Abstract
After somitogenesis, skeletal muscle precursors elongate into muscle fibers that anchor to the somite boundary, which becomes the myotome boundary. Fibronectin (Fn) is a major component of the extracellular matrix in both boundaries. Although Fn is required for somitogenesis, effects of Fn disruption on subsequent muscle development are unknown. We show fn knockdown disrupts myogenesis. Muscle morphogenesis is more disrupted in fn morphants than in a mutant where initial somite boundaries did not form, aei/deltaD. We quantified this disruption using the 2D Wavelet-Transform Modulus Maxima method, which uses the variation of intensity in an image with respect to the direction considered to characterize the structure in a cell lattice. We show that fibers in fn morphants are less organized than in aei/deltaD mutant embryos. Fast- and slow-twitch muscle lengths are also more frequently uncoupled. These data suggest fn may function to regulate fiber organization and limit fast-twitch muscle fiber length.
INTRODUCTION
During vertebrate development, transient structures called somites give rise to skeletal musculature, vertebrae, cartilage, tendon, and dermis (Brent and Tabin, 2002; Buckingham et al., 2003; Kalcheim and Ben-Yair, 2005). Following somite formation, muscle fibers derived from somitic cells form and attach to the basement membrane. Force generated by muscle fiber contraction is transduced through the basement membrane, to tendons, and finally to the skeletal system. Muscle diseases such as the muscular dystrophies can result when various components of this force relay are disrupted (Kanagawa and Toda, 2006; Schessl et al., 2006). Thus, both somite formation and myotome differentiation are critical for the development of a functional locomotor system.
The relative simplicity of the zebrafish musculoskeletal system makes it an ideal model for study of muscle fiber and tendon morphogenesis. In zebrafish, slow- and fast-twitch muscle fibers are spatially segregated (Devoto et al., 1996). Slow-twitch muscle cells are specified by Hedgehog signaling and are initially located directly adjacent to the midline (Blagden et al., 1997; Du et al., 1997; Lewis et al., 1999). After somite formation, slow-twitch muscle cells migrate laterally through the fast muscle domain to generate the most lateral layer of muscle (Devoto et al., 1996). The lateral migration of slow-twitch muscle cells not only correlates with fast-twitch muscle cell elongation and attachment (Cortes et al., 2003), but is also necessary and sufficient to trigger timely fast muscle cell elongation (Henry and Amacher, 2004).
The elongation, fusion, and attachment of somitic cells to the somite boundary results in formation of myotomes, segmentally reiterated groups of specified muscle fibers. The somite boundary is now called a myotome boundary and it separates the myotomes. In teleosts, myotome boundaries give rise to the myotendenous junction (MTJ) (Long et al., 2002). This junction is functionally equivalent to the mammalian MTJ (Gemballa and Vogel, 2002; Summers and Koob, 2002). The MTJ is crucial for muscle physiology because it is the primary site of force transmission from muscle to the skeletal system. Currently, the cellular and molecular networks that mediate muscle fiber elongation and attachment to the MTJ/tendon are incompletely understood.
The extracellular matrix (ECM) is critical for somite formation during embryogenesis in zebrafish, Xenopus, chick and mouse (George et al., 1993; Georges-Labouesse et al., 1996; Winklbauer and Keller, 1996; Julich et al., 2005; Koshida et al., 2005; Kragtorp and Miller, 2006; Rifes et al., 2007). However, less is known about roles for cell-matrix adhesion proteins during muscle morphogenesis in vivo. Fibronectin (Fn) is highly concentrated at both initial somite boundaries as well as myotome boundaries, suggesting that Fn may play a role in muscle development (Crawford et al., 2003). Fn is a high molecular weight, dimeric, three domain glycoprotein that is a major component of ECM. Fibrillar Fibronectin links cells to the ECM via Integrin cell-surface receptors and can also bind to heparin, collagen, fibrin, and fibulin (Vakonakis and Campbell, 2007). Although it is not clear how Fn fibrillogenesis proceeds, cellular tension and interdomain interactions are important (Morla and Ruoslahti, 1992; Ingham et al., 1997; Mao and Schwarzbauer, 2005). The ability of Fn to be stretched up to fourfold by living cells (Erickson, 2002) may be particularly relevant for dynamic cell movements that occur during development.
