Abstract
The human promyelocytic cell line THP-1 expresses high level of HLA class II (HLA-II) molecules after IFN-γ treatment. Here, we report a variant of THP-1 that does not express HLA-II after IFN-γ. The variant's HLA-II phenotype is constant over time in culture and it is not related to a defective IFN-γ-signalling pathway. Transfection of CIITA, the HLA-II transcriptional activator, under the control of a cytomegalovirus promoter rescues high level of HLA-DR surface expression in the variant indicating that the biosynthetic block resides in the expression of CIITA and not in the CIITA-dependent transactivation of the HLA-II promoters. Treatment of the variant with 5-azacytidine (5-aza), which inhibits CpG methylation, restores inducibility of HLA-II by IFN-γ both at transcriptional and phenotypic level and antigen presenting and processing function of the variant. DNA studies demonstrate that the molecular defect of the THP-1 variant originates from the methylation of the CIITA promoter IV. Furthermore, treatment with 5-aza produces a substantial demethylation of CIITA promoter IV and a significant increase of IFN-γ-dependent HLA-II expression in another myelomonocytic cell line, U937. Therefore hyper-methylation of CIITA promoter IV may be a relevant mechanism of epigenetic control preventing HLA-II IFN-γ inducibility in the myelomonocytic cell lineage.
Keywords: 5-azacytidine, gene regulation, MHC class II, transcription factors
Introduction
MHC class II (MHC-II) molecules, designated HLA class II (HLA-II) molecules in human, play a key function in triggering the adaptative immune response by presenting antigenic peptides to CD4+ T helper cells. HLA-II molecules are expressed constitutively in dendritic cells (DCs), B cells and cells of the monocyte/macrophage lineage; these cells are cumulatively referred to as professional antigen-presenting cells (APCs). Moreover, cytokines, mainly IFN-γ, may further increase or de novo induce HLA-II in macrophage-like cells or in cells of extrahemopoietic origin (1).
Expression of HLA-II genes is mainly regulated at the transcriptional level. Triggering of HLA-II transcription is mediated by the non-DNA-binding specific co-activator CIITA which is required for both constitutive and inducible expression (2). CIITA transcription is driven by multiple promoters (pI, pIII and pIV) of the activator of immune response-1 (AIR-1) gene (3) that function in a cell type-specific manner, each promoter directing the expression of an unique first exon. pI is active primarily in DC; pIII is mainly responsible for directing CIITA constitutive expression in B cells but it is also involved in the IFN-γ-inducible expression. Finally, p-IV is the principal promoter for the IFN-γ-inducible expression (4).
IFN-γ signalling pathway promotes the expression of >200 genes (5) and p-IV transcription requires the cooperative binding of IFN regulatory factor-1 (IRF-1), upstream stimulatory factor-1 and phosphorylated signal transduction and activation of transcription-1 (6).
Given its role on the homeostasis of immune response, much attention has also been focussed on the expression of HLA-II in disease and particularly in those disease states, such as cancer, in which the immune response appears to be hampered.
In tumour cells, HLA-II expression is known to be associated with a better prognosis for colorectal cancer (7) and breast cancer (8). Moreover, in ovarian carcinoma (9) and lymphoma (10), positive correlation between tumour-infiltrating lymphocytes and the amount of HLA-II in tumour cells has been documented. Conversely, HLA-II expression has been linked to disease progression and poor prognosis in melanoma (11). In mice, several reports suggest that forced expression of MHC-II in cancer cells leads to increased immunogenicity and loss of tumorigenicity (12–14). Moreover, it has been the frequently observed loss of HLA-II IFN-γ inducibility in cancer cells when compared with the normal cellular counterpart (15, 16).
Epigenetic modifications influencing MHC-II expression have been frequently observed during tumour progression. These modifications include both DNA hyper-methylation and histone deacetylation and may result in the silencing of MHC-II promoter and/or of AIR-1 promoter regions (17).
The human promyelocytic tumour cell line THP-1 constitutively expresses low amount of HLA-DR that is highly increased after IFN-γ stimulation (18, 19). However, the generation of variants refractory to IFNg-induced HLA-II expression was not an uncommon finding in our THP-1 cell cultures. This prompted us to investigate in detail the molecular basis of the restricted loss of inducibility. We found that it was strongly correlated to the specific methylation of the AIR-1 pIV. Possible implications of this finding, particularly in relation to tumour heterogeneity and progression, are discussed.
Methods
Cell cultures and treatments
The THP1-L clone was isolated from the parental THP-1 cells (THP1-pc) on the basis of its unresponsiveness to IFN-γ in terms of HLA class II induction. The authentication of the THP1-pc cell line and its subclones was performed by short tandem repeat (STR) DNA profiling (20). The DNA typing methodology involves simultaneous amplification of six STR loci (D18S51, D21S11, D8S1179, vWF, FGA and TH01) and the amelogenin gene in a multiplex PCR. The STR profiles obtained for THP-1 sublines under investigation were identical; furthermore, the STR profiling of THP1-pc resulted identical to those previously published (20). THP1-pc and U937 myelomonocytic cell lines, as well as the human B cell line Raji, were cultured in RPMI medium supplemented with 10% foetal bovine serum and glutamine without antibiotics. All cell lines were demonstrated to be mycoplasma free by PCR and ELISA. Immature dendritic cells (iDCs) were obtained by established procedures (21) and were cultured in RPMI medium as above. ACN and IMR5 human neuroblastoma cell lines were cultured in DMEM. THP1-pc, THP1-L and U937 cells were treated daily with the hypomethylating agent 5-azacytidine (5-aza) (SIGMA, Milan, Italy) at 0.3–1 μM for 5–7 days. Trycostatin A (TSA) (SIGMA, Milan), a histone deacetylase inhibitor, was used at 300 nM concentration for 2 days. In some experiments, the cells were treated sequentially with TSA and 5-aza. After drug treatment, cells were stimulated with 150 U ml−1 of IFN-γ (kindly provided by Cantone, Roussel Pharma, Milan, Italy) for the indicated times.
pcDNA3/CIITA (kindly provided by J. Ting, Chapel Hill, NC, USA) was transfected in THP-1 by electroporation using the BIORAD gene pulser apparatus (250 V, 350 μF). Stable bulk transfectant was selected in 1.2 mg ml−1 G418 as described (22).
