic lesions [12,13]. The latter has a special significance for non-genotoxic agents, considering the fact that many carcinogens presented to regulatory agencies today act via non-genotoxic mode of action [14].
Peroxisome proliferators, a structurally diverse group of chemicals and therapeutic agents, are one of the most extensively studied classes of non-genotoxic carcinogens [15]. Long-term exposure to these agents results in the development of liver tumors in male and female mice and rats [16,17]. Peroxisome proliferators are thought to cause liver cancer in rodents via a complex mode of action which involves activation of the peroxisome proliferator-activated receptor α(PPARα) which leads to changes in transcription of many metabolism genes, increase in size and amount of peroxisomes in liver parenchymal cells, increased hepatocellular proliferation, suppression of apoptosis, and secondary oxidative stress leading to DNA damage [18]. In addition, recent evidence suggests that epigenetic events, such as progressive global hypomethylation of liver DNA, decrease in trimethylation of histone H4 lysine 20 and H3 lysine 9, and a gradual loss of cytosine methylation in major and minor satellites and other repetitive elements may play a role in the mechanism of the carcinogenic action of a model peroxisome proliferator WY-14,643 [19].
Since it has been suggested that a carcinogenic potency of the chemical may be related to its effects on DNA methylation patterns in target tissue [20], better understanding of the molecular effects leading to dysregulation in maintenance of genome methylation may provide important clues into both timing and causality of carcinogenesis. Several mechanisms for the carcinogenic agent-induced genomic hypomethylation have been suggested. These include alterations in cellular one-carbon metabolism [7,11,12], changes in expression and/or activity of DNA methyltransferases [21], increased cell proliferation [7,9], and DNA damage [22]. Since little is known about how non-genotoxic rodent liver carcinogens cause changes in DNA methylation, this study was conducted to define the underlying mechanisms of DNA hypomethylation by comparing the effects of WY-14,643 and DEHP. We show that accumulation of DNA single-strand breaks coupled to the increase in cell proliferation and diminished DNA methyltransferase expression may explain genomic hypomethylation during peroxisome proliferator-induced liver carcinogenesis and that the extent of loss of DNA methylation may be related to the carcinogenic potential of the chemical agent.
2. Materials and methods
2.1. Animals and diets
The studies detailed herein were performed using archived liver tissue samples (stored at −80 °C) from previously reported animal studies [23]. Briefly, male F344 rats, 10 weeks of age, were obtained from Charles River Breeding Laboratories (Raleigh, NC). Rats were housed in polycarbonate cages in a facility with automatically controlled temperature (22 °C), humidity (50%) and light (12 h light/12 h dark cycle). Rats were randomly assigned to three dietary exposure groups beginning at 12 weeks of age: WY-14,643 (0.1% (w/w), ChemSyn Science Labs, Lenexa, KS), DEHP (1.2% (w/w), Eastman Chemical Company, Kingsport, TN), or control. All diets were mixed using NIH-07 diet (Ziegler Brothers, Gardners, PA). Diets were analyzed for chemical after mixing and used if the actual concentration was within 20% of target concentration. Following 22 weeks of ad libitum feeding, rats were killed by exsanguinations following deep isoflurane anesthesia. The livers were excised and frozen immediately in liquid nitrogen, and stored at −80 °C for subsequent analyses. All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee.
2.2. Assessment of epigenetic changes and DNA strand breaks in liver
The extent of global DNA methylation was evaluated using a radiolabeled [3H]dCTP extension assay as described elsewhere [24]. The methylation status of the promoter region of the glutathione-s-transferase placental form (Gstp) gene was determined by the methylation-sensitive PCR-based assay as previously described [25]. Specifically, genomic DNA was isolated and digested with methylation-sensitive restriction endonuclease AciI, HpaII, or BstUI followed by PCR amplification of a 168 bp fragment of the Gstp promoter region. The recovery of PCR product is varied directly with the extent of AciI-, HpaII-, or BstUI-induced DNA breaks at unmethylated CCGC, CCGG, or CGCG sites, respectively. The quantitative aspect of the procedure was verified by a linear increase in the PCR product recovery with increasing cycle number and DNA template concentration. The results were reproduced in two independent experiments with all samples.
The determination of s-adenosylmethionine (SAM) and s-adenosylhomocysteine (SAH) content in liver tissue extracts was performed by a HPLC method with coulometric electrochemical detection as previously described [26]. DNA strand breaks were detected using random oligonucleotide-primed synthesis assay as described previously [27].