Recent studies of cell-matrix adhesion proteins during zebrafish somite formation have focused on Integrins, heterodimeric transmembrane receptors that bind to extracellular matrix proteins. Integrin α5 is required for initial somite formation and/or boundary maintenance (Julich et al., 2005; Koshida et al., 2005). Although fn1 mutants as well as fn1+fn3 morphants show disrupted somite formation and/or maintenance (Julich et al., 2005; Koshida et al., 2005), discrete requirements for Fn in muscle morphogenesis have not been identified. Because of the complexity of cell-ECM interactions, it is critical to systematically assess roles for individual Integrins and ligands during multiple stages of muscle morphogenesis. Integrin α5β1 is a receptor for not only Fn, but also osteopontin, fibrillin, L1, thrombospondin, and ADAM family members. Conversely, Integrin α5 is only one of six known Integrin receptors for Fn (Humphries et al., 2006). Thus, although it is clear that cell-matrix adhesion is required for somite formation, discrete and mechanistic roles for multiple different Integrin family members and their potential ligands have not been identified.
We find that Fn knockdown disrupts muscle morphogenesis. Slow-twitch fibers show irregular lengths and spacing. Fast-twitch fibers in fn 1+3 morphants are abnormally long and cross the MTJ. The disruption of muscle development in fn 1+3 morphants is distinctly more severe than the disruption of muscle development in a mutant where initial somites do not form. In aei/deltaD mutant embryos, initial somite boundary formation is disrupted but MTJ formation does occur by 26 hours post fertilization (hpf). Although the MTJs are not regularly spaced or V-shaped (van Eeden et al., 1998; Henry et al., 2005; Julich et al., 2005), they do persist throughout both slow and fast-twitch muscle domains. In contrast, fast-twitch fibers in fn morphants are more likely to cross the MTJ respected by slow-twitch muscle fibers. One quantitative assessment of structure is anisotropy. The 2D Wavelet-Transform Modulus Maxima (WTMM) method is a multifractal image analysis formalism that is perfectly suited to quantitatively characterize different types of anisotropic signatures (Arneodo et al., 2000; Arneodo et al., 2003; Khalil et al., 2006). The strength of the anisotropic signature of slow-twitch muscle fibers in fn morphant embryos is significantly lower than in aei/deltaD mutant embryos. This result indicates that slow-twitch muscle fibers in aei/deltaD mutant embryos exhibit less random structure than slow-twitch fibers in fn morphant embryos. Taken together, these results indicate that Fn plays a discrete role in regulating muscle morphogenesis in addition to being critical for normal somite boundary formation.
MATERIALS AND METHODS
Zebrafish husbandry and mutant alleles
Zebrafish embryos were obtained from natural spawnings of adult fish kept at 28.5°C on a 16 h light/8 h dark cycle and were staged according to (Kimmel et al., 1995). The allele of after eight/deltaD (aeitr233) has been previously described (Holley et al., 2000).
Morpholino Injections
Morpholino-modified antisense oligonucleotides (MOs) were synthesized by Gene-Tools, LCC. The fn1 MO is as previously described: 5′-ttttttcacaggtgcgattgaacac-3′ (Trinh and Stainier, 2004). The fn3 MO1 and fn3 MO2 are as previously described: fn3 MO1 5′-tactgactcacgggtcattttcacc-3′ and fn3 MO2 5′-gcttctggctttgactgtatttcgg-3′ (Julich et al., 2005). Morpholinos were dissolved to a stock concentration of 50 ng/nl in sterile water. A combined MO mixture was injected into the embryos at the 1–2 cell stage using an ASI pressure injector (ASI Systems).
In situ hybridization and immunocytochemistry
Whole mount in situ hybridization was performed as previously described (Jowett, 1999). F59 was utilized to visualize myosin fibers as previously described (Crow and Stockdale, 1986; Devoto et al., 1996). The Fibronectin antibody (Ab-10, catalogue number RB-077-A0) was obtained from Lab Vision and the β-catenin antibody was obtained from Sigma (C7207). Alexa Fluor 488 and 546 phalloidin were obtained from Molecular Probes. For antibody staining, embryos were fixed in 4% paraformaldehyde (PFA) for 4 hours at room temperature and incubated in block (5% BSA, 1% DMSO, 1% Triton X-100, 0.2% saponin in PBS) for 1 hour. Staining was conducted in PBDT (1% BSA, 1% DMSO, 1% Triton X-100 in PBS). For visualization of actin using phalloidin staining, embryos were fixed as above, followed by permeabilization in 2% Triton X-100/PBS for 1.5 hours and incubation in 1:20 Alexa-Fluor 488 or 543 conjugated phalloidin (Invitrogen) for 1 hour. Embryos were then rinsed overnight prior to proceeding with antibody staining.
Imaging
Images were acquired using a Zeiss ApoTome running on a Zeiss Axio Imager Z1. Embryos were mounted in 80% glycerol/20% PBS with 1.5 coverslips and visualized using a 20x objective. The resolution thus achieved was 400 nm per pixel. Image quality was optimized by averaging 4–5 frames. Staining longevity for ApoTome images was improved by using SlowFade reagent (Invitrogen). Images were processed in Adobe Photoshop (only linear modifications to brightness/contrast were made) and collated in Adobe Illustrator.