Cell surface phenotyping
Cell surface expression was assessed by indirect immunofluorescence and flow cytometry. The mAbs used were D1-12 (HLA-DR), B7/21 (HLA-DP), B9.12 (HLA-ABC) and 84H10 (ICAM-1, CD54) (the latter kindly provided by A. Poggi, Genova, Italy). An irrelevant, isotype-matched antibody was used as negative control. Flow cytometry analysis of 5-aza-treated cells was performed only on living cells (>70% total input events) by propidium iodide exclusion.
Real-time quantitative reverse transcription–PCR
Total RNA was prepared using RNA clean (Hybaid, Ashford, UK) following the manufacturer's instructions. Synthesis of cDNA was performed using murine leukaemia virus reverse transcriptase (Promega, Milan, Italy). Retrotranscription was performed after treatment of total RNA with RNAse-free RQ1 DNAse (Promega) following the manufacturer's instructions. The PCR was performed on iCycler iQ Real-Time PCR Detection System (Bio-Rad, Milan, Italy) and the amplification product was detected using the fluorescence reporter dye SYBR Green I. All reagents in the PCR were provided in the iQ SYBR Green Supermix (Bio-Rad). The Primer Express Software was used to design primers.
The cDNAs from human cells were amplified using the following primers: total CIITA forward 5′-CCTGCTGTTCGGGACCTAAA-3′, reverse 5′-GGATCCGCACCAGTTTGG-3′; DRA forward 5′-CTCTTCTCAAGCACTGGGAGTTT-3′, reverse 5′-ATGAAGATGGTCCCAATAATGATG-3′; IRF-1 forward 5′-ATGCCCATCACTCGGATGC-3′, reverse 5′-CCCTGCTTTTATCGGCCTG-3′ (23); AIR-1-pI, forward 5′-CACTCTGCTCCATGAGCCT-3′, reverse 5′-CAAATCCAGGTCTCCTTTCTCTAGTAGG-3′; AIR-1-pIII forward 5′-TGGGATTCCTACACAATGCGTT-3′, reverse 5′-ACACTGTGAGCTGCCTTGGG-3′; AIR-1-pIV forward 5′-GCGGCCCCAGAGCTGG-3′, reverse 5′-GAAGCTCCAGGTAGCCACCTTCTA-3′ (24).
Two housekeeping genes were used with similar results: glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse 5′-CATGGGTGGAATCATATTGGAA-3′ and RNA pol-II forward 5′-GACAATGCAGAGAAGCTGG-3′, reverse 5′-GCAGGAAGACATCATCATCC-3′.
For each transcript under investigation, the procedure of quantification was validated by analysis of a standard curve consisting in a serial dilution of a positive control cDNA. For each sample, PCR data value (threshold cycle) was normalized to the corresponding value obtained for the GAPDH or the RNA pol-II, the housekeeping transcripts chosen as control. The normalized PCR data value obtained in the untreated cells was defined as the arbitrary unit (value 1). All other normalized PCR data values were then expressed as fraction or multiple of the arbitrary unit.
Analysis of the methylation status of the AIR-1-pIV
Semi-quantitative restriction analysis.
Purified human genomic DNA was digested with HindIII and aliquots were subsequently cleaved with HpaII, MspI or HhaI. The analysis of methylation status of AIR-1-pIV was then performed as described (25) by using the primers: forward 5′-GGTTGGACTGAGTTGGAGAGAAACAGAGAC-3′; reverse 5′- TGCCGCGGCCCCAGAGCTGGCGGGAGGGAG- 3′. By this analysis, two CpG duplets were evaluated (−238 and −77, with respect to beginning of transcription from AIR-1 p-IV).
Quantitative pyrosequencing analysis.
Pyrosequencing (26) is a sequence-by-synthesis analysis of short genomic sequences for the fast and quantitative estimation of methylation density. Briefly, after the standard bisulphite modification to convert the unmethylated C into T (27), 2 μl of the modified DNA was amplified with primers specifically designed with the Assay Design Software for Pyrosequencing (PSQ Assay Design 1.0, Biotage, Uppsala, Sweden) to avoid CpG sites, in order to amplify the target sequence irrespective of its methylation status. The amplified DNA was then subjected to pyrosequencing analysis utilizing a primer designed with the same dedicated software. The primers used to generate the amplicon for the pyrosequencing analysis were: forward 5′biotinylated-GGATGTTTTAGGTAGTTGGGATGT3′, reverse 5′-CAATTCTTTTTCCCTTTCACTT-3′ (45 PCR cycles, T annealing 58°C and the length of the amplicon is 127 bp). The sequencing primer was 5′-CCTTTCACTTTCTTTCTACA-3′ and the sequence to analyze was 5′-ACRCTAAACTCRAACCAAACTATAAAAC-3′. The sequencing reaction was performed with PyroGold reagent kit PSQ 96MA (Biotage) according to the manufacturer's instructions using a PSQ 96MA instrument (Biotage). By this analysis, two CpG duplets were evaluated (−86 and −77, with respect to beginning of transcription from AIR-1 p-IV). Values are expressed as percentage of methylation at the −86 and −77 sites.
Antigen processing and presentation assay
Both THP1-H and THP1-L, treated with 5-aza for 4 days, were analysed for their antigen processing and presentation capacity following IFN-γ stimulation. Briefly, cells were treated with IFN-γ 150 U ml−1 for 6 h and then extensively washed out to remove traces of the cytokine. Thereafter, APCs were dispersed in a flat-bottomed microtiter plate at a concentration of 1 × 104 cells per well and cultured in presence of 5 μg ml−1 purified protein derivative (PPD) or 1 μg ml−1 peptide 11 (aa 91–108 of Ag85 protein) (28) for 8 h. Finally, following preliminary titration experiments for optimal APC/T ratio, 2 × 104 cells per well of an established T cell line specific for peptide 11 and restricted by HLA-DR2 haplotype expressed in THP-1 were added for 20 h. Activation of T cells was assessed by quantitative measurement of the secreted IFN-γ by ELISA.