2.3. Western blot analysis of protein expression and histone modifications
The liver protein levels of DNA methyltransferase 1 (DNMT1), proliferating cells nuclear antigen (PCNA), c-Myc, and β-actin were determined by Western immunoblotting. Briefly, liver tissue lysates were prepared by homogenization of 50 mg of tissue in 500 μl of lysis buffer (50 mM Tris–HCl, pH 7.4; 1% NP-40; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EDTA; 1 mM PMSF; 1 μg/ml each aprotinin, leupeptin, pepstatin; 1 mM Na3 VO4, 1 mM NaF), sonication, and incubation at 4 °C for 30 min, followed by centrifugation at 10,000 × g at 4 °C for 20 min. Protein concentration was determined by the Bradford assay (Pierce, Rockford, IL). Extracts containing equal quantities of proteins were separated by SDS-PAGE on 8 or 10% polyacrylamide gels and transferred to PVDF membranes (Millipore, Billerica, MA). Membranes were probed with primary antibodies against DNMT1 (1:500; Abcam, Cambridge, MA), PCNA (1:300; Santa Cruz Biotechnology, Santa Cruz, CA), c-Myc (1:200; Santa Cruz Biotechnology), or β-actin (1:500; Santa Cruz Biotechnology). Alkaline phosphatase-coupled donkey anti-rabbit secondary antibodies were used for visualization. Signals were quantified using ImageQuant 5.1 software (Molecular Dynamics, Sunnyvale, CA) and normalized relative to β-actin. The status of histone H3 lysine 9 trimethylation (H3K9me3) and histone H4 lysine 20 trimethylation (H4K20me3) was determined by Western blot analysis as described previously [19].
2.4. Statistical analysis
Results are presented as mean ± S.D. and were assessed by one-way ANOVA, using treatment as the fixed factor. P-values < 0.05 were considered significant.
3. Results
3.1. DNA methylation status
Rapid and progressive hypomethylation of DNA has been reported to occur in the livers of mice fed a WY-14,643-containing diet both acutely [28], and chronically for up to 5 months [19]. In this study, we aimed to determine whether similar effects can be observed in other species susceptible to hepatocarcinogenesis by WY-14,643 (i.e., rats), as well as to compare the effects of a potent, such as WY-14,643 [29], and weak, such as DEHP [29], hepatocarcinogens. Dietary feeding of F344 rats with a WY-14,643-containing diet for 5 months resulted in substantial hypomethylation of genomic DNA in the liver at CCGG sequences (Fig. 1, top panel). In contrast, administration of a DEHP-containing diet for 5 months had no appreciable effect on the methylation status of hepatic DNA.
Fig. 1.
Long-term administration of WY-14,643, but not DEHP leads to hypomethylation of genomic DNA in rat liver. DNA methylation status was assessed in livers of control (white bars), DEHP- (grey bars), or WY-14,643- (black bars) treated rats using [3H]dCTP extension assay after digestion of genomic DNA with methylation-sensitive restriction endonuclease HpaII (top panel), BssHII (middle panel), or AscI (bottom panel) as detailed in Section 2. Data is presented as mean ± S.D. (n = 5) relative to control for each group and asterisks indicate significant difference from the age-matched control rats.
Since the majority of cytosine methylation in mammalian cells resides mainly in the GC-rich regions of repetitive elements [30] and alterations in methylation of these regions have been mechanistically linked to tumor formation [7,31,32], we assessed the methylation status of the GC-rich domains following exposure to WY-14,643 and DEHP. The long-term exposure to WY-14,643, but not DEHP, resulted in the hypomethylation of the GC-rich DNA regions, evident from an increase in [3H]dCTP incorporation into BssHII- or AscI-digested DNA (Fig. 1, middle and bottom panels).
Global DNA hypomethylation, in addition to hypomethylation of repetitive sequences, is frequently associated with the hypomethylation of normally methylated CpG islands in gene promoters [33]. Indeed, the results of our previous study demonstrated that loss of global DNA methylation during rat hepatocarcinogenesis was accompanied by hypomethylation of normally methylated-specific CpG sites in the promoter region of the Gstp gene [34]. Because of this association, we assessed the status of Gstp promoter methylation in the livers of control rats and rats exposed to WY-14,643 or DEHP (Fig. 2). WY-14,643, but not DEHP, caused a substantial loss of cytosine methylation at AciI and HpaII sites normally methylated in the Gstp promoter, as evidenced by a decrease in the PCR product recovery after pre-treatment of DNA with methylation-sensitive restriction endonucleases AciI or HpaII.