Qualitative boundary analysis
Individual boundaries were analyzed to determine if fast fibers were the same length as slow fibers. Embryos were stained with F59 to visualize slow muscle and phalloidin to visualize fast muscle. A z-series was taken and used to assess fast versus slow fiber length. Boundaries posterior to somite 22 were not included in the analysis of fn1+3 morpholino-injected embryos because somite 22 was the average posterior limit of segmentation defects in fast muscle fibers. Because the two sides of the embryos, dorsal and ventral, frequently differed, each side was assessed individually.
Quantification of Fn levels, segment parameters, muscle fiber lengths, and statistics
The Zeiss software modules Inside4D and Automeasure were utilized to measure morphometric parameters of segments and muscle fibers. Because of the nature of the stains used, the software was unable to automatically segment discrete boundaries and muscle fibers. Thus, all measurements were done in the “interactive processing of the 3D mask” page. To quantify segment lengths, areas, and perimeters, myotome boundaries in projected images were traced interactively. To quantify muscle fiber lengths, muscle fibers in projected images were traced interactively.
The Automeasure module was also utilized to measure average intensities for the dose-dependent decrease in Fn protein after morpholino injection.
A two sample T-test was performed using SYSTAT. * denotes p < 0.05 and ** denotes p < 0.01.
Characterizing Anisotropy with the 2D WTMM method
The 2D WTMM (wavelet transform modulus maxima maxima) method is used to assign a number that quantifies how random, disordered or unstructured the morphological shape of objects in an image are. In brief, the WTMM analysis consists of filtering an image with the gradient of a smoothing function (i.e. a wavelet) at a given size scale a. Places within the image where the intensity variation is maximal are given by the wavelet-transform modulus maxima (i.e. the WTMM). These are visible as edge detection lines in Fig. 4 (the sea green lines). Next, the positions of maximal intensity variation along the WTMM (i.e. along the sea green lines) are identified. These are the WTMM Maxima (or WTMMM) vectors and are shown as the red spots in Fig. 4. At these nodes, the direction where the signal has the sharpest variation is calculated and is visible as the yellow arrows. An arrow that points upward has an angle of π/2 and an arrow that points down has an angle of −π/2. For each size scale a, the anisotropy factor Fa is then calculated from the probability density function, Pa(A), of the angles A of the WTMMM vectors. Fa is defined in such a way that isotropy (random structure) has a value of Fa = 0 and any value of Fa > 0 will quantify departure from isotropy. A randomly structured image will have angles pointing in all directions and a low anisotropy factor. A more organized structure will have more arrows pointing in the same direction and a stronger anisotropic signature. Thus, this formalism objectively provides a quantitative assessment of morphological structure.
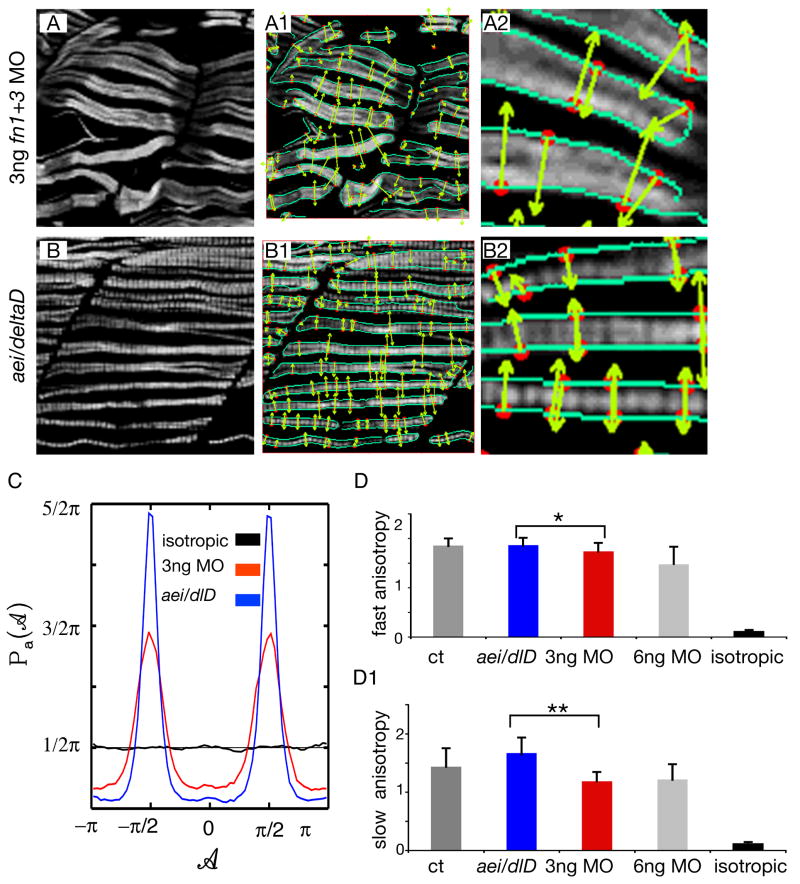
Figure 4. Muscle fibers in 3 ng fn1+3 morphant embryos are more randomly structured than fibers in aei/deltaD mutant embryos.