Results
The THP1-L variant does not express HLA class II molecules after stimulation with IFN-γ
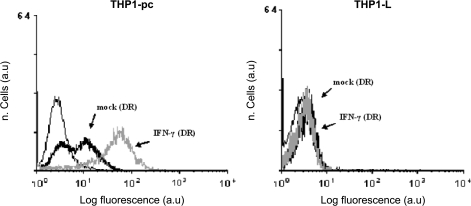
During the phenotypic characterization of various sublines of the myelomonocytic cell line THP-1, we have observed a divergent level in HLA-II (HLA-DR) cell surface expression after IFN-γ treatment. One of these sublines was further cloned by limiting dilution and the majority of clones displayed an HLA-II-negative phenotype after IFN-γ treatment. A subclone, designated THP1-L (L for low), characterized by undetectable expression of HLA-II, both in constitutive conditions and after IFN-γ stimulation (see Fig. 1), was selected for further analysis. The HLA-II phenotype of THP1-L was stable with time in culture and it is still stable after >100 cell divisions. THP1-L growth characteristics are similar to those of THP-1 parental cells, with the distinctive difference that the variant grows non-adherent to the plastic (data not shown). The analysis by informative DNA polymorphic markers confirmed that THP1-L is a true variant of the THP-1 parental cell line (data not shown).
Fig. 1.
HLA-DR phenotype of parental THP-1 and of THP1-L. Indirect immunofluorescence analysis by flow cytometry by using FITC-conjugated anti-mouse IgG secondary antibody. Cell surface expression in untreated cells (mock) or after 48 h in presence of 150 U ml−1 of IFN-γ. Cells stained with an isotype-matched antibody were used as negative control (thin line). Results are expressed in abscissa as log fluorescence value in arbitrary units. The results shown are from one representative experiment of four with similar results.
To assess whether unresponsiveness to IFN-γ treatment was due to a generalized defect in the IFN-γ signalling pathway, the cell surface expression of HLA class I and ICAM-1 molecules, known to be up-regulated by IFN-γ, was also studied. The results (see Table 1) indicate that both HLA class I and ICAM-1 are strongly up-regulated in the THP1-L variant demonstrating that the IFN-γ signal transduction pathway is fully functional for other gene products and therefore the defect is restricted to HLA class II cell surface induction. This was further confirmed by quantitative reverse transcription (RT)–PCR analysis and western blotting of IRF-1 mRNA and its protein product, respectively, know to be up-regulated by IFN-γ as well (data not shown).
Table 1.
Phenotypic analysis of IFN-γ signal transduction pathway in THP-1 cells
| Cells | HLA class I |
ICAM-1 |
||||||
| Mock |
IFN-γ |
Mock |
IFN-γ |
|||||
| %a | m.f.b | % | m.f. | % | m.f. | % | m.f. | |
| THP1-pc | 100 | 128 | 100 | 253 | <1 | 0.3 | 82 | 43 |
| THP1-L | 98 | 132 | 100 | 220 | <2 | 0.3 | 65 | 35 |
Percent of positive cells.
Mean fluorescence value; negative controls were the cells incubated with isotype-matched non-specific mAb. The mean fluorescence value of negative controls was always ≤0.3.
The THP1-L HLA-II-negative phenotype is due to lack of transcription of the AIR-1-encoded CIITA transactivator. Rescuing by 5-aza
To assess whether the absence of IFN-γ-dependent HLA-II expression in THP1-L was due to transcriptional or post-transcriptional events, HLA-II DRA mRNA was measured by quantitative RT–PCR. Results showed barely detectable levels of class II transcripts, 100-fold less than the parental THP-1 cells (THP1-pc), which remained unchanged after IFN-γ (Fig. 2A, third and fourth white column from the left, respectively; compare with THP1-pc, first and second white columns from the left). The lack of HLA-II transcripts after IFN-γ in THP1-L cell line could be due either to a mutational event or to an epigenetic mechanism involving key elements of the HLA-II gene expression pathway and possibly the CIITA-encoding AIR-1 gene that is the direct target of the IFN-γ stimulation. The rather frequent isolation of subclones carrying the phenotype of THP1-L suggested that an epigenetic phenomenon was more likely. Thus, to further characterize the molecular mechanisms responsible of HLA-II lack of induction after IFN-γ in the THP1-L variant, cells were treated with drugs that inhibit either DNA methylases (5-aza) or histone deacetylases (TSA) and then stimulated with IFN-γ.
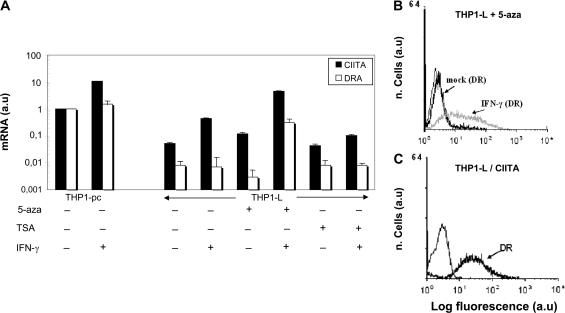
Fig. 2.
