Abstract
Olfaction depends on the differential activation of olfactory receptor neurons (ORNs) and on the proper transmission of their activities to the brain. ORNs select individual receptors to express, and they send axons to particular targets in the brain. Little is known about the molecular mechanisms underlying either process. We have identified a new Drosophila POU gene, pdm3, that is expressed in ORNs. Genetic analysis shows that pdm3 is required for odor response in one class of ORNs. We find that pdm3 acts in odor receptor expression in this class, and that the odor response can be rescued by the receptor. Another POU gene, acj6, is required for receptor expression in the same class, and we find a genetic interaction between the two POU genes. The results support a role for a POU gene code in receptor gene choice. pdm3 is also expressed in other ORN classes in which it is not required for receptor expression. For two of these classes, pdm3 is required for normal axon targeting. Thus, this mutational analysis, the first for a POU class VI gene, demonstrates a role for pdm3 in both of the processes that define the functional organization of ORNs in the olfactory system.
Keywords: POU gene, odor receptor, axon targeting, Drosophila, maxillary palp, antenna
Introduction
Each olfactory receptor neuron (ORN) makes two remarkable choices that underlie the sense of smell. ORNs select individual odor receptor genes to express, a process that determines which odors they detect, and they send axons to particular targets in the brain, which determines what behaviors are elicited by the odors. The molecular mechanisms underlying both choices are largely unknown.
ORNs in Drosophila are contained within two organs, the antenna and the maxillary palp. The odor response spectrum of an ORN class is conferred by the expression of one or a small number of odor receptor (Or) genes (Dobritsa et al., 2003; Hallem et al., 2004; Goldman et al., 2005). The organization of ORN classes is stereotyped, and depends on the proper selection of individual Or genes from among a large repertoire. ORNs send axons to individual glomeruli, which are spheroidal modules in the antennal lobe of the brain. ORNs that express the same receptor converge on the same glomerulus (Gao et al., 2000; Vosshall et al., 2000).
In Drosophila, the expression of particular Or genes, and hence the odor specificity of the ORN, has recently been found to depend on a regulatory code of cis-acting elements (Ray et al., 2007, 2008). Transcription factors that act in the process of receptor gene expression include abnormal chemosensory jump 6 (Acj6), a POU transcription factor. The acj6 gene was identified by a defect in olfactory behavior (McKenna et al., 1989; Ayer and Carlson, 1991; Clyne et al., 1999b). In a null mutant of acj6, some maxillary palp ORNs respond normally to odors, some are present but have lost response to all odors, and some undergo changes in odor specificity (Clyne et al., 1999b). Correspondingly, acj6 is required for the expression of a subset of Or genes (Clyne et al., 1999a).
In the present study, we identify a new POU gene, pdm3, which is a POU gene of class VI. We find that pdm3 is expressed in ORNs. In the first study of a POU VI mutant in any organism, we find that loss of pdm3 results in loss of odor response in a class of ORNs in the maxillary palp. Correspondingly, the defective ORNs lose Or gene expression. The phenotypes can be rescued with a cDNA representing either pdm3 or the affected Or gene, indicating a surprising degree of specificity to the odor-sensitivity phenotype. We demonstrate a genetic interaction between pdm3 and acj6, and ORNs can be divided into three categories, those that depend on both of these POU genes, those that depend on acj6 alone, and those that depend on neither for the acquisition of odor specificity. These results are consistent with a combinatorial code of POU genes. pdm3 is expressed in ORN classes in which it is not required for odor response, and in at least two of these classes pdm3 is required for normal axon targeting. pdm3 thus functions in both of the processes that dictate the precise organization of the Drosophila olfactory system: the expression of individual odor receptors and the targeting of individual glomeruli.
Materials and Methods
Drosophila stocks.
The pdm3 mutant was obtained from the Bloomington Stock Center (PBac {WH}pdm3f00828) (Thibault et al., 2004) and contains a 10 kb insertion of the piggyBac{WH} transposon (for location, see Fig. 1B). The insertion location was verified using inverse PCR and DNA sequencing of the surrounding genomic DNA. Homozygous adults show reduced mobility and a darkened cuticle, and have a shortened life span, with an average adult survival time of 4 d. To maximize yield of pdm3 mutant adults, culture vials were passed every 3–4 d, and a Kimwipe was inserted into vials when larvae grew to the third instar stage. pdm3 mutants were then collected immediately after eclosion and placed in fresh vials for 1–2 d at room temperature.
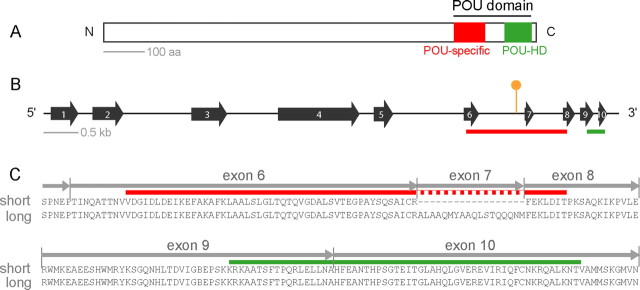
Figure 1.
The structure of pdm3. A, The POU domain, with the POU-specific domain (red) and POU homeodomain (green) indicated. B, The genomic organization of pdm3. The transposon insertion (orange symbol) is located in an intron between two exons that encode the POU-specific domain (red bar). C, Optional splicing of exon 7. The region of the POU domain indicated by a dashed red bar is excluded from the short splice form but included in the long splice form.
As a control for the genetic background, we generated flies carrying a precise excision of the piggyBac insertion. pdm3 mutant flies were crossed with flies carrying piggyBac transposase, white-eyed flies were recovered, and stocks were established. We confirmed that the excision was precise by cloning and sequencing the pdm3 gene region from genomic DNA. Control flies were treated in the same manner as mutant flies The Df stock was also obtained from the Bloomington Stock Center (Df(2R)Exel6058) (Parks et al., 2004). The acj6 mutant used in this study is the acj66 allele, shown to be a null allele by Clyne et al. (1999b).
Reverse transcription-PCR.
In an initial reverse transcription (RT)-PCR, we amplified the C-terminal half of the pdm3 transcript, which includes the POU domain. The following primers were used with cDNA from Canton-S heads as template: 5′-AATGAACCCACCATCAACCAG-3′, 5′-GCTCTAGACTATACCATTCCCTTGGACATC-3′ (Yale Keck Oligo Synthesis). The PCR products were cloned using TOPO TA Cloning (Invitrogen) and sequenced.
Electrophysiology.