A, B: F59 denotes slow-twitch fibers, side views, anterior left, dorsal top. Note that fibers in 3 ng fn1+3 morphant embryos appear slightly more disorganized than those in aei/deltaD mutant embryos. The maxima chains (A1, A2, B1, B2, green edge detection lines) are defined by the positions where the Wavelet-Transform Modulus is locally Maximum (i.e. the WTMM) in the direction A of the gradient vector. The WTMM maxima (WTMMM) (A1, A2, B1, B2, red dots) indicate locally the direction where the signal has the sharpest variation (A1, A2, B1, B2, green arrows).
C: The WTMMM vector angle pdfs are displayed for the aei/deltaD mutants (blue curve), the fn1+3 morphants (red curve), as well as for the isotropic fBm surfaces analyzed for calibration purposes (black curve fluctuating around π/2). Also shown is the flat 1/2π curve that would be obtained for a purely theoretical isotropic process (flat pointed line at 1/2π). Note the stronger peaks in aei/detaD mutant embryos.
D–D1: The anisotropy factor, an indication of organized structure, of fast-twitch fibers in fn morphant embryos is not only significantly lower than in control embryos, but is also significantly lower than in aei/deltaD mutant embryos. In contrast, the anisotropy factor of fast-twitch fibers in aei/deltaD mutant embryos is not significantly different from control embryos. These results indicate that fast-twitch fibers in fn morphant embryos not only appear more disorganized, but are quantitatively more disorganized than in aei/deltaD mutant embryos. The anisotropy of slow-twitch fibers in fn morphant embryos is also significantly lower than in control embryos and aei/deltaD mutant embryos. * denotes p < 0.05, ** denotes p < 001. For comparison, the anisotropy factor obtained from the analysis of the isotropic fBm images (0.12 +/− 0.02) is also shown.
The 2D WTMM method is a multifractal image analysis formalism introduced in (Arneodo et al., 2000), where the different dilations of the analyzing wavelet reveal quantitative roughness information at every length scale considered. By considering two wavelets that are, respectively, the partial derivatives with respect to x and y of a 2D smoothing Gaussian function, the Wavelet Transform (WT) is thus the gradient vector of the analyzed image smoothed by dilated versions of the Gaussian filter. A very efficient way to perform point-wise regularity analysis is to use the Wavelet Transform Modulus Maxima (WTMM) (Mallat and Hwang, 1992; Mallat and Zhong, 1992). At a given scale a, the WTMM are defined by the positions where the Wavelet Transform Modulus is locally maximum in the direction A of the gradient vector. When analyzing rough surfaces, these WTMM lie on connected chains called maxima chains (Arneodo et al., 2000), as shown in Figure 4, panels A1, B1, green lines. One only needs to record the position of the local maxima of the gradient along the maxima chains together with the angle A at the corresponding locations. At each scale a, the wavelet analysis thus reduces to store those WTMM maxima (WTMMM) only (red dots in Figure 4, panels A1, B1). They indicate locally the direction where the signal has the sharpest variation (green arrows in Figure 4, panels A1, B1).
An image having an anisotropic signature means that the intensity variation in the image will differ according to the direction considered. Such images having an anisotropic signature can be easily characterized from the directional information provided by the continuous 2D Wavelet Transform (Khalil et al., 2006). This is done by considering, at all size scales a, the probability density functions (pdfs), Pa(A), of the angles, A, associated to each WTMMM vector (see Figure 4 C). A flat pdf indicates unprivileged random directions of sharpest intensity variation (i.e. isotropy), while any departure from a flat distribution is interpreted as the signature of anisotropy. For the present study, a strong anisotropic signature is interpreted as a strongly structured cell lattice.
Anisotropy Factor
In order to obtain quantitative information from the angle pdfs, they are compared to a theoretical flat distribution representing an ideal isotropic signature (see Figure 4 C). The anisotropy factor, Fa, defined for each value of the scale parameter a, is given by the area between the curve corresponding to the observed pdfs and a flat distribution:
Therefore Fa has been defined in such a way that a theoretically isotropic surface will have a value of Fa = 0, while any value greater than 0 quantifies a departure from isotropy. For the present study, however, given the fact that all cells have roughly the same width, only one size scale is considered for the wavelet analysis. That size scale was determined by the average measured width of the cells analyzed, which is ~ 2 μm.