Treatment with 5-aza restores CIITA and consequent HLA-DR inducibility by IFN-γ in THP1-L. (A) Quantitative analysis by real-time RT–PCR of CIITA and DRA transcript. THP1-L cells were treated with 5-aza for 5–7 days or with TSA for 2 days. Freshly prepared 5-aza was added to cultures every day. Thereafter, THP1-L and THP1-pc were treated with IFN-γ for 2 days and total RNA was extracted. The housekeeping normalized data value obtained in mock THP1-pc was defined as the arbitrary unit (value 1). All other normalized PCR data values were then expressed as fraction or multiple of the arbitrary unit (mRNA a.u.) in the ordinate in a log scale. Bars represent the standard deviation from the mean of three distinct experiments. (B) THP1-L cells treated with 5-aza for 5 days were then cultured in absence (mock) or in presence of 150 U ml−1 IFN-γ (IFN-γ) for 2 days and analysed for HLA-DR surface expression by indirect immunofluorescence and flow cytometry. The results shown are from one representative experiment of three with similar results. (C) THP1-L cells were transfected with a CIITA expression vector under the control of CMVp. Stable bulk transfection was obtained under G418 selection and further subcloned. HLA-DR surface expression (DR) was evaluated by indirect immunofluorescence and flow cytometry. Results refer to a representative clone of 10 with similar DR phenotype.
Assessment of HLA-DRA transcripts and of CIITA transcripts was then performed. CIITA-specific mRNA was very low in the THP1-L variant (Fig. 2A, third black column from the left) and although it increased after IFN-γ treatment, it barely reached 50% of the level of CIITA mRNA expressed in unstimulated parental THP-1 cells (compare fourth black column with first black column). This amount of CIITA was insufficient to activate detectable transcription of HLA-II DRA gene (Fig. 2A, compare third and fourth white columns). Interestingly, although treatment with 5-aza did not significantly increase the steady-state level of CIITA transcripts by itself, it modified dramatically the response to IFN-γ, as the cells expressed 8- to 10-fold more CIITA transcripts than unstimulated THP-1 parental cells and close to 40–45% of the amount expressed in the IFN-γ-treated THP-1 parental cells (Fig. 2A, compare sixth black column with second black column). As a result, transcription of HLA class II genes (Fig. 2A, sixth white column) and corresponding cell surface expression of HLA-DR molecules (Fig. 2B) were observed. On the contrary, treatment with TSA followed by IFN-γ did not significantly result in the activation of transcription of the AIR-1 gene and by consequence of the HLA-II genes (Fig. 2A, last two series of columns).
That the block in HLA-II expression was due to lack of CIITA transcription and not to defect in mechanisms downstream to CIITA expression was further confirmed by transfection of CIITA under cytomegalovirus (CMV) promoter in THP1-L cells. Indeed, transgenic expression of CIITA rescued high HLA-DR expression in THP1-L transfectant (Fig. 2C).
These results indicate that epigenetic events acting on the AIR-1 gene transcription that are partially counteracted by 5-aza, but not TSA, are responsible of the HLA-II-uninducible phenotype of the THP1-L variant.
Hyper-methylation of AIR-1 promoter IV prevents IFN-γ-dependent CIITA expression in THP1-L variant
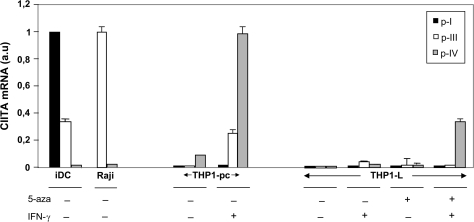
The CIITA-encoding AIR-1 gene can be transcribed from three functional promoters, pI, pIII and pIV. In order to determine which promoter was specifically involved in the silencing of the AIR-1 transcription in THP1-L variant and in the rescuing by the combined treatment of 5-aza and IFN-γ, quantitative RT–PCR was performed using promoter-specific primers. As shown in Fig. 3, pI, which is strongly expressed in iDCs (Fig. 3, first black column from the left) is neither expressed in THP-1 parental cells (THP1-pc) nor in the THP1-L variant. This is true irrespective of the treatment of parental or THP1-L variant with 5-aza, IFN-γ or with both 5-aza and IFN-γ. Similarly, pIII which is strongly expressed in B cells (Fig. 3, Raji cells first column) and well expressed in iDC is not expressed in THP1-pc and THP1-L variant cell lines. After treatment with IFN-γ, pIII is up-regulated in THP1-pc, indicating and confirming that pIII is sensitive to the cytokine also in inducible myelomonocytic cells (4). On the other hand, pIII is barely, if any, up-regulated in the THP1-L variant treated with IFN-γ, and pre-treatment with 5-aza does not modify this uninducible behaviour.
Fig. 3.
Quantitative real-time RT–PCR analysis of promoter-specific CIITA transcripts in various cells. THP1-L cells were treated with 5-aza for 5 days and then either left untreated or incubated with IFN-γ for 2 days. THP1-pc, untreated or similarly incubated with IFN-γ, iDCs and the B cell Raji were also analysed. For AIR-1 p-I, the housekeeping normalized PCR data value obtained in iDC was defined as the arbitrary unit (value 1). For AIR-1 p-III, the housekeeping normalized PCR data value obtained in Raji was defined as the arbitrary unit (value 1). For AIR-1 p-IV, the housekeeping normalized PCR data value obtained in THP1-pc treated with IFN-γ was defined as the arbitrary unit (value 1). The results shown are from one representative experiment of two with similar results.
A different situation is observed, instead, when the expression of pIV is analysed in the various experimental conditions. pIV is already expressed, although at low levels, in THP1-pc (Fig. 3, THP1-pc, third column) and this correlates with the low but distinctive expression of HLA-DR molecules (see Fig. 1). As expected, after IFN-γ treatment, pIV is strongly induced in THP1-pc (Fig. 3, THP1-pc, sixth column). Interestingly, and of particular relevance, the combined treatment with 5-aza and IFN-γ, but not the single treatment with either reagent, strongly up-regulates the expression of pIV in the THP1-L variant (Fig. 3, THP1-L, last column.). These results indicate that pIV is the promoter specifically involved in the silencing of the AIR-1 transcription in THP1-L variant.