Extracellular single-unit recordings were performed as described by de Bruyne et al. (2001). Odorants were diluted 1:100 in paraffin oil unless otherwise indicated, and 50 μl were pipetted onto a filter disc within the neck of a Pasteur pipette. A P-1000 blue micropipette tip was placed over the larger end of the pipette. The small tip of the Pasteur pipette cartridge containing the odorant was placed into a hole in a glass tube through which a purified air stream (32 ml/s) passed continuously in the direction of the fly, and the odor stimulus was delivered via a 0.5 s pulse of charcoal-filtered air (3.2 ml/s) through the pipette cartridge. All odor cartridges were used a maximum of three times. Responses were quantified by counting the number of spikes within the 0.5 s odor stimulus interval, and subtracting the number of spikes in a 0.5 s interval before the stimulus.
Pdm3 antibody production.
A recombinant GST-Pdm3 antigen was produced, which included Pdm3 amino acids 411–766 fused to the C terminus of the GST polypeptide. The following primers were used to amplify this region from pdm3 cDNA (catalog #EDM1133-6872894, clone #AT16994; Open Biosystems) as template: 5′-GGGAGAGGGAAAGGGAGAGAGAAC-3′, 5′-GCTCTAGACTACTGCATGTGGGAAACACTCGATG-3′ (Invitrogen; XbaI site indicated by boldface type). This region was chosen because it does not contain common protein motifs such as the POU domain. The product was cloned into pGEX6P-1 (GE Healthcare) and transformed into BL21 cells, which were grown at 37°C until A600 reached 0.8. The fusion protein was purified from the sonicate of this culture using glutathione Sepharose 4B beads (GE Healthcare) according to the manufacturer's instructions. It was stored at 4°C and sent to Cocalico Biologicals, where it was used to inoculate rats. The polyclonal antibody was used at a 1:100 dilution.
RNA in situ hybridization and immunohistochemistry.
RNA in situ hybridizations were performed as described previously (Goldman et al., 2005). To synthesize the pdm3 RNA probe, a pdm3 cDNA clone in the vector pOTB7 (Open Biosystems) was used as template to amplify the gene, and all subsequent steps were performed according to the manufacturer's instructions (Roche).
Adult labella were dissected and immunostained as follows, modified from Laissue (1999) and Thorne (2004). Thirty to forty heads were dissected, placed into 500 μl of fix solution (4% paraformaldehyde in 1× PBS + 0.2% Triton X-100), and fixed for 3 h on wet ice. The heads were rinsed twice in wash solution (1× PBS + 0.2% Triton X-100), then washed for 1 h with four changes of wash solution on a rotator. The fixed heads were either stored at 4°C for up to 2 weeks or used immediately in immunostaining experiments as follows. Wash solution was replaced with blocking solution (1% BSA or 3% NGS in 1× PBS + 0.2% Triton X-100), and labella were dissected in blocking solution, then blocked for 1 h at room temperature on a rotator. Primary antibody diluted in blocking solution [anti-Pdm3 at 1:100, anti-green fluorescent protein (GFP) (Invitrogen rabbit anti-GFP, A6455) at 1:50, and anti-Elav (Hybridoma Bank mouse anti-Elav) at 1:10] was applied to the sample (500 μl per sample) and incubated overnight at 4°C on a rotator. The primary antibody mix was then saved for two more stainings, and the sample was washed three times for ≥30 min at room temperature. Secondary antibody diluted in blocking solution (1:250) was used to stain samples for 4 h at room temperature, in the dark and on a rotator. The secondary mix was discarded, and the sample was again washed in three 30 min washes at room temperature. Most of the wash solution was removed, and one to two drops of Vectashield Mounting Medium (Vectashield H-1000) were applied. Mounted slides were either stored at 4°C for a maximum of 2 d or immediately viewed and photographed using a Bio-Rad MRC 1024 confocal microscope. Adult brains were dissected and immunostained as previously described by Wu and Luo (2006). Slides were viewed on a Bio-Rad MRC 1024 confocal microscope.
In RNA hybridization/immunostaining double stainings, first the RNA in situ hybridization experiments were performed as described by Goldman et al. (2005), followed by antibody staining as described above.
Transgenes.
To generate UAS-pdm3 cDNA constructs, the commercially available pdm3 cDNA (Open Biosystems) was used as template to amplify the long pdm3 cDNA splice form, with the following primers, which include restriction enzyme sites: 5′-GTACAGATCTATGGAGATCACGGACGGC-3′ (BglII site indicated by boldface type), 5′-ATGCGCGGCCGCCTATACCATTCCCTTGGACATC-3′ (NotI site indicated by boldface type). The PCR product was cloned into pGEM-T Easy (Promega). The short form was made by using two BsmI (New England Biolabs) sites that flank the alternatively spliced region to swap the long splice form DNA out of the construct and replace it with short splice form DNA, which had been cloned previously into the pCRII TOPO vector (Invitrogen) in initial RT-PCR analysis. Clones were sequenced, and those clones with the correct sequence were swapped into pUAST (Invitrogen) using the restriction enzymes BglII and NotI (New England Biolabs).
For all experiments using GFP, a UAS-mCD8-GFP construct was used (Lee and Luo, 1999), and the GFP was detected with the anti-GFP antibody.
Results
Identification of a new Drosophila POU gene, pdm3
We identified a new POU gene by searching the Drosophila genome sequence with POU domain sequences from all six POU gene classes [for review of POU classes, see Ryan and Rosenfeld (1997)]. The new gene belongs to class VI, and we named it pdm3 (POU domain motif 3) by analogy to the previously named pdm1 and pdm2 genes. This is the first POU class VI gene in Drosophila, and we found no other members of this class in the fly genome.
The POU domain of pdm3 consists of a POU-specific domain, a POU-homeodomain, and a linker region that separates the two (Fig. 1A). It is located in the C-terminal part of the gene, as in other POU genes of class VI. pdm3 spans ∼8 kb and contains 10 exons (Fig. 1B). The predicted POU domain is the most highly conserved region of the gene, showing 70–90% identity with other class VI POU proteins and 40–50% identity with POU proteins of other classes (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
In initial RT-PCR experiments, we amplified pdm3 from adult head RNA. Two splice forms were amplified, henceforth called the “long” and “short” forms. They differ in that the long form includes exon 7, which is missing from the short form (Fig. 1B,C). This exon encodes 18 aa that interrupt the POU domain. An optional exon has likewise been found at the same location in Rpf-1, a human ortholog of pdm3 (Zhou et al., 1996), and other class VI POU genes (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
pdm3 is expressed in ORNs
The fruit fly detects odors with two types of olfactory organ, the antenna and the maxillary palp (Fig. 2A). In situ hybridization experiments showed that pdm3 is expressed in a subset of cells in the antenna (Fig. 2B). Expression was also detected in the maxillary palp, where we performed a double-label experiment to determine whether expression was neuronal. We found that cells that express pdm3 RNA are labeled by an antibody against Elav, which labels the nuclei of differentiated neurons (Fig. 2C).