Construction of simulated isotropic surfaces for calibration purposes
Following the standard procedures presented in (Arneodo et al., 2000; Khalil et al., 2006), fractional Brownian motion (fBm) isotropic surfaces were generated. Two-dimensional fBm’s are processes with stationary zero-mean Gaussian increments that are statistically invariant under isotropic dilations. The isotropic fBm surfaces were generated by the so-called Fourier transform filtering method (Ffm) (Peitgen et al., 1988). This particular synthesis method is used because of the simplicity of its implementation. Indeed it amounts to a fractional integration of a 2D “white noise” and therefore it is expected to reproduce quite faithfully the isotropic scaling invariance properties.
RESULTS AND DISCUSSION
Fn is required for normal muscle morphogenesis
Although it has been shown that Fn is required for initial somite boundary formation and maintenance, (Julich et al., 2005; Koshida et al., 2005) subsequent requirements of Fn in muscle morphogenesis have not yet been elucidated. There are two fibronectin genes in zebrafish, nat/fn1 and fn3 (Trinh and Stainier, 2004; Julich et al., 2005; Koshida et al., 2005). Although fn3 is more highly expressed in somites as well as presomitic mesoderm, fn1 is expressed in the tailbud and posterior presomitic mesoderm (Julich et al., 2005; Koshida et al., 2005). Thus, both fn1 and fn3 are expressed in muscle precursor cells.
Morpholinos (MOs) against fn1 and fn3 have been well characterized. Injection of MOs against both fn genes (750 μM fn1 MOs1+2 + 500μM fn3 MO2) disrupts somite formation throughout the anterior-posterior axis (Julich et al., 2005). We find a correlation between the amount of morpholino injected and the levels of Fn protein observed in vivo. Fn protein expression is inhibited in embryos injected with 6 ng fn1+3 MOs (Fig. 1B, D, 12/16 embryos injected with 6 ng fn1+3 MOs had no Fn staining, 4/16 had very weak Fn staining). Injection of 3 ng of morpholinos against fn1 + fn3 results in slightly reduced Fn as assayed by immunostaining (Fig. 1C, D, 25/25 embryos injected with 3 ng fn1+3 morpholinos had qualitatively reduced Fn protein in whole mount immunostaining). In these morphants, Fn protein levels are slightly lower in the anterior of the embryo and slightly higher in the posterior of the embryo (Fig. 1C, green and white arrows, respectively). This may reflect reduced translational inhibition by the MOs through time. Thus, whole mount immunostaining indicates a dose-dependent correlation (Fig. 1D) between MOs and Fn protein concentration at segment boundaries. MOs are frequently not fully penetrant and utilizing different amounts of MOs can correspond to an allelic series (Heasman, 2002; Dahm and Geisler, 2006; Riedel-Kruse et al., 2007). We exploited the dose-dependent reduction in Fn protein to ask if there is a corresponding dose-dependent disruption of muscle morphogenesis.
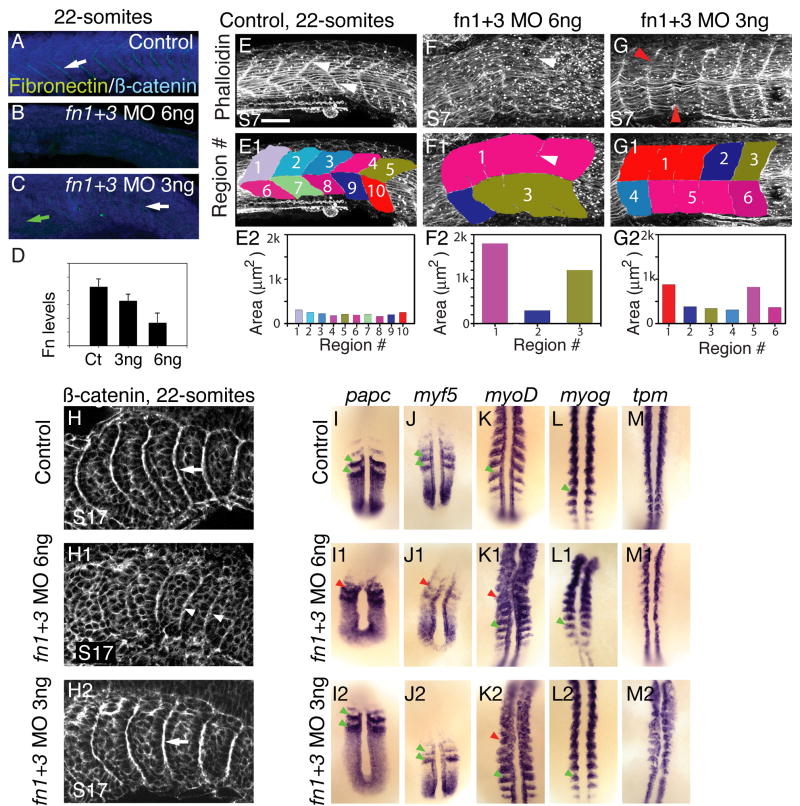
Figure 1. A reduction in Fn levels disrupts formation of anterior somites.