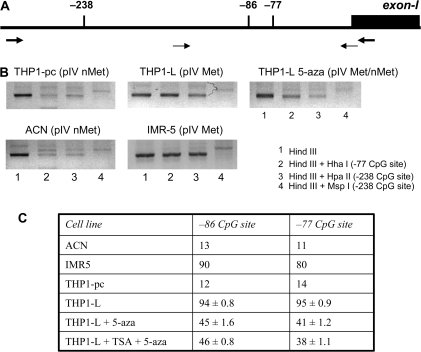
The fact that pre-treatment with 5-aza restores specifically the response of AIR-1 pIV to IFN-γ suggests that the epigenetic event at the basis of the lack of expression of HLA class II genes in the THP1-L variant may be due to methylation of pIV. To investigate this point, the state of pIV methylation in THP1-L and in the THP-1 parental cells before and after treatment with 5-aza was analysed. The results of semi-quantitative PCR analysis (Fig. 4B) are informative for two CpG dinucleotide sites in HpaII/MspI or HhaI restriction sites, mapping within the AIR-1-pIV (−238 and –77, Fig. 4A). pIV is unmethylated in THP-1 parental cells [THP1-pc (pIV nMet)]. In contrast, pIV is mostly methylated in the THP1-L variant [THP1-L (pIV Met)]. When the THP1-L cells were treated with 5-aza, pIV was modified towards mostly an unmethylated status (THP1-L 5-aza pIV Met/nMet).
Fig. 4.
Hyper-methylation of AIR-1 pIV as the major cause of lack of CIITA expression in response to IFN-γ. (A) Schematic representation of the AIR-1 p-IV region. Thick arrows indicate the positions of the primers used to analyse CpG sites by semi-quantitative restriction analysis; thin arrows indicate the positions of the primers used to analyse CpG sites by quantitative pyrosequencing. CpG sites under investigation are shown as vertical bars: sites −238 (HpaI MspI) and −77 (HhaI) were studied by restriction analysis; sites −86 and −77 were studied by quantitative pyrosequencing. (B) Semi-quantitative analysis of two CpG sites by restriction enzyme digestion. THP1-pc (pIV nMet), THP-1 parental cells with a non-methylated pIV; THP1-L (pIV Met), the THP1 variant with a fully methylated pIV; THP1-L 5-aza (pIV Met/nMet), the THP1-L variant treated with 5-aza showing a significantly reduced methylation of pIV. IMR5 and ACN are two neuroblastoma cell lines chosen as control of methylated (Met) and non-methylated (nMet) AIR-1 p-IV, respectively (15). The results shown are from one representative experiment of two with similar results. (C) Quantitative analysis of two CpG sites by pyrosequencing. ACN, IMR5 and THP1-pc were analysed in duplicates (mean value), THP1-L, THP1-L + aza and THP1-L TSA + aza were analysed in triplicate (mean value ± SD). Values are expressed as percentage of methylation at the two sites.
Two CpG dinucleotide sites in p-IV (−86 and −77) were further studied by quantitative pyrosequencing (Fig. 4C). This recently introduced methodology (26) allows accurate and quantitative de novo sequencing of the methylation state of each CpG, showing reproducible variations of methylation in contiguous CpGs, irrespective of their methyl-sensitive restriction endonuclease pattern. The results confirmed and extended the finding obtained by the semi-quantitative restriction analysis. The proportion of methylated CpG sites was >90% in THP1-L and dropped to <45% after 5-aza treatment.
It has been reported that the treatment with TSA may produce an ‘open state’ of the chromatin and this effect may increase the efficacy of demethylating drugs (29). However, this seems not to be the case with pIV CIITA in THP-L, since the combined treatment with 5-aza and TSA does not produce further demethylation around the sites analysed (Fig. 4C).
Taken together, these data strongly suggest that methylation of AIR-1 p-IV is a major cause for the defective CIITA expression in THP1-L after IFN-γ induction.
5-aza treatment rescues the capacity of IFN-γ to induce HLA-II molecules in another human promyelocytic cell line, U937
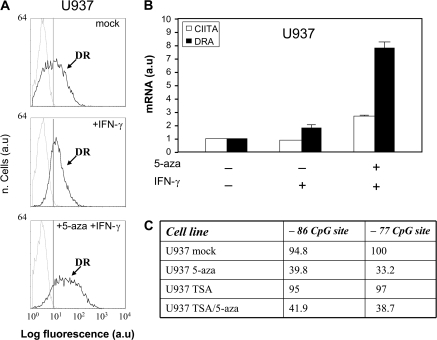
In order to assess whether hyper-methylation of AIR-1 p-IV could account for impaired HLA class II expression in other promyelocytic cell lines, the analysis was extended to U937 cell line, as U937 cells present very low HLA class II constitutive expression and low HLA-II inducibility by IFN-γ (Fig. 5A) (30). The HLA-II cell surface phenotype correlates with a low amount of constitutive HLA-DR transcript expression that is weakly increased after IFN-γ (Fig. 5B). Interestingly, pre-treatment with 5-aza results in a 5-fold increase of HLA-II-specific transcripts and corresponding cell surface molecules that correlated (Fig. 5B) with a distinctive increase of CIITA transcript over the level detected in the mock-treated cells or in cells treated with IFN-γ alone (see Fig. 5B). The pyrosequencing analysis focussed on the two CpG sites −86 and −77 of AIR-1 pIV demonstrates, even in this case, that the sites are heavily methylated in U937 and that the treatment with 5-aza significantly decreases the methylation status (Fig. 5C). Similarly to what it was found for THP1-L cells, no synergistic effect of 5-aza and TSA was found in U937.
Fig. 5.