Fig. 2.
Long-term administration of WY-14,643, but not DEHP, leads to hypomethylation of the Gstp gene promoter in rat liver. (A) Partial structure of the rat Gstp promoter region extending from bases +60 to +98 (accession no. L29427) with location of analyzed BstUI, AciI, and HpaII sites. We reported previously that these sites are methylated in normal liver tissue but lose their methylation in pre-neoplastic livers and in liver tumors [34]. (B) Methylation-sensitive PCR-based assay was used to assess site-specific methylation in Gstp promoter region. A decrease in PCR product recovery after pre-treatment of DNA with restriction enzymes AciI or HpaII indicates hypomethylation of the promoter, while BstUI site remains methylated. Representative agarose gel images are shown. (C) Quantitation of the restriction fragments following digestion with methylation-specific enzymes. Data is presented as mean ± S.D. (n = 5) relative to control for each group and asterisks indicate significant difference from the age-matched control rats.
3.2. Histone H3K9 and H4K20 methylation
When WY-14,643 was fed in the diet to mice, trimethylation of histone H4K20 and H3K9 was significantly decreased in liver as early as 1 week of treatment and persisted for the duration of the experiments for up to 5 months [19]. Interestingly, in this study conducted in the rat at comparable dose and duration of treatment, no changes in the extent of trimethylation of histones H3K9 and H4K20 was observed in the liver in animals exposed to either DEHP or WY-14,643 (Fig. 3).
Fig. 3.
Trimethylation of histones H3K9 and H4K20 in rat liver is not affected by long-term administration of WY-14,643 or DEHP. Histone proteins were separated by SDS-PAGE from livers of control (white bars), DEHP- (A, grey bars), or WY-14,643- (B, black bars) treated rats and subjected to immunoblotting using specific antibodies against H3Kme3 and H4K20me3 as detailed in Section 2. Representative Western immunoblot images are shown. Quantitation of histone trimethylation is presented as mean ± S.D. (n = 5) relative to control for each group.
3.3. SAM and SAH content
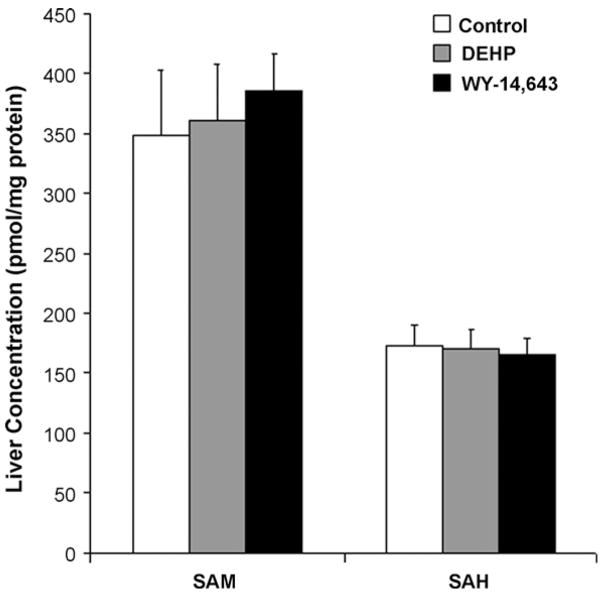
One of the main factors assuring the proper maintenance of DNA methylation is the cellular availability of methyl groups [11]. To determine whether WY-14,643-induced hypomethylation of DNA is associated with alterations in the cellular one-carbon metabolic pathways, hepatic content of SAM and SAH was measured in the livers of WY-14,643- and DEHP-treated and control rats. Fig. 4 shows that neither WY-14,643, nor DEHP had an effect on the intracellular concentrations of SAM and SAH in rat liver.
Fig. 4.
Levels of SAM and SAH in rat liver are note affected by long-term administration of WY-14,643 or DEHP. SAM and SAH were assessed in livers of control (white bars), DEHP- (A, grey bars), or WY-14,643- (B, black bars) treated rats as detailed in Section 2. Data is presented as mean ± S.D. (n = 5) for each group.