A–D: Side views, anterior left, dorsal top, of 22-somite embryos stained for Fn (green) and β-catenin (blue) to visualize all cells. Somite 10, or the approximate region of somite 10, is to the left.
In control embryos (A), Fn is concentrated at segment boundaries (white arrow). Injection of 6ng fn1+3 MOs results in a strong reduction Fn protein (B). In embryos injected with 3 ng fn1+3 MOs, Fn is reduced anteriorly (C, green arrow). Posteriorly, Fn concentration at boundaries is stronger, but not as strong as control posterior boundaries (white arrow). D: Quantification of levels of Fn in 3 ng and 6 ng morphants (y axis is arbitrary units). Fn protein is reduced in 3ng fn1+3 morphants and strongly reduced in 6 ng fn1+3 morphants.
E–G: Side views, anterior left, dorsal top, of 22-somite embryos stained with phalloidin to outline cells. Somite 7, or the approximate region of somite 7 is to the left. In control embryos, segment boundaries are clearly visible (white arrowheads, E). Segment boundaries were interactively traced (E1, see Materials and Methods). The myotomes in control embryos are approximately the same size (E2). In 6 ng fn1+3 MO-injected embryos, anterior myotome boundary formation is severely disrupted (F, F1, white arrowheads indicate a tiny myotome boundary in the original data and the same image with myotome regions overlaid) and myotomes areas are large and irregular. Anterior myotomes are less disrupted in 3 ng fn1+3 MO-injected embryos (G, red arrowheads indicate where boundaries should be). Scale bar 50μm.
H: Side views, anterior left, dorsal top, of 22-somite embryos stained with β-catenin, somite 17 or the approximate region thereof is on the left. In control embryos somite boundaries are seen as bright white lines (H, white arrow). In embryos injected with 6ng of fn1+3 MOs imperfect posterior somite boundaries form (H1, white arrowheads). Robust somite boundaries form in the posterior of 3ng fn1+3 morphant embryos (H2, white arrow).
I–M: Dorsal views, anterior top, of 18-somite embryos. Analysis of mRNA expression correlates with the morphological analysis in that injection of 3 ng fn1+3 MOs disrupts anterior, but not posterior, patterning. Note that gene expression patterns in the posterior presomitic mesoderm such as papc and myf5 is segmental in both control and 3 ng fn1+3 MO-injected embryos (I, I2, J, J2, green arrowheads) but the segmental pattern is slightly disrupted in 6 ng fn1+3 MO-injected embryos (I1,J1, red arrowheads). myoD is expressed in stripes in control (K, green arrowhead) embryos throughout the A-P axis. Striped expression of myoD is disrupted in the anterior of 3 and 6 ng fn1+3 MO-injected embryos (K1, K2, red arrowheads) but is segmental posteriorly (K1, K2, green arrowheads). The expression patterns of myogenin (myog) and tropomyosin (tpm) are similiar, but levels of myog and tpm in the posterior of 6 ng fn1+3 morphants are qualitatively reduced.
We do observe a dose-dependent correlation between Fn protein levels and segmentation, which is quantifiable by measuring myotome area. Myotome formation is a dynamic process that proceeds anteriorly to posteriorly. Thus, in 22-somite stage embryos, myotomes have formed in anterior segments but somite formation is ongoing in the posterior of the embryo. Myotome boundaries were interactively traced utilizing the Zeiss Axiovision Automeasure module (see Materials & Methods). Because the dorsal and ventral halves of the myotome in fn1+3 morphant embryos frequently differ, we assayed the dorsal and ventral halves separately. In the trunk of 22-somite control embryos, the boundaries between segments are clearly visible (Fig. 1 E arrowheads, somite 7 is to the left) and segment area is consistent among myotomes (Fig. 1 E1, E2). This regular area reflects the organized, segmentally reiterated nature of control myotomes.
Few segment boundaries form in 6 ng fn1+3 knockdowns (Fig. 1 F). Both slow-twitch and fast-twitch fibers are highly disorganized and full-length myotome boundaries are not clear. Interactive tracing of all visible myotome boundaries (Fig. 1 F, F1, white arrowheads point to partial boundaries) highlights the disrupted nature of myotome morphogenesis. No boundaries transverse the entire dorsal half of the myotome, resulting in a very large region (Fig. 1 F1, light purple region numbered 1). Comparison of the areas of the 3 regions within this embryo highlights the inconsistent shapes of myotomes (Fig. 1 F2).