Expression of HLA-DR and CIITA in U937 cells incubated with IFN-γ before and after treatment with 5-aza and analysis of the AIR-1 pIV methylation status. (A) HLA-DR cell surface phenotype of U937 cells as assessed by immunofluorescence and flow cytometry. Thin histograms represent the negative controls after each treatment (staining with irrelevant isotype-matched antibody); mock, U937 cells untreated; +IFN-γ, U937 cells treated with IFN-γ for 48 h; +5-aza + IFN-γ, U937 cells treated with 5-aza for 5 days and then with IFN-γ for 48 h. Results are expressed in abscissa as log fluorescence value in arbitrary units. The results shown are from one representative experiment of four with similar results. (B) Quantitative real-time RT–PCR analysis of CIITA and DRA transcripts. U937 cells were treated as in (A) and total RNA was extracted. The data values obtained for CIITA and DRA transcripts were normalized with respect to the value of the housekeeping gene transcript of choice in mock-treated U937 cells and defined as the arbitrary unit (value 1). All other normalized PCR data values were then expressed as fraction or multiple of the arbitrary unit. Bars represent the standard deviation of the mean of three distinct experiments. (C) Quantitative analysis of −86 and −77 CpG sites by pyrosequencing (see schematic representation of AIR-1 p-IV, Fig. 4A). U937 cells were treated as in (A) and analysed in duplicate. Values are expressed as mean percentage of methylation at the two sites.
Functional correlate of AIR-1 pIV demethylation in THP1-L cells
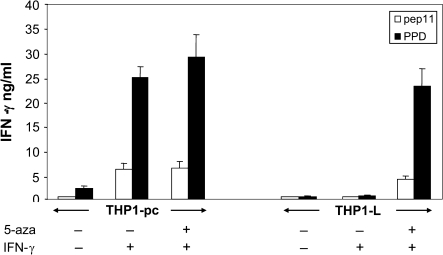
In order to investigate whether the demethylation of pIV induced by 5-aza, which results in the restored response to IFN-γ and consequent expression of HLA-II molecules, rendered myelomonocytic cells competent APCs, THP1-L cells were incubated with either the whole protein PPD or its derived immunodominant peptide pep11. Presentation of pep11 is restricted by the HLA-DR2 allele, which is specifically expressed by the THP-1 cells. The APC function was assessed by the capacity of the THP1-L cells treated with 5-aza and IFN-γ to stimulate an HLA-DR2-restricted T cell clone specific for pep11. Results show that 5-aza- and IFN-γ-treated THP1-L variant not only presents the pep11 but also process the PPD protein to produce and present the relevant peptide to T cells (Fig. 6); moreover, the processing and presentation capacity of THP1-L cells is very similar to the one expressed by the THP-1 parental cell line treated with IFN-γ or with 5-aza and IFN-γ, indicating that all steps downstream the expression of CIITA are not only phenotypically but also functionally conserved in the THP1-L variant.
Fig. 6.
HLA-DR2-restricted, antigen-specific T cell activation assay using THP-1 as APC. THP1-L was treated with 5-aza for 4 days; after this period, both THP1-L and THP1-pc cells were treated with IFN-γ for 6 h and then extensively washed to remove traces of the cytokine. Cells were pulsed with antigen (either peptide pep11 or whole PPD protein) and then mixed with a pep11-specific CD4+ T cell clone in flat-bottomed microtiter plate at a 2:1 stimulator/responder ratio. Activation of T cell was quantitatively assessed after 20 h of co-culture by measuring the amount of IFN-γ secreted by ELISA.
Discussion
It is well established that IFN-γ up-regulates HLA-II expression via the transcriptional activation of CIITA and particularly through a specific activation of the AIR-1 pIV (2).
The results presented in this report unambiguously show that spontaneous variants of the myelomonocytic cell line THP-1 can lose the IFN-γ-dependent expression of HLA-II molecules because of epigenetic events correlated with the methylation of promoter IV (pIV) of the AIR-1 gene encoding the MHC class II transactivator CIITA. In addition, the THP1-L variant analysed in detail in this study has lost also the constitutive expression of HLA-II genes present in the parental THP-1 cells, suggesting that the epigenetic block may affect the function of a more extended region of the AIR-1 promoter. The conclusion that AIR-1 pIV methylation was the major element associated with lack of HLA-II inducibility after IFN-γ in the THP1-L variant came from several observations: (i) the de novo presence of a hyper-methylated pIV promoter region, absent in the THP1 parental cells, as assessed by two distinct assays (the semi-quantitative RT–PCR followed by enzyme restriction analysis and the more refined pyrosequencing analysis); (ii) the reversion from a methylated to a partially methylated status of the pIV promoter after treatment with the demethylating agent 5-aza, which was accompanied by the reconstitution of an IFN-γ-dependent HLA-II-positive phenotype; (iii) the fact that a normal HLA-II expression was observed after stable transfection of CIITA cDNA under the control of an CMVp.
It was important to find that the variant did not lose the capacity to activate or up-regulate expression of other genes such as HLA class I genes or ICAM-1 gene in response to IFN-γ. Therefore, methylation of pIV can be a specific way to generate an HLA-II-negative phenotype without affecting the IFN-γ activation pathway in its entirety.
This idea is further substantiated by the finding that U937, an other promyelocytic cell line whose treatment with IFN-γ does not result in up-regulation of CIITA transcripts and consequent HLA-II expression, can be induced to express CIITA and consequent HLA-II expression by IFN-γ after pre-treatment with the demethylating agent 5-aza.
Similarly to the THP1-L variant, U937 cell line displays an almost fully methylated pIV, which becomes significantly hypo-methylated after 5-aza treatment. Moreover, as previously reported (22), transfection of CIITA under the control of the CMVp rescues high level of HLA-DR surface expression in U937, indicating that the biosynthetic block resides in the expression of CIITA and not in the CIITA-dependent transactivation of the HLA class II promoters. Thus, hyper-methylation of AIR-1 pIV is not a rare event in promyelocytic cells or at least in their representative tumour cell lines.
Do these data bear importance for understanding physiopathologic conditions associated with lack of HLA-II expression?