3.4. Expression of DNMT1 and PCNA proteins
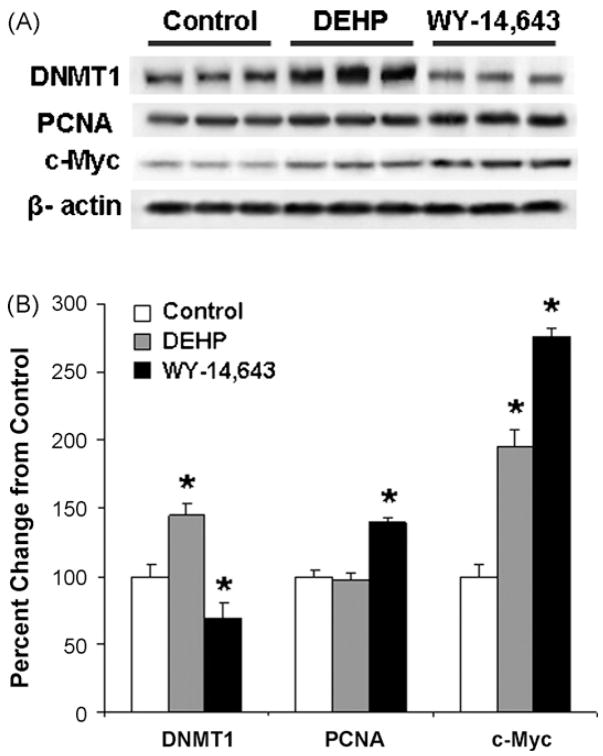
DNMT1 is the main cellular enzyme responsible for the maintenance of DNA methylation patterns in somatic mammalian cells and disruption in its activity and/or expression may lead to alterations in DNA methylation [21]. Therefore, we assessed the effect of long-term exposure to WY-14,643 and DEHP on the levels of the DNMT11 protein in rat liver. Fig. 5 shows that long-term continuous exposure of F344 rats to WY-14,643 resulted in a decrease in the protein levels of DNMT1. In contrast, exposure to DEHP led to up-regulation of DNMT1.
Fig. 5.
Western blot analysis of DNMT1, PCNA, and c-Myc proteins in the liver of control rats and rats exposed to DEHP or WY-14,643. Liver tissue lysates were prepared from livers of control (white bars), DEHP- (grey bars), or WY-14,643-(black bars) treated rats and subjected to immunoblotting using specific antibodies against DNMT1, PCNA, c-Myc and β-actin as detailed in Section 2. (A) Representative Western immunoblot images. (B) Quantitative analysis of the protein levels after normalization to β-actin. Data is presented as mean ± S.D. (n = 5) relative to control for each group and asterisks indicate significant difference from the age-matched control rats.
Exposure or Fisher 344 rat to WY-14,643 or DEHP for 22 weeks in identical doses is known to lead to about 60% increase in liver to body weight ratio [35]. Fig. 5 shows that long-term exposure to WY-14,643 resulted in an increase in liver PCNA protein expression by 40% as compared to control animals. In contrast, PCNA expression was not affected by DEHP.
The observed up-regulation of the PCNA in the livers of WY-14,643-treated rats and the results of recent findings by Dominguez-Sola et al. [36] suggesting that c-Myc has a direct role in the control of DNA replication, prompted us to investigate expression of c-Myc protein in the livers of rats exposed to WY-14,643 and DEHP. Fig. 5 shows that long-term administration of WY-14,643 resulted in the pronounced 2.7-fold up-regulation of c-Myc in the rat liver as compared to control rats. The level of c-Myc was also increased, albeit to a lesser extent, in DEHP-exposed rats.
3.5. DNA damage
In addition to the availability of methyl group donors, functioning of DNA methyltransferases, and cell proliferation, another important and frequently overlooked factor that determines the proper maintenance of the DNA methylation patterns is the integrity of genomic DNA. When integrity of DNA is compromised, particularly caused by oxidation, methylation efficiency of cytosine residues is thought to be impacted [37,38]. Indeed, oxidative stress and oxidative damage to DNA are recognized as one of the mechanisms for the carcinogenic effects of peroxisome proliferators [39]. We have previously shown in the same tissue samples as those used for this study [23] that WY-14,643, a potent carcinogen, increased expression of several base excision DNA repair enzymes, a marker of persistent oxidative damage, in a dose- and time-dependent manner. At the same time, DEHP, a much less potent carcinogen [29], had much weaker effect when compared with WY-14,643. Therefore, in this study we measured the number of 3′OH DNA ends (single-strand breaks) in liver genomic DNA, lesions that may be generated by DNA damaging agents, endonucleases, or during DNA repair, to further characterize the effects of WY-14,643 and DEHP on DNA. A 40% increase in [3H]dCTP incorporation into DNA isolated from the livers of rats fed a WY-14,643-containing diet was observed (Fig. 6) while DEHP had no effect as compared to control levels.