Although myotome boundaries form in the anterior of 22-somite 3ng fn morphant embryos, boundary formation is disrupted. In particular, some fast-twitch muscle fibers are aberrantly long and traverse multiple myotomes (Fig. 1 G, red arrowheads point to unusually long fast fibers). The presence of aberrantly long fast muscle fibers results in a myotome boundary that does not persist throughout the dorsal-ventral extent of the musculature. Interactive tracing of myotome boundaries reveals the irregular nature of myotome boundary formation: whereas some regions exhibit relatively normal areas (Fig. 1 G1, G2, regions 2,3,4,6), other regions are unusually large (regions 1,5).
These data indicate that muscle morphogenesis is disrupted in embryos with reduced or abrogated levels of Fn protein. One confounding factor is that myotomes are derived from somites and somite patterning in Fn deficient embryos is also disrupted (Julich et al., 2005; Koshida et al., 2005). The somite boundary is known to stop elongating myoblasts (Henry et al., 2005). Thus, it is difficult to distinguish whether defects in muscle morphogenesis are due to: (1) a requirement of Fn during muscle morphogenesis or (2) an indirect consequence of disrupted somite boundary formation. In order to asses a possible role for Fn subsequent to somite formation, during muscle morphogenesis, we asked if somites formed when levels of Fn protein were not obliterated, but instead reduced. Therefore, we examined early segmentation in 6 ng morphants, compared with 3 ng morphants.
Elimination of Fn protein (6 ng fn1+3 MOs) disrupts somite boundary formation throughout most of the anterior-posterior axis (Fig. 1 H1, white arrowheads indicate slightly disrupted posterior somite boundaries). Molecular analysis also indicates that somite patterning is disrupted when Fn is eliminated. In 18-somite control embryos, the cell-cell adhesion gene papc and the myogenic regulatory factor myf5 are expressed in two stripes that correspond to the primordia of somites 19 and 20 (Fig. 1 I, J, green arrowheads). Segmental expression of papc and myf5 is disrupted in the primordia of somites 19 and 20 in 6 ng morphants (Fig. 1 I1, J1). Although myoD is robustly expressed in 6 ng fn1+3 morphant embryos (Fig. 1 K1), posterior myogenin (myog) and tropomyosin (tpm) expression are qualitatively reduced (Fig. 1 L1, M1). Analysis of somite boundary morphogenesis also indicates that segmentally reiterated somite border formation is disrupted in 6ng fn1+3 morphant embryos. Thus, inhibition of Fn severely disrupts somite boundary morphogenesis and patterning.
Reduction, but not abrogation, of Fn protein via injection of 3 ng fn1+3 morpholinos results in disrupted segmentation in the anterior of 22-somite embryos but morphologically normal posterior somites form (Fig. 1 H2). Molecular analysis also indicates that reduction of Fn disrupts anterior but not posterior somite formation. Both papc and myf5 are segmentally expressed in the primordia of somites 19 and 20 in 18-somite 3 ng fn1+3 morphant embryos (Fig. 1 I2, J2, green arrowheads). myoD, myogenin (myog) and tropomyosin (tpm) are also segmentally expressed in the posterior of 18-somite 3 ng fn1+3 morphant embryos (Fig. 1 K2, L2, M2). Taken together, these data indicate that although anterior somite formation and/or maintenance is disrupted in 3 ng fn1+3 morphant embryos, both patterning and somite boundary formation in posterior somites occur relatively normally.
Fast-twitch and slow-twitch muscle fiber lengths are uncoupled in Fn morphant embryos
The above results indicate that although anterior somite formation is disrupted in 3ng fn morphant embryos, posterior somites do form. Analysis of the posterior limit of defects (PLD) through time indicates that anterior segmentation is consistently more disrupted than posterior segmentation. We defined the PLD as being the point along the anterior-posterior axis at which segmentally reiterated boundaries formed. The PLD does not indicate the point at which perfect boundaries form, but rather the point at which robust, consistent boundaries form (Fig. 2 A). The PLD in both 3ng and 6ng fn1+3 morphants is relatively consistent through 48 hpf (Fig. 2 B). This indicates segmentation is more robust in the posterior trunk and tail of fn morphant embryos.