Figure 2.
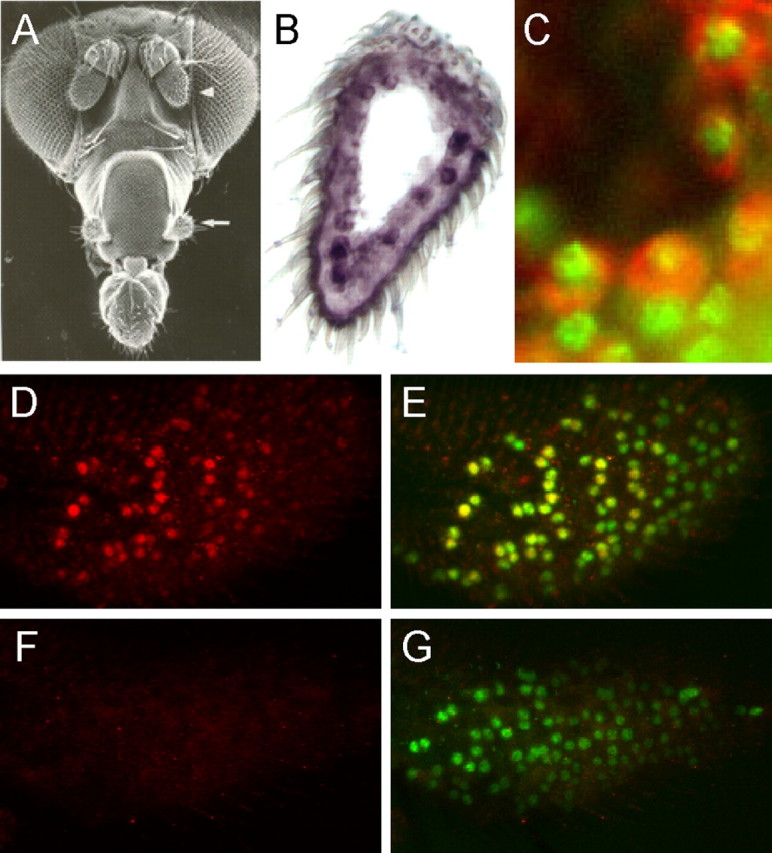
pdm3 is expressed in neurons of the olfactory system. A, Drosophila olfactory organs. The third segment of the antenna is indicated by an arrowhead, and the maxillary palp is indicated by an arrow [reprinted with permission from Carlson (1996), his Fig. 1]. B, pdm3 in situ hybridization to the antenna shows expression in a subset of cells, in blue. C, pdm3-expressing cells of the maxillary palp, stained by an RNA probe in red, are also stained in the nucleus by anti-Elav (green), a marker for mature Drosophila neurons. D, E, A control maxillary palp stained with anti-Pdm3 in red (D, E) and anti-Elav in green (E). F, G, A pdm3 maxillary palp stained with anti-Pdm3 in red (F, G) and anti-Elav in green (G). The nuclear anti-Pdm3 staining visible in neurons of the control maxillary palp is lost in the mutant. The control used here and in the remainder of this study is a precise excision of the transposon insertion.
To characterize the expression of pdm3 in more detail, we then generated an antibody, using as antigen a portion of the protein that did not contain POU domain sequences. The antibody labeled cells in the maxillary palp (Fig. 2D). All or almost all of these Pdm3+ cells are Elav+ (Fig. 2E). Many, but not all, of the Elav+ cells are Pdm3+. In the Pdm3+ Elav+ cells, the labeling appeared coincident. Together, our results show that pdm3 is expressed in the nuclei of a subset of maxillary palp neurons.
We obtained a mutant line that contains a transposon insertion in pdm3 (Thibault et al., 2004). The insertion lies within an intron between two exons that encode the POU domain (Fig. 1B). This insertion eliminated Pdm3 expression, indicating specificity of the antibody (Fig. 2F). The insertion did not markedly reduce the number of Elav+ cells (Fig. 2G), suggesting that the loss of pdm3 does not cause a loss of neurons.
A defect in odor response in the pdm3 maxillary palp
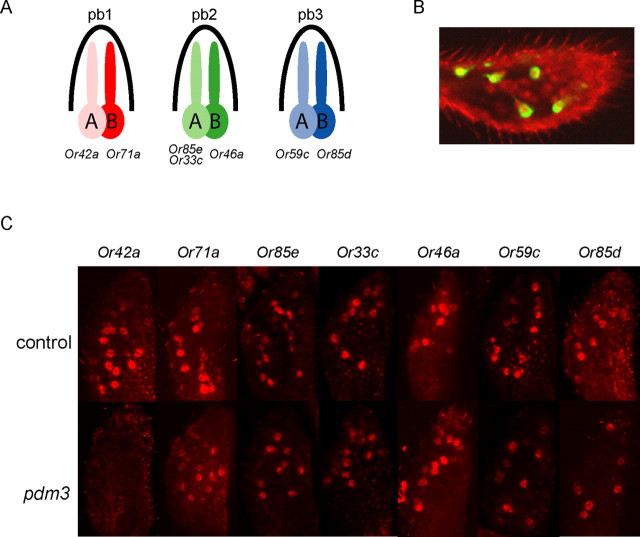
To determine whether pdm3 is required for the function of ORNs, we measured ORN responses to odors using single-unit electrophysiology. The antenna and maxillary palp contain a number of functional types of sensilla, as defined by electrophysiological and molecular analysis (de Bruyne et al., 1999, 2001; Couto et al., 2005; Goldman et al., 2005). Each sensillum type contains up to four ORNs, usually two, which are combined in a stereotyped configuration. The maxillary palp is the simpler organ, in that it contains only three functional types of sensilla, pb1, pb2, and pb3, each of which contains a pair of ORNs: pb1A and pb1B; pb2A and pb2B; pb3A and pb3B. Odorants pass through pores in the sensillum walls, traverse the internal lymph, and activate the ORNs. Seven Or genes are expressed in the maxillary palp: of the six ORN classes, each expresses one odor receptor, except that one class, pb2A, expresses two.