In physiologic conditions, the silencing of pIV through methylation may be a suitable way to avoid expression of HLA-II antigens in tissues, such as trophoblast, that are refractory to IFN-γ induction of CIITA (31, 32), although this may not be always and/or only due to pIV methylation (25). This would favour the maternal immune unresponsiveness against the foetus. Similarly, in pathological conditions, as in cancer, the capacity to negatively modulate HLA-II expression by AIR-1 promoter methylation may offer the tumour cells the advantage of escaping the anti-tumour immune response, particularly in case of tumours originated from the myelomonocytic cell lineage which includes the classical APCs. It is intriguing that during neoplastic transformation of the myelomonocytic cells the phenotypic characteristics of the cancer cells often mimic the ones observed in the THP1-L variant and in the U937 cells described here. Indeed, loss of HLA class II expression in myelomonocytic tumour cells has been frequently found and a recent study has shown that the distinctive methylation of the AIR-1 pIV was associated with the HLA class II-negative phenotype in 7 of 32 cases of primary acute myeloid leukaemia (33). Thus, the finding that our THP1-L variant and U937 cells do not express CIITA because of methylation of pIV may reflect the tendency of myeloid tumour cells to accumulate specific epigenetic modifications more than a mere in vitro artefact due to long-term culture (34, 35).
Previous extensive phenotypic studies on a wide variety of histological distinct tumour cell lines, including T cell lines, some myelomonocytic cell lines and gastrointestinal cell lines, have shown a correlation between HLA-II-negative, IFN-γ-uninducible phenotype and aberrant methylation in the CIITA pIV region (16, 33). However, with few exceptions, experimental approaches such as demethylation of CIITA pIV by treatment with 5-aza, followed by treatment with IFN-γ were not pursued in detail to demonstrate the causative association between methylation of CIITA pIV, lack of CIITA expression and consequent lack of HLA-II expression. More importantly, previous studies have not experimentally addressed the crucial issues of the real functional importance of the negative epigenetic regulation of HLA-II expression in tumour cells, and particularly in tumour cells of the myelomonocytic cell lineage, in terms of antigen processing and presentation capacity of these altered cells.
Therefore, we decided to focus our attention on the epigenetic regulation of CIITA and HLA-II expression in myelomonocytic tumour cells because these cells are normally HLA-II inducible by IFN-γ and they are key cells in triggering activation of the adaptive immune system, due to their antigen processing and presentation function for HLA-II-restricted CD4 T cells.
As previously reported (19), IFN-γ-induced THP1 cells are an efficient APC for HLA-II-restricted antigen processing and presentation. Interestingly, the treatment of THP1-L with 5-aza rescued the IFN-γ-dependent APC processing and presentation function. Thus, the comparative molecular and functional study of THP-1 parental cells and its THP1-L isogenic variant, together with the analysis of an additional myelomonocytic cell line, U937, provided us with the unprecedented demonstration that CIITA pIV methylation was the cause of lack of expression of CIITA, consequent lack of expression of HLA-II molecules and, importantly, abolished APC function in myelomonocytic tumour cells. Since CIITA-mediated expression of MHC class II molecules by murine tumour cells can result in acquisition of strong APC capacity in vitro and, more importantly, in potent CD4-dependent immune rejection of the tumour in syngeneic hosts (13, 14), it is reasonable that loss of HLA-II expression and APC capacity in myelomonocytic-derived tumour cells may indeed contribute to tumour escape from immune control as well.
Moreover, it has been reported that DNA hyper-methylation and/or histone deacetylation in cancer cell may silence not only MHC class II but also accessory/co-stimulatory molecule and, importantly, tumour-associated antigen expression (36). Within this frame, our findings may bear importance in envisaging the use of epigenetic modifying drugs for increasing the immunogenicity of myeloid cancer.
Funding
Italian National Institute of Health (ISS), National Research Project on AIDS n° 40F.1 and 45G.1 (to R.S.A.); Fondi di Ateneo per la Ricerca 2006–2007 (to A.D.L.B. and R.S.A.); Department of Clinical and Biological Sciences, University of Insubria, targeted grant on ‘Diabetes’ (to A.D.L.B. and R.S.A.); Italian Ministry of Health, Ordinary Project (to S.F.); Associazione Italiana per la Ricerca sul Cancro grant (to M.R. and S.F.).
Glossary
Abbreviations
- AIR-1
activator of immune response gene-1
- APC
antigen-presenting cell
- 5-aza
5-azacytidine
- CMVp
cytomegalovirus promoter
- DC
dendritic cell
- IRF-1
IFN regulatory factor-1
- PPD
purified protein derivative
- RT
reverse transcription
- STR
short tandem repeat
- TSA
trycostatin A
References
- 1.Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu. Rev. Immunol. 1993;11:403. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 2.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 2001;19:331. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 3.Accolla RS, Jotterand-Bellomo M, Scarpellino L, Maffei A, Carra G, Guardiola J. aIr-1, a newly found locus on mouse chromosome 16 encoding a trans-acting activator factor for MHC class II gene expression. J. Exp. Med. 1986;164:369. doi: 10.1084/jem.164.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 6.Muhlethaler-Mottet A, Di Bernardino W, Otten LA, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 7.Lovig T, Andersen SN, Thorstensen L, et al. Strong HLA-DR expression in microsatellite stable carcinomas of the large bowel is associated with good prognosis. Br. J. Cancer. 2002;87:756. doi: 10.1038/sj.bjc.6600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheen-Chen SM, Chou FF, Eng HL, Chen WJ. An evaluation of the prognostic significance of HLA-DR expression in axillary-node-negative breast cancer. Surgery. 1994;116:510. [PubMed] [Google Scholar]