Fig. 6.
Effect of long-term administration of WY-14,643 and DEHP on DNA strand breaks in the liver of control rats and rats exposed to DEHP or WY-14,643. The extent of DNA strand breaks was measures by assessing the number of 3′OH DNA ends (breaks) in genomic DNA prepared from livers of control (white bars), DEHP-(grey bars), or WY-14,643- (black bars) treated rats as detailed in Section 2. Data is presented as mean ± S.D. (n = 5) relative to control for each group and the asterisk indicates significant difference from the age-matched control group.
4. Discussion
Both genotoxic and non-genotoxic environmental agents that can cause liver cancer in rodents have been shown to lead to changes in DNA methylation and histone modifications [9]. Indeed, genomic hypomethylation is regarded as one of the causal events in hepatocarcinogenesis [10,20,40] and several mechanisms, such as the availability of methyl group donors, activity and/or expression of DNA methyltransferases, increased cell proliferation, and DNA damage, have been proposed to lead to a progressive loss of DNA methylation [9]. However, even though time-dependent changes in global and region-specific DNA methylation patterns have been reported to occur as a result of treatment with classical rodent liver carcinogens [19,41,42], little is known about the causality of such epigenetic changes.
Our current work shows that long-term (22 weeks) administration of WY-14,643 to rats results in pronounced changes in DNA methylation, especially in hypomethylation of GC-rich sequences, in the liver. In contrast, exposure to DEHP had no effect on the methylation status of hepatic DNA. This data not only demonstrates that WY-14,643 causes loss of DNA methylation in two species, rat (this study) and mouse [19], which are sensitive to hepatocarcinogenesis by peroxisome proliferators, but it also suggests that the carcinogenic potency of the compounds within this class may be linked to their ability to elicit epigenetic effects. Currently, there are two identified mechanism by which loss of genomic methylation may contribute to tumorigenesis. Specifically, (a) loss of DNA methylation may compromise genomic integrity via chromatin decondensation, the induction of centromere and telomere abnormalities, chromosome segregation defects [43], and (b) DNA hypomethylation may promote tumor development by activation and transposition of endogenous retroviral elements [44,45]. This could result in a variety of genomic instability events including cis-and trans-insertional mutagenesis, unequal homologous recombination, rearrangements, and segmental duplications leading to deletions and duplications [32,44,45]. The causal role of these lesions, as integral part of neoplastic transformation in etiology of cancer, including liver cancer, is now commonly accepted [46].
The differences in carcinogenic potency among chemicals that belong to a class of peroxisome proliferators have been studied extensively. While it has been suggested that the extent of induction of peroxisomes in rodent liver may be a good predictor of hepatocarcinogenesis [47], it was also shown that the degree of peroxisome proliferation does not correlate well with tumorigenicity [29]. On the other hand, cell proliferation rates [29], and liver oxidative DNA damage [23] have been shown to correlate well both with the time, dose and carcinogenic potency of peroxisome proliferators. Thus, our data which shows profound differences in the effects on DNA methylation between WY-14,643 and DEHP suggests that epigenetic markers may also serve to examine the potency of the rodent liver non-genotoxic carcinogens. However, it should also be noted that while aberrant methylation in tumors may be a useful prognostic marker in the clinic [10,11], it may not be a sensitive early predictor of the carcinogenicity of environmental agents in animal studies. Specifically, our data shows lack of effects with DEHP, a known albeit weaker than WY-14,643 [29] rodent hepatocarcinogen, which suggests that epigenetic markers alone may be prone to false negative observations.
Importantly, this study also assessed the underlying mechanisms associated with abnormalities in DNA methylation during peroxisome proliferator-induced hepatocarcinogenesis. Several studies suggested a link between DNA hypomethylation during carcinogenesis and aberrations in the intracellular levels of SAM and/or SAH [6,11]. This is especially true for carcinogenesis associated, at least in part, with alterations in one-carbon metabolism caused by endogenous (polymorphisms in one-carbon-related genes, or deficiency in vitamin B12/folic acid), or exogenous (arsenic, etc.) factors. Our data shows that WY-14,643-induced loss of DNA methylation occurred in the absence of changes in the intracellular levels of SAM and SAH in the liver. This suggests that loss of genomic methylation during peroxisome proliferator-induced liver carcinogenesis is not due to lack of availability of methyl donors.