We found that of the six ORN classes of the maxillary palp, one is severely defective in pdm3. The pb1A ORN, which in wild type responds to a pulse of E2-hexenal with a train of action potentials, does not respond in pdm3 (Fig. 3A). pb1A is present and yields spontaneous action potentials, but does not respond to the odorant.
Figure 3.
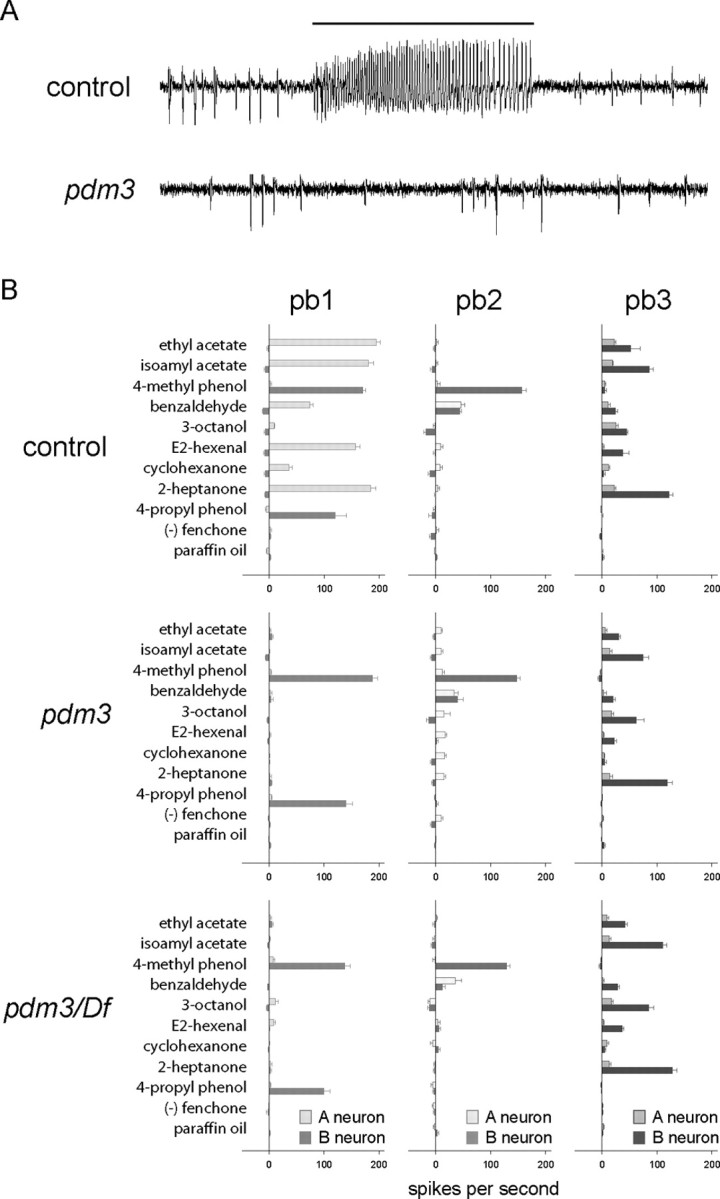
An olfactory defect in the pdm3 maxillary palp. A, Electrophysiological responses from control and mutant pb1 sensilla. Spikes, or action potentials, of large amplitude are produced by the A neuron; spikes of small amplitude are produced by the B neuron. The control pb1A neuron responds to a 0.5 s stimulus of E2-hexenal, indicated by the bar. Although both neurons are present in the mutant pb1 sensillum, neither responds to the odor stimulus. B, Responses of neurons in spikes per second (n ≥ 11; error bars represent SEM). pb1 sensilla were identified by the response of the neighboring ORN in the sensillum to 4-propyl phenol.
Systematic measurement of neuronal responses, measured in action potentials/s, after stimulation with a diverse panel of odorants, revealed that pb1A failed to respond to any tested odorant. In contrast, in the same sensillum, the neighboring pb1B ORN responded normally to 4-methyl phenol and 4-propyl phenol, its most effective odorants. Neurons of the pb2 and pb3 sensilla gave very similar responses in pdm3 and control animals (Fig. 3B).
As a more stringent test of the role of pdm3 in ORN response, we tested the pdm3 mutation in heterozygous combination with a deletion for the region. We found that in this pdm3/Df heterozygote, all responses were very similar to those of the pdm3 homozygote, indicating that the pdm3 insertion is a null allele (Fig. 3B).
The olfactory phenotype is not limited to the maxillary palp. We examined the large basiconic sensilla of the antenna and found that one of the four ORNs in the ab1 sensillum, ab1A, is present but has lost odor response (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Limited analysis of the ORNs in ab2 and ab3 sensilla revealed no abnormalities. The detection of an olfactory phenotype in some antennal ORNs is consistent with our detection of pdm3 expression in a subset of antennal cells (Fig. 2B).
A defect in odor receptor gene expression
Because pb1A is present in pdm3 and yields spontaneous action potentials, but does not respond to odors, we wished to test the hypothesis that pdm3 affects expression of its odor receptor. Or42a is the odor receptor that is expressed in pb1A (Goldman et al., 2005); a receptor-to-neuron map of the maxillary palp is shown in Figure 4A.
Figure 4.
Gene expression in the pdm3 maxillary palp. A, A receptor-to-neuron map of the maxillary palp. B, pdm3 is expressed in the pb1A neuron class. pb1A neurons were labeled in green using Or42a-GAL4; UAS-mCD8GFP, and pdm3-expressing cells were labeled in red with the Pdm3 antibody. All labeled pb1A neurons are labeled with the Pdm3 antibody. C, RNA in situ hybridizations for each of the maxillary palp Or genes in control and mutant.
As a first test of this hypothesis, we asked whether Pdm3 is expressed in pb1A in wild type. A double-label experiment showed that Pdm3 is in fact expressed in Or42a-expressing cells of wild type (Fig. 4B). We then found that Or42a expression is undetectable in pdm3 (Fig. 4C). In contrast, the neighboring pb1B cell shows normal expression of its receptor gene, Or71a, and all other maxillary palp odor receptor genes are expressed in pdm3 (Fig. 4C). Thus these expression studies coincide precisely with the functional studies: the one class of ORN that has lost function is the one class of ORN that has lost expression of a receptor gene.
Rescue of olfactory response by Or42a and pdm3
To determine whether the loss of pb1A odor response is attributable solely to loss of Or42a expression or whether other essential components are lost as well, we drove expression of Or42a in pb1A neurons of pdm3 using the GAL4-UAS system. We tested response to the two odorants that elicit the strongest response from pb1A and found that responses to both were completely or largely restored when Or42a was driven by Elav-GAL4 (Fig. 5A,B). These results suggest a surprising degree of specificity to the pdm3 phenotype at the molecular level.