- 9.Matsushita N, Ghazizadeh M, Konishi H, Araki T. Association of ovarian tumor epithelium coexpressing HLA-DR and CA-125 antigens with tumor infiltrating cytotoxic T lymphocytes. J. Nippon Med. Sch. 2003;70:40. doi: 10.1272/jnms.70.40. [DOI] [PubMed] [Google Scholar]
- 10.Rimsza LM, Roberts RA, Miller TP, et al. Loss of major histocompatibility class II expression in non-immune-privileged site diffuse large B-cell lymphoma is highly coordinated and not due to chromosomal deletions. Blood. 2004;103:4251. doi: 10.1182/blood-2005-04-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Duinen SG, Ruiter DJ, Broecker EB, et al. Level of HLA antigens in locoregional metastases and clinical course of the disease in patients with melanoma. Cancer Res. 1988;48:1019. [PubMed] [Google Scholar]
- 12.Armstrong T, Clements V, Matin B, Ting J, Ostrand-Rosenberg S. Major histocompatibility complex class II-transfected tumor cells present endogenous antigen and are potent inducers of tumor-specific immunity. Proc. Natl Acad. Sci. USA. 1997;94:6886. doi: 10.1073/pnas.94.13.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meazza R, Comes A, Orengo AM, Ferrini S, Accolla RS. Tumor rejection by gene transfer of the MHC class II transactivator in murine mammary adenocarcinoma cells. Eur. J. Immunol. 2003;33:1183. doi: 10.1002/eji.200323712. [DOI] [PubMed] [Google Scholar]
- 14.Mortara L, Castellani P, Meazza R, et al. CIITA-induced MHC class II expression in mammary adenocarcinoma leads to a Th1 polarization of the tumor microenvironment, tumor rejection, and specific antitumor memory. Clin. Cancer Res. 2006;12:3435. doi: 10.1158/1078-0432.CCR-06-0165. [DOI] [PubMed] [Google Scholar]
- 15.Croce M, De Ambrosis A, Corrias MV, et al. Different levels of control prevent interferon-gamma-inducible HLA-class II expression in human neuroblastoma cells. Oncogene. 2003;22:7848. doi: 10.1038/sj.onc.1207054. [DOI] [PubMed] [Google Scholar]
- 16.Satoh A, Toyota M, Ikeda H, et al. Epigenetic inactivation of class II transactivator (CIITA) is associated with the absence of interferon-gamma-induced HLA-DR expression in colorectal and gastric cancer cells. Oncogene. 2004;23:8876. doi: 10.1038/sj.onc.1208144. [DOI] [PubMed] [Google Scholar]
- 17.Wright KL, Ting JP. Epigenetic regulation of MHC-II and CIITA genes. Trends Immunol. 2006;27:405. doi: 10.1016/j.it.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Tsuchiya S, Yamabe M, Yamaguci Y, Kabayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int. J. Cancer. 1980;26:171. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 19.De Lerma Barbaro A, Tosi G, Valle MT, et al. Distinct regulation of HLA class II and class I cell surface expression in the THP-1 macrophage cell line after bacterial phagocytosis. Eur. J. Immunol. 1999;29:499. doi: 10.1002/(SICI)1521-4141(199902)29:02<499::AID-IMMU499>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 20.Masters JR, Thomson JA, Dely-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc. Natl Acad. Sci. USA. 2001;98:8012. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1994;179:1109. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Lerma Barbaro A, Procopio FA, Mortara L, Tosi G, Accolla RS. The MHC class II transactivator (CIITA) mRNA stability is critical for the HLA class II gene expression in myelomonocytic cells. Eur. J. Immunol. 2005;35:603. doi: 10.1002/eji.200425378. [DOI] [PubMed] [Google Scholar]
- 23.De Ambrosis A, Casciano I, Croce M, et al. An interferon-sensitive response element is involved in constitutive caspase-8 gene expression in neuroblastoma cells. Int. J. Cancer. 2007;120:39. doi: 10.1002/ijc.22173. [DOI] [PubMed] [Google Scholar]
- 24.Hornell TM, Beresford GW, Bushey A, Boss JM, Mellins ED. Regulation of the class II MHC pathway in primary human monocytes by granulocyte-macrophage colony-stimulating factor. J. Immunol. 2003;171:2374. doi: 10.4049/jimmunol.171.5.2374. [DOI] [PubMed] [Google Scholar]
- 25.Holtz R, Choi JC, Petroff MG, Piskurich JF, Murphy SP. Class II transactivator (CIITA) promoter methylation does not correlate with silencing of CIITA transcription in trophoblasts. Biol. Reprod. 2003;69:915. doi: 10.1095/biolreprod.103.017103. [DOI] [PubMed] [Google Scholar]
- 26.Dupont JM, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 2004;333:119. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valle MT, Megiovanni AM, Merlo A, et al. Epitope focus, clonal composition and Th1 phenotype of the human CD4 response to the secretory mycobacterial antigen Ag85. Clin. Exp. Immunol. 2001;123:226. doi: 10.1046/j.1365-2249.2001.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin S. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 30.Jayaram Y, Buckle AM, Hogg N. The Fc receptor, FcRI, and other activation molecules on human mononuclear phagocytes after treatment with interferon-gamma. Clin. Exp. Immunol. 1989;75:414. [PMC free article] [PubMed] [Google Scholar]
- 31.Morris AC, Spangler WE, Boss JM. Methylation of class II trans-activator promoter IV: a novel mechanism of MHC class II gene control. J. Immunol. 2000;164:4143. doi: 10.4049/jimmunol.164.8.4143. [DOI] [PubMed] [Google Scholar]
- 32.van den Elsen PJ, van der Stoep N, Datema G, Vietor HE. Transcriptional control of MHC genes in fetal trophoblast cells. J. Reprod. Immunol. 2001;52:129. doi: 10.1016/s0165-0378(01)00115-2. [DOI] [PubMed] [Google Scholar]
- 33.Morimoto Y, Toyota M, Satoh A, et al. Inactivation of class II transactivator by DNA methylation and histone deacetylation associated with absence of HLA-DR induction by interferon-gamma in haematopoietic tumour cells. Br. J. Cancer. 2004;90:844. doi: 10.1038/sj.bjc.6601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 35.Jones P, Wolkowicz MJ, Rideout WM, et al. De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proc. Natl Acad. Sci. USA. 1990;87:6117. doi: 10.1073/pnas.87.16.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigalotti L, Coral S, Fratta E, et al. Epigenetic modulation of solid tumors as a novel approach for cancer immunotherapy. Semin. Oncol. 2005;32:473. doi: 10.1053/j.seminoncol.2005.07.005. [DOI] [PubMed] [Google Scholar]