Another crucial factor determining the proper maintenance of DNA methylation patterns is related to the function of DNA methyltransferases [48]. We show that WY-14,643-induced global, GC-rich region-specific, and gene-specific hypomethylation of DNA were associated with a decrease in DNMT1 protein levels, especially relative to DNA replication (i.e., concurrent increase in PCNA levels in WY-14,643-treated rats). However, the extensive loss of DNA methylation detected in the present study is likely not due to a decrease in expression of DNMT1 alone. This suggestion is supported by the evidence that DNMT1-deficient mice, despite reductions in DNMT1, are able to maintain normal levels of DNA methylation [49]; however, the ability of DNMT1 to preserve proper methylation status depends on DNA replication rates [48] and may be disturbed by an elevated cell proliferation, especially under conditions when integrity of DNA is compromised through DNA damage.
Peroxisome proliferator-induced cell proliferation and oxidative damage to DNA in rodent liver are thought to be key events in the carcinogenesis mode of action. However, recent evidence indicates that peroxisome proliferation, cell proliferation and oxidative stress are not directly responsible for the development of peroxisome proliferator-induced liver cancer [50]. Our expected observations that WY-14,643 caused induction of PCNA and lead to an increase in DNA single-strand breaks suggest that DNA damage, together with increased proliferation, may have an effect on DNA methylation patterns. DNA damage plays a central role of in cytosine demethylation [51] and our findings of lower CpG methylation in DNA from WY-14,643-treated rats indicate that DNA single-strand breaks, in addition to increased cell proliferation, are essential prerequisites associated with the loss of DNA methylation during peroxisome proliferator-induced carcinogenesis.
In conclusion, this study provides further evidence that epigenetic alterations are crucial components in peroxisome proliferator-induced liver carcinogenesis. More importantly, we show that the degree of hepatic DNA methylation is related to the carcinogenic potential of the toxicant and is likely a result of accumulating DNA damage, increased cell proliferation and altered expression of DNA methyltransferase DNMT1.
Acknowledgments
This work was supported in part by the Postgraduate Research Program administered by the Oak Ridge Institute for Science and Education (V.T. and T.B.) and by the National Institutes of Health grants ES12686, ES11391, and ES11660.
Footnotes
Note: The views expressed in this paper do not necessarily represent those of the U.S. Food and Drug Administration.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 4.Rivenbark AG, Coleman WB. The use of epigenetic biomarkers for pre-clinical detection of hepatocellular carcinoma: potential for non-invasive screening of high-risk populations. Clin Cancer Res. 2007;13:2309–2312. doi: 10.1158/1078-0432.CCR-07-0086. [DOI] [PubMed] [Google Scholar]
- 5.Guerrero-Preston R, Santella RM, Blanco A, Desai M, Berdasco M, Fraga M. Global DNA hypomethylation in liver cancer cases and controls—a phase I pre-clinical biomarker development study. Epigenetics. 2007;2:223–226. doi: 10.4161/epi.2.4.5214. [DOI] [PubMed] [Google Scholar]
- 6.Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci USA. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watson RE, Curtin GM, Doolittle DJ, Goodman JI. Progressive alterations in global and GC-rich DNA methylation during tumorigenesis. Toxicol Sci. 2003;75:289–299. doi: 10.1093/toxsci/kfg190. [DOI] [PubMed] [Google Scholar]
- 8.Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pogribny IP, Rusyn I, Beland FA. Epigenetic aspects of genotoxic and non-genotoxic hepatocarcinogenesis—studies in rodents. Environ Mol Mutagen. 2008;49:9–15. doi: 10.1002/em.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvisi DF, Simile MM, Ladu S, Pellegrino R, De VM, Pinna F, Tomasi ML, Frau M, Virdis P, De Miglio MR, Muroni MR, Pascale RM, Feo F. Altered methionine metabolism and global DNA methylation in liver cancer: relationship with genomic instability and prognosis. Int J Cancer. 2007;121:2410–2420. doi: 10.1002/ijc.22940. [DOI] [PubMed] [Google Scholar]