Figure 5.
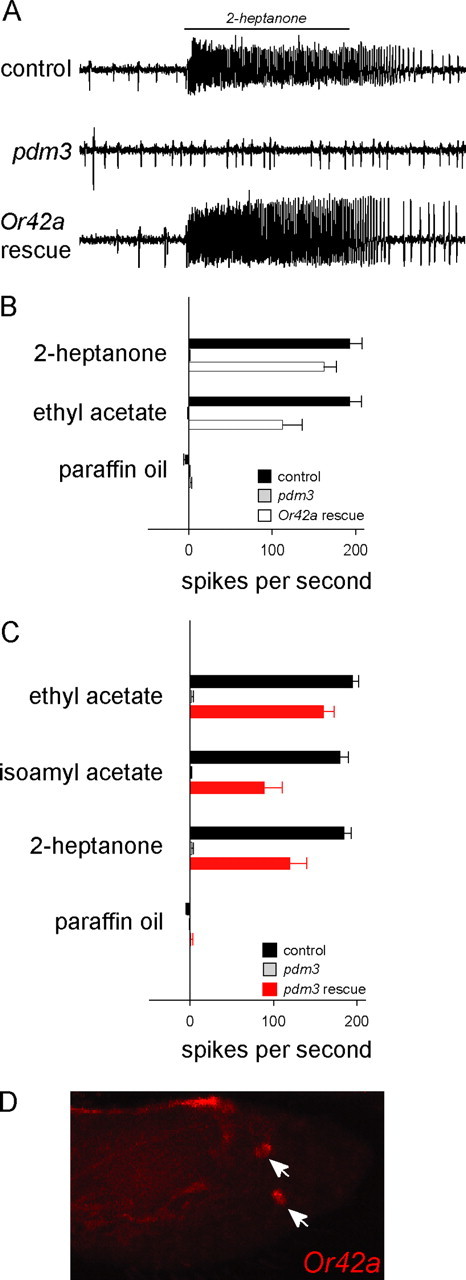
Rescue of pb1A odor response. A, The response to a 0.5 s pulse of 2-heptanone is restored by expression of Or42a in the pdm3 mutant. Or42a was expressed in the pdm3 background using C155(elav)-GAL4; UAS-Or42a. B, Rescue of pb1A responses (white) to the two odors that activate pb1A the most strongly. The large-amplitude spikes of the A neuron were counted. n = 6; error bars represent SEM. C, Rescue with a pdm3 cDNA, using acj6-GAL4 and UAS-pdm3 cDNA (long isoform) in the mutant background. n = 6. D, In situ hybridization shows expression (arrows) of Or42a in maxillary palps of acj6-GAL4/w; pdm3, UAS-pdm3.
We next investigated the ability of pdm3 splice forms to rescue the phenotype. Of the two splice forms, we amplified only the long form from adult maxillary palps in RT-PCR experiments (data not shown). When we drove expression of a long form cDNA using the GAL4-UAS system, we were able to rescue the phenotype of pb1A completely or largely in 7 of 29 pb1 sensilla, tested with three odorants (Fig. 5C). Expression of Or42a was also restored in a number of cells (Fig. 5D). In this experiment, we used an acj6-GAL4 driver (Bourbon et al., 2002), which drives expression in all or almost all maxillary palp ORNs. We were unable to construct a faithful driver for pdm3, whose 5′ end is separated from the next annotated upstream gene by a long region; it is possible that a pdm3 driver might restore the pb1A phenotype in a greater fraction of pb1 sensilla. We also drove expression of a short form cDNA and found rescue in 1 of 19 pb1 sensilla tested (data not shown).
We note that in addition to the rescued pb1A cells, we also found a number of pb1A cells that acquired an odor response profile different from that of wild type. Further testing revealed that this odor response profile is very similar to that conferred by a larval odor receptor, Or85c (supplemental Fig. S3A, available at www.jneurosci.org as supplemental material) (Kreher et al., 2005). Moreover, Or85c expression is detected in several cells of the “rescued” maxillary palp (supplemental Fig. S3B, available at www.jneurosci.org as supplemental material). The proportion of cells with this response profile is greater when pdm3 is driven with a promoter that is expected to drive later expression in ORNs (supplemental Fig. S3C, available at www.jneurosci.org as supplemental material). Further testing beyond the scope of this study would be required to determine whether these results reflect the presence of different pdm3 interaction partners at different times at the promoters of Or42a and Or85c.
A genetic interaction between pdm3 and acj6 in the pb1A neuron
Because pb1A is affected by both pdm3 and another POU gene, acj6 (Clyne et al., 1999b), we investigated the relationship between them. Although the concept of a combinatorial code of transcription factors has been previously invoked in discussion of receptor gene choice in Drosophila (Ray et al., 2007, 2008), the interaction of two transcription factors within the same ORN has not yet been examined in Drosophila.
We found first that neither POU gene appears to regulate the other in the maxillary palp: pdm3 is expressed in a null mutant of acj6, and acj6 is expressed in a null mutant of pdm3 (Fig. 6B,C). We then found that although both pdm3 and acj6 are fully recessive, the transheterozygote shows a reduced response of pb1A (Fig. 6D). The response of pb1B is normal in this genotype. These results indicate a genetic interaction between the two POU genes in pb1A.
Figure 6.
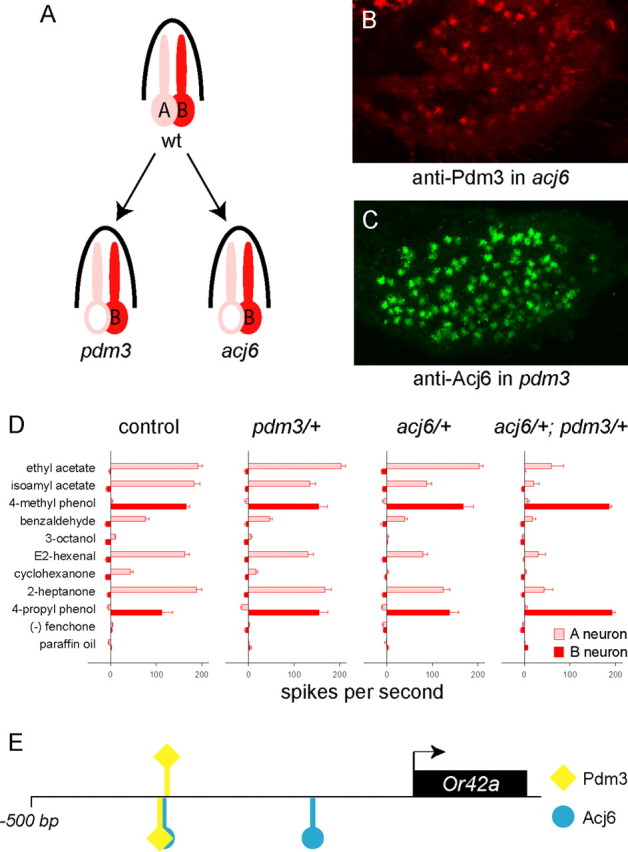
A genetic interaction between pdm3 and acj6 in pb1A. A, The pb1A neuron is affected by both pdm3 and acj6. B, Anti-Pdm3 immunostaining of acj66 maxillary palps. C, Anti-Acj6 immunostaining of pdm3 maxillary palps. D, pb1A responses are reduced in the transheterozygote. pb1A responses of each single heterozygote appear comparable with control responses. n ≥ 7; error bars = SEM. E, Putative Pdm3 and Acj6 binding sites upstream of Or42a. One putative Pdm3 binding site overlaps with an Acj6 site, and another lies immediately adjacent.
Interestingly, two putative Pdm3 binding sites, i.e., sequences corresponding to binding sites for rat and zebrafish orthologs [(Brn-5) and (Pou[c]), respectively], lie near an Acj6 site (L. Bai and J. R. Carlson, unpublished observations), ∼320 bp upstream of Or42a. One putative Pdm3 site overlaps with the Acj6 site, and the other is immediately adjacent to it (Fig. 6E). POU genes have been shown to heterodimerize on DNA (Voss et al., 1991), and this cluster of POU sites may bring Pdm3 and Acj6 in a position to do so upstream of a gene whose expression depends on both.
Pdm3 is expressed in four of six ORN classes in the maxillary palp
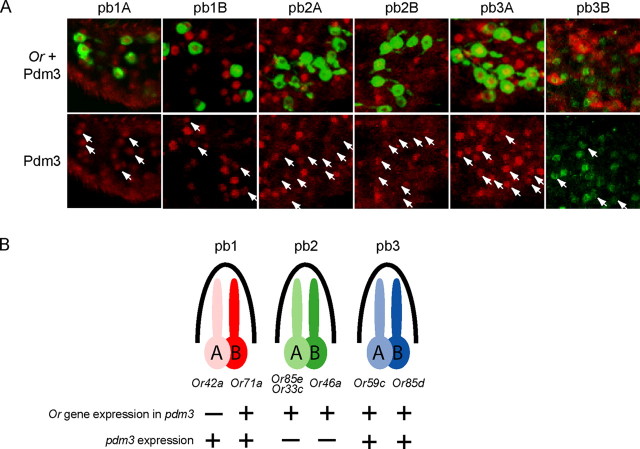
From our initial analysis of pdm3 expression in the maxillary palp (Fig. 2D), it is clear that the number of cells expressing pdm3 is greater than the number of pb1A cells. To determine in which ORN classes pdm3 is expressed, we performed a series of double-label experiments using ORN class-specific probes and an anti-Pdm3 antibody. We found that Pdm3 is expressed in four of the six maxillary palp ORN classes: pb1A, pb1B, pb3A, and pb3B (Fig. 7A,B). Expression was not detected in pb2A or pb2B. The expression of pdm3 in three ORN classes that do not require its expression for odor response suggested the possibility that pdm3 might have a different function in these cells.
Figure 7.
pdm3 is expressed in four maxillary palp ORN classes. A, The pb1A, pb1B, pb2A, pb2B, and pb3A neuron classes were labeled in green using the GAL4 system to drive GFP, and in these cases pdm3-expressing cells were labeled in red using the Pdm3 antibody. The pb3B neuron class was labeled in red using an Or85d in situ hybridization probe, and in this case pdm3-expressing cells were labeled in green with the Pdm3 antibody. Arrows in the bottom panels indicate the location of ORNs that are labeled in the top panels. Anti-Pdm3 staining overlaps with that of pb1A, pb1B, pb3A, and pb3B, but not with that of pb2A and pb2B. B, Summary. One Or gene is affected in pdm3, and pdm3 is expressed in four maxillary palp ORN classes.
pdm3 is required for axon targeting of two ORN classes
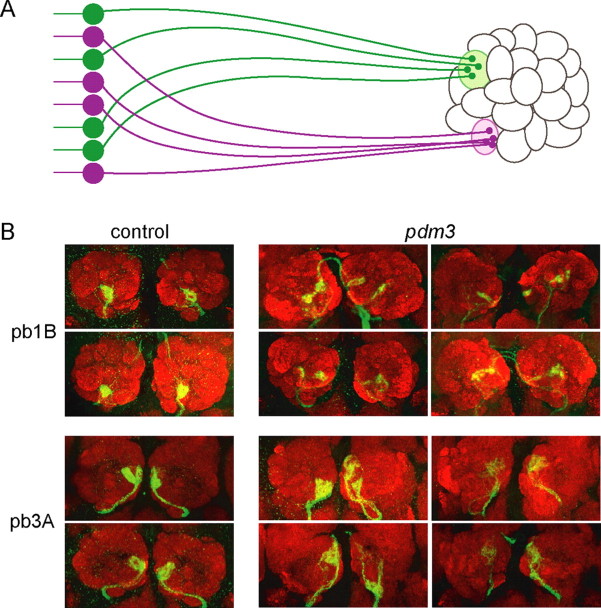
ORNs send axons to the antennal lobe of the brain, and axons of an individual ORN class converge precisely on an individual unit of the antennal lobe called a glomerulus (Fig. 8A) (Gao et al., 2000; Vosshall et al., 2000; Couto et al., 2005; Fishilevich and Vosshall, 2005). Thus, a spatial map of olfactory information is created in the antennal lobe by the stereotyped pattern of connectivity. There are 43 glomeruli in the antennal lobe, and both receptor-to-ORN and ORN-to-glomerulus maps have been constructed (Hallem et al., 2004; Couto et al., 2005; Fishilevich and Vosshall, 2005; Goldman et al., 2005). A number of genes required for normal ORN axon targeting have been identified (Komiyama and Luo, 2006).
Figure 8.
pdm3 has effects on ORN axon targeting. A, ORN axons of a given class, represented in a common color (green or purple), project to the same glomerulus of the antennal lobe. B, Targeting of the pb1B (green, Or71a-GAL4; UAS-GFP) and pb3A (green, Or59c-GAL4; UAS-GFP) ORN classes are affected by pdm3. Neuropil is stained in red with anti-nc82.