- 12.Moggs JG, Goodman JI, Trosko JE, Roberts RA. Epigenetics and cancer: implications for drug discovery and safety assessment. Toxicol Appl Pharmacol. 2004;196:422–430. doi: 10.1016/j.taap.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Preston RJ. Epigenetic processes and cancer risk assessment. Mutat Res. 2007;616:7–10. doi: 10.1016/j.mrfmmm.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Schwetz BA. Thoughts on carcinogenesis testing—the FDA today. Drug Metab Rev. 2000;32:211–214. doi: 10.1081/dmr-100100573. [DOI] [PubMed] [Google Scholar]
- 15.Peraza MA, Burdick AD, Marin HE, Gonzalez FJ, Peters JM. The toxicology of ligands for peroxisome proliferator-activated receptors (PPAR) Toxicol Sci. 2006;90:269–295. doi: 10.1093/toxsci/kfj062. [DOI] [PubMed] [Google Scholar]
- 16.Rusyn I, Peters JM, Cunningham ML. Modes of action and species-specific effects of di-(2-ethylhexyl)phthalate in the liver. Crit Rev Toxicol. 2006;36:459–479. doi: 10.1080/10408440600779065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez FJ, Shah YM. PPARalpha: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246:2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 18.Klaunig JE, Babich MA, Baetcke KP, Cook JC, Corton JC, David RM, Deluca JG, Lai DY, McKee RH, Peters JM, Roberts RA, Fenner-Crisp PA. PPAR-alpha agonist-induced rodent tumors: modes of action and human relevance. Crit Rev Toxicol. 2003;33:655–780. doi: 10.1080/713608372. [DOI] [PubMed] [Google Scholar]
- 19.Pogribny IP, Tryndyak VP, Woods CG, Witt SE, Rusyn I. Epigenetic effects of the continuous exposure to peroxisome proliferator WY-14,643 in mouse liver are dependent upon peroxisome proliferator-activated receptor alpha. Mutat Res. 2007;625:62–71. doi: 10.1016/j.mrfmmm.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman JI, Watson RE. Altered DNA methylation—a secondary mechanism involved in carcinogenesis. Annu Rev Pharmacol Toxicol. 2002;42:501–525. doi: 10.1146/annurev.pharmtox.42.092001.141143. [DOI] [PubMed] [Google Scholar]
- 21.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a pre-cancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 22.James SJ, Pogribny IP, Pogribna M, Miller BJ, Jernigan S, Melnyk S. Mechanisms of DNA damage, DNA hypomethylation, and tumor progression in the folate/methyl-deficient rat model of hepatocarcinogenesis. J Nutr. 2003;133:3740S–3747S. doi: 10.1093/jn/133.11.3740S. [DOI] [PubMed] [Google Scholar]
- 23.Rusyn I, Denissenko MF, Wong VA, Butterworth BE, Cunningham ML, Upton PB, Thurman RG, Swenberg JA. Expression of base excision repair enzymes in rat and mouse liver is induced by peroxisome proliferators and is dependent upon carcinogenic potency. Carcinogenesis. 2000;21:2141–2145. doi: 10.1093/carcin/21.12.2141. [DOI] [PubMed] [Google Scholar]
- 24.Pogribny IP, Yi P, James SJ. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem Biophys Res Commun. 1999;262:624–628. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- 25.Pogribny IP, Pogribna M, Christman JK, James SJ. Single-site methylation within the p53 promoter region reduces gene expression in a reporter gene construct: possible in vivo relevance during tumorigenesis. Cancer Res. 2000;60:588–594. [PubMed] [Google Scholar]
- 26.Melnyk S, Pogribna M, Pogribny IP, Yi P, James SJ. Measurement of plasma and intracellular s-adenosylmethionine and s-adenosylhomocysteine utilizing coulometric electrochemical detection: alterations with plasma homocysteine and pyridoxal 5′-phosphate concentrations. Clin Chem. 2000;46:265–272. [PubMed] [Google Scholar]
- 27.Basnakian AG, James SJ. Quantification of 3′OH DNA breaks by random oligonucleotide-primed synthesis (ROPS) assay. DNA Cell Biol. 1996;15:255–262. doi: 10.1089/dna.1996.15.255. [DOI] [PubMed] [Google Scholar]
- 28.Ge R, Wang W, Kramer PM, Yang S, Tao L, Pereira MA. Wy-14,643-induced hypomethylation of the c-myc gene in mouse liver. Toxicol Sci. 2001;62:28–35. doi: 10.1093/toxsci/62.1.28. [DOI] [PubMed] [Google Scholar]