We examined the axon targeting of two ORN classes that express pdm3 but that do not require pdm3 for odor response: pb1B, which expresses the Or71a receptor, and pb3A, which expresses Or59c. We found that in a pdm3 mutant, the axonal projections of both ORN classes are abnormal (Fig. 8B). In the wild type, in both cases the projections converge on a single, discrete glomerulus in each antennal lobe. In pdm3, the convergence is less precise and the boundaries of the targeted areas are less discrete. In some cases, the axon tracts appear to bifurcate, with labeled axons occupying more than one region. In most cases, the axons appear to terminate in the general vicinity of the wild-type target, but not in the precise positions of the wild-type glomerular targets. In summary, pdm3 has effects on ORN axon targeting, in addition to its role in specifying odor response and receptor gene expression.
Discussion
The development of the olfactory system depends on two intriguing processes. ORNs must select which odor receptors to express and which glomeruli to target in the brain. Little is known about the molecular mechanisms underlying either process.
We have identified a new Drosophila POU gene, pdm3, that acts in both receptor gene expression and axonal targeting. Pdm3 is a POU transcription factor of class VI. Vertebrate members of class VI are expressed in brain and spinal cord (Andersen et al., 1993; Johansen et al., 1993; Okamoto et al., 1993; Wey et al., 1994; Zhou et al., 1996). Notably, a mouse ortholog of pdm3, Emb, is expressed in the olfactory bulb (Wey et al., 1994).
The mechanism of ORN fate determination has been elegantly examined in Caenorhabditis elegans. The AWA, AWB, and AWC cells express multiple olfactory receptors and sense distinct but overlapping sets of odors (Bargmann et al., 1993). The specification of AWA fate requires lin-11, a LIM homeodomain gene (Sarafi-Reinach et al., 2001). lin-11 regulates ODR-7, a nuclear hormone receptor that promotes expression of AWA-specific genes and represses AWC-specific genes (Sengupta et al., 1994). The Aristaless-related homeodomain gene alr-1 is also required for specification of AWA (Melkman and Sengupta, 2005). ceh-37, an Otx homeodomain gene, regulates lim-4, another LIM homeobox gene, to promote AWB neuron fate (Sagasti et al., 1999; Lanjuin et al., 2003). Another Otx gene, ceh-36, is required for AWC neuron fate (Lanjuin et al., 2003).
The first olfactory phenotype we detected for pdm3 was its defective odor response in an electrophysiological assay. The phenotype is striking in its specificity, at both the cellular and molecular levels. At the cellular level, only one ORN class in the maxillary palp, pb1A, was defective in a null mutant of pdm3. The pb1B neuron, which neighbors pb1A in the same sensillum, and the other four ORN classes all appeared to yield normal odor responses. At the molecular level, the pb1A physiological phenotype appeared to be attributable largely if not completely to the loss of a single gene: Or42a, the odor receptor gene normally expressed in this neuron. Moreover, Or42a is the only maxillary palp Or gene affected by pdm3.
Although we were surprised by the specificity of the molecular and cellular phenotypes, the specificity of one phenotype is in good agreement with the specificity of the other. The cascade of molecular steps between the presentation of an odor molecule and the production of an action potential is poorly understood but presumably requires the agency of a number of different genes. It seems likely, however, that the signaling pathway used by one maxillary palp ORN class is the same as that used by the others, with the exception of the odor receptor. Thus the ability to rescue the pb1A odor response with its receptor gene alone supports a model in which the rest of the signaling pathway does not depend on pdm3. In this case, pdm3 would affect odor response only in those maxillary palp ORNs whose receptor genes it affects, i.e., only in pb1A.
The specificity of pdm3 is in contrast to that of acj6, which affects the odor response of four ORN classes in the maxillary palp (Clyne et al., 1999b). Moreover, in acj6 some of the unresponsive ORNs were not detected electrophysiologically, either because of a defect that made them physiologically silent or because they were absent. Thus acj6 appears to affect more ORNs and to affect some of them more severely than pdm3. We note the formal possibility that pdm3 acts in some ORNs other than pb1A but is functionally redundant in them.
The requirement of both pdm3 and acj6 in pb1A allowed us to investigate whether they act together or independently. We found a genetic interaction between these two POU genes in our analysis of pb1A odor response. The location of overlapping putative binding sites for both transcription factors upstream of Or42a suggests the possibility of a biochemical interaction between them. Heterodimerization, one possible means of interaction, has been shown previously for other POU proteins (Voss et al., 1991). The genetic interaction and coexpression of acj6 and pdm3 in ORNs is in contrast to the relationship between acj6 and another POU gene, drifter, in projection neurons (PNs), the postsynaptic partners of ORNs. Although both acj6 and drifter act in PNs, they are expressed in mutually exclusive subsets of PNs.
A major problem in olfactory system biology is how individual ORNs select which of a large family of odor receptor genes to express. There are 60 Or genes in the genome of Drosophila melanogaster (Clyne et al., 1999a; Vosshall et al., 1999; Robertson et al., 2003). The expression of particular Or genes, and hence the odor specificity of the ORN, has recently been found to depend on a regulatory code of cis-acting elements. Positive and negative regulatory elements named Dyad-1 and Oligo-1 are required for the selection of Or genes in the correct olfactory organ (Ray et al., 2007). Within the maxillary palp, additional elements act positively to promote expression of individual Or genes in a subset of ORN classes, whereas other elements act negatively to restrict expression of individual Or genes to a single ORN class (Ray et al., 2007, 2008). We also found evidence that a combinatorial code of transcription factors underlies the problem of receptor gene choice (Ray et al., 2007). Transcription factors that act in this process include Lozenge, a Runx domain-containing protein that is required for the expression of two Or genes (Ray et al., 2007), and Scalloped, which mediates repression (Ray et al., 2008). Another subset of maxillary palp Or genes depends on the POU gene acj6.
Our current finding that two POU genes, acj6 and pdm3, are required in the same ORN for receptor gene expression suggests that the combinatorial action of POU genes may be an important part of such a code. The Or genes of the maxillary palp can be divided into three classes: those that require both POU genes (Or42a), those that require only acj6 (Or85e, Or33c, Or46a, and Or59c), and those that require neither (Or71a and Or85d). Heterodimer formation and alternative splicing could expand the number of components that act in selecting individual receptors from the entire family of 60 Or genes in the entire olfactory system, including the ORNs of the antenna.