- 29.Marsman DS, Cattley RC, Conway JG, Popp JA. Relationship of hepatic peroxisome proliferation and replicative DNA synthesis to the hepatocarcinogenicity of the peroxisome proliferators di(2-ethylhexyl)phthalate and (4-chloro-6-(2,3-xylidino)-2-pyrimidinylthio) acetic acid (Wy-14,643) in rats. Cancer Res. 1988;48:6739–6744. [PubMed] [Google Scholar]
- 30.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carnell AN, Goodman JI. The long (LINEs) and the short (SINEs) of it: altered methylation as a precursor to toxicity. Toxicol Sci. 2003;75:229–235. doi: 10.1093/toxsci/kfg138. [DOI] [PubMed] [Google Scholar]
- 32.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation-induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 33.Kaneda A, Tsukamoto T, Takamura-Enya T, Watanabe N, Kaminishi M, Sugimura T, Tatematsu M, Ushijima T. Frequent hypomethylation in multiple promoter CpG islands is associated with global hypomethylation, but not with frequent promoter hypermethylation. Cancer Sci. 2004;95:58–64. doi: 10.1111/j.1349-7006.2004.tb03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinmetz KL, Pogribny IP, James SJ, Pitot HC. Hypomethylation of the rat glutathione s-transferase pi (GSTP) promoter region isolated from methyl-deficient livers and GSTP-positive liver neoplasms. Carcinogenesis. 1998;19:1487–1494. doi: 10.1093/carcin/19.8.1487. [DOI] [PubMed] [Google Scholar]
- 35.Cattley RC, Glover SE. Elevated 8-hydroxydeoxyguanosine in hepatic DNA of rats following exposure to peroxisome proliferators: relationship to carcinogenesis and nuclear localization. Carcinogenesis. 1993;14:2495–2499. doi: 10.1093/carcin/14.12.2495. [DOI] [PubMed] [Google Scholar]
- 36.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 37.Cerda S, Weitzman SA. Influence of oxygen radical injury on DNA methylation. Mutat Res. 1997;386:141–152. doi: 10.1016/s1383-5742(96)00050-6. [DOI] [PubMed] [Google Scholar]
- 38.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 39.Rusyn I, Asakura S, Pachkowski B, Bradford BU, Denissenko MF, Peters JM, Holland SM, Reddy JK, Cunningham ML, Swenberg JA. Expression of base excision DNA repair genes is a sensitive biomarker for in vivo detection of chemical-induced chronic oxidative stress: identification of the molecular source of radicals responsible for DNA damage by peroxisome proliferators. Cancer Res. 2004;64:1050–1057. doi: 10.1158/0008-5472.can-03-3027. [DOI] [PubMed] [Google Scholar]
- 40.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao L, Wang W, Li L, Kramer PK, Pereira MA. DNA hypomethylation induced by drinking water disinfection by-products in mouse and rat kidney. Toxicol Sci. 2005;87:344–352. doi: 10.1093/toxsci/kfi257. [DOI] [PubMed] [Google Scholar]
- 42.Bachman AN, Phillips JM, Goodman JI. Phenobarbital induces progressive patterns of GC-rich and gene-specific altered DNA methylation in the liver of tumor-prone B6C3F1 mice. Toxicol Sci. 2006;91:393–405. doi: 10.1093/toxsci/kfj155. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capellà G, Ribas M, Peinado MA. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–8468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 44.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 46.Coleman WB, Tsongalis GJ. Molecular mechanisms of human carcinogenesis. EXS. 2006;96:321–349. doi: 10.1007/3-7643-7378-4_14. [DOI] [PubMed] [Google Scholar]
- 47.Reddy JK, Moody DE, Azarnoff DL, Rao MS. Di-(2-ethylhexyl)phthalate: an industrial plasticizer induces hypolipidemia and enhances hepatic catalase and carnitine acetyltransferase activities in rat and mice. Life Sci. 1976;18:941–945. doi: 10.1016/0024-3205(76)90412-4. [DOI] [PubMed] [Google Scholar]
- 48.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trasler J, Deng L, Melnyk S, Pogribny I, Hiou-Tim F, Sibani S, Oakes C, Li E, James SJ, Rozen R. Impact of DNMT1 deficiency, with and without low folate diets, on tumor numbers and DNA methylation in Min mice. Carcinogenesis. 2003;24:39–45. doi: 10.1093/carcin/24.1.39. [DOI] [PubMed] [Google Scholar]
- 50.Yang Q, Ito S, Gonzalez FJ. Hepatocyte-restricted constitutive activation of PPARα induces hepatoproliferation but not hepatocarcinogenesis. Carcinogenesis. 2007;28:1171–1177. doi: 10.1093/carcin/bgm046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA. 2006;103:11112–11117. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]