We found that pdm3 is also required for axon targeting of pb1B and pb3A cells. Interestingly, acj6 is also required for axon targeting of these ORNs (Komiyama et al., 2004), but we do not know whether the two POU proteins act together on common transcriptional target genes or independently on different genes required for axonal wiring.
The relationship between receptor expression and ORN axon targeting has been a topic of great interest. In vertebrates, there is evidence that the odor receptor acts in both processes (Mombaerts et al., 1996; Wang et al., 1998; Feinstein and Mombaerts, 2004). In Drosophila, the receptor does not appear to be required for normal axon targeting (Dobritsa et al., 2003; Wang et al., 2003). Likewise, the effects of pdm3 on receptor expression and axon targeting appear to be separable, in that pb1B and pb3A show apparently normal receptor gene expression but abnormal targeting. A related issue is the relationship between ORN activity and axon targeting. In vertebrates, there is evidence that ORN activity is necessary for the establishment or maintenance of correct ORN axon targeting (Zhao and Reed, 2001; Imai et al., 2006; Serizawa et al., 2006). We were unable to ask whether in pdm3 the lack of odor response in pb1A correlates with a failure in axon targeting, for lack of a pb1A GAL4-driver that functions normally in the absence of pdm3. However, we found that the normal odor responses of pb1B and pb3A ORNs are not sufficient for normal axon targeting.
This is the first mutational analysis of a class VI POU gene and demonstrates the essential role that pdm3 plays in the development of a highly complex and precisely organized sensory system. Further study of pdm3 may uncover critical roles in other systems as well.
Footnotes
This work was supported by a National Institutes of Health (NIH) Training Grant 2T32GM007499-31, by a National Research Service Award to A.L.T., and by NIH grants to J.R.C. We thank Lei Bai and Tong-Wey Koh for critical reading of this manuscript.
References
- Andersen B, Schonemann MD, Pearse RV, 2nd, Jenne K, Sugarman J, Rosenfeld MG. Brn-5 is a divergent POU domain factor highly expressed in layer IV of the neocortex. J Biol Chem. 1993;268:23390–23398. [PubMed] [Google Scholar]
- Ayer RK, Jr, Carlson J. acj6: a gene affecting olfactory physiology and behavior in Drosophila. Proc Natl Acad Sci U S A. 1991;88:5467–5471. doi: 10.1073/pnas.88.12.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Bourbon H-M, Gonzy-Treboul G, Peronnet F, Alin M-F, Ardourel C, Benassayag C, Cribbs D, Deutsch J, Ferrer P, Haenlin M, Lepesant J-A, Noselli S, Vincent A. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- Carlson JR. Olfaction in Drosophila: from odor to behavior. Trends Genet. 1996;12:175–180. doi: 10.1016/0168-9525(96)10015-9. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999a;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Certel SJ, de Bruyne M, Zaslavsky L, Johnson WA, Carlson JR. The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU-domain transcription factor. Neuron. 1999b;22:339–347. doi: 10.1016/s0896-6273(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Clyne PJ, Carlson JR. Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yuan B, Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat Neurosci. 2000;3:780–785. doi: 10.1038/77680. [DOI] [PubMed] [Google Scholar]
- Goldman AL, Van der Goes van Naters W, Lessing D, Warr CG, Carlson JR. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45:661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Johansen T, Moens U, Holm T, Fjose A, Krauss S. Zebrafish pou[c]; a divergent POU family gene ubiquitously expressed during embryogenesis. Nucleic Acids Res. 1993;21:475–483. doi: 10.1093/nar/21.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Luo L. Development of wiring specificity in the olfactory system. Curr Opin Neurobiol. 2006;16:67–73. doi: 10.1016/j.conb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Carlson JR, Luo L. Olfactory receptor neuron axon targeting: intrinsic transcriptional control and hierarchical interactions. Nat Neurosci. 2004;7:819–825. doi: 10.1038/nn1284. [DOI] [PubMed] [Google Scholar]
- Kreher SA, Kwon JY, Carlson JR. The molecular basis of odor coding in the Drosophila larva. Neuron. 2005;46:445–456. doi: 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–552. [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- McKenna M, Monte P, Helfand SL, Woodard C, Carlson J. A simple chemosensory response in Drosophila and the isolation of acj mutants in which it is affected. Proc Natl Acad Sci U S A. 1989;86:8118–8122. doi: 10.1073/pnas.86.20.8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkman T, Sengupta P. Regulation of chemosensory and GABAergic motor neuron development by the C. elegans Aristaless/Arx homolog alr-1. Development. 2005;132:1935–1949. doi: 10.1242/dev.01788. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Wakamiya M, Noji S, Koyama E, Taniguchi S, Takemura R, Copeland NG, Gilbert DJ, Jenkins NA, Muramatsu M. A novel class of murine POU gene predominantly expressed in central nervous system. J Biol Chem. 1993;268:7449–7457. [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Ray A, van der Goes van Naters W, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van der Goes van Naters W, Carlson JR. A regulatory code for neuron-specific odor receptor expression. PLoS Biol. 2008;6:1069–1083. doi: 10.1371/journal.pbio.0060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan AK, Rosenfeld MG. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 1999;13:1794–1806. doi: 10.1101/gad.13.14.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafi-Reinach TR, Melkman T, Hobert O, Sengupta P. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development. 2001;128:3269–3281. doi: 10.1242/dev.128.17.3269. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Colbert HA, Bargmann CI. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell. 1994;79:971–980. doi: 10.1016/0092-8674(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila. Curr Biol. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Voss JW, Wilson L, Rosenfeld MG. POU-domain proteins Pit-1 and Oct-1 interact to form a heteromeric complex and can cooperate to induce expression of the prolactin promoter. Genes Dev. 1991;5:1309–1320. doi: 10.1101/gad.5.7.1309. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Wey E, Lyons GE, Schäfer BW. A human POU domain gene, mPOU, is expressed in developing brain and specific adult tissues. Eur J Biochem. 1994;220:753–762. doi: 10.1111/j.1432-1033.1994.tb18676.x. [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Zhao H, Reed RR. X inactivation of the OCNC1 channel gene reveals a role for activity-dependent competition in the olfactory system. Cell. 2001;104:651–660. doi: 10.1016/s0092-8674(01)00262-8. [DOI] [PubMed] [Google Scholar]
- Zhou H, Yoshioka T, Nathans J. Retina-derived POU-domain factor-1: a complex POU-domain gene implicated in the development of retinal ganglion and amacrine cells. J Neurosci. 1996;16:2261–2274. doi: 10.1523/JNEUROSCI.16-07-02261.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]