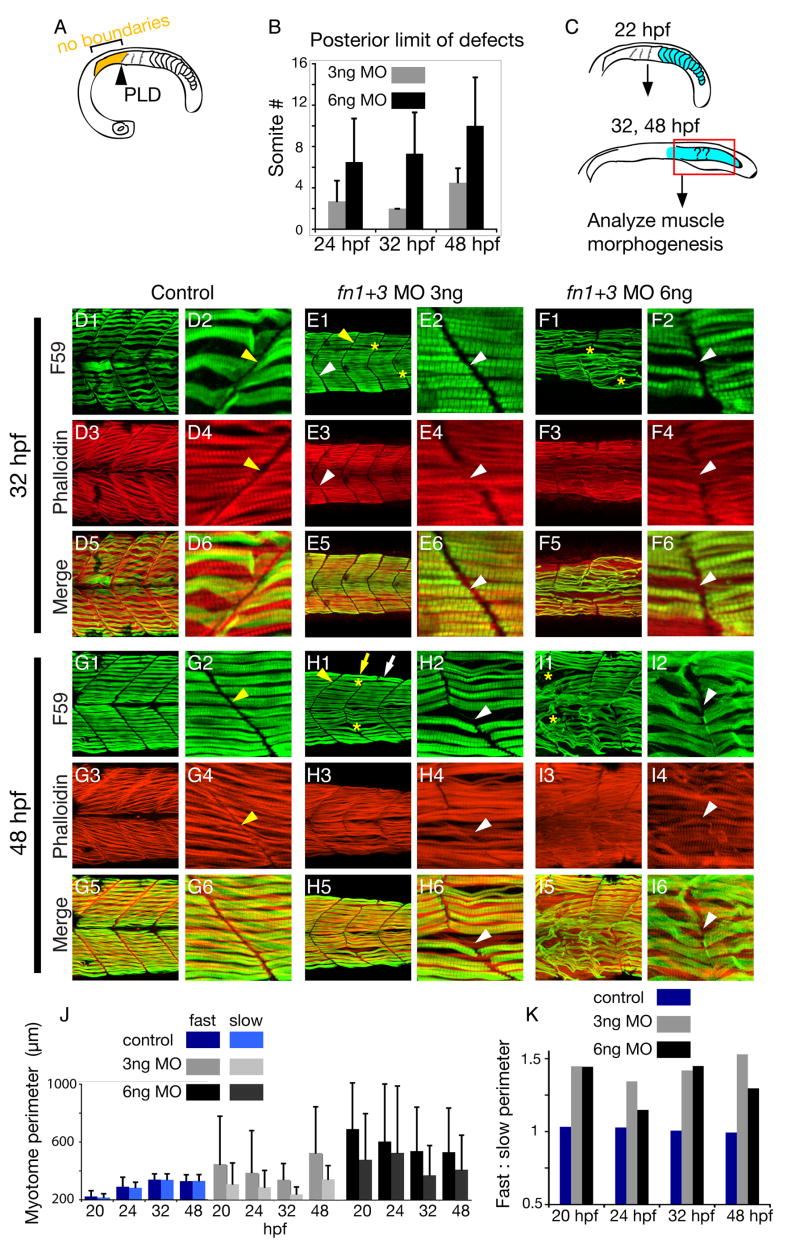
Figure 2. Uncoupling of fast- and slow-twitch muscle morphogenesis in fn morphants.
A: Cartoon showing the qualitative definition of the posterior limit of defects (PLD) that was used in this study. The PLD was considered to be the anterior most somite at which segmentally reiterated somite boundaries (albeit disrupted) were observed.
B: Graph depicting posterior limit of boundary defects of embryos injected with 3 or 6 ng fn 1+3 MOs. Morphant myotome defects are examined at three time points: 24 (n = 11 embryos), 32 (n = 20 embryos), and 48 hpf (n = 13 embryos). Average PLD ± standard deviation is shown.
C: Cartoon depicting the region of analysis in the embryo. Because posterior somites form in 3 ng fn1+3 morphant embryos, subsequent muscle morphogenesis was analyzed only in the posterior region (> somite 8) of 32 and 48 hpf embryos.
D–I: Side views, anterior left, dorsal top, of embryos stained with F59 (green) to visualize heavy chain myosin (slow-twitch muscle) and phalloidin (red) denoting filamentous actin (fast-twitch muscle).
D, G: In control embryos, MTJ boundaries are observed in slow-twitch and fast-twitch muscle (D, G, yellow arrowheads). When the two domains are overlaid, it is clear that the fast-twitch domain mirrors the slow-twitch domain (D5, D6). At 48 hpf, control embryos exhibit similar muscle morphology, with clear slow- and fast-twitch myotome boundaries and the two domains line up (G5, G6).
E, H: Uncoupling of MTJ boundaries in the fast-and slow-twitch muscle domains in 3 ng fn1+3 morphant embryos. Although there are gaps in the slow-twitch domain (E1, H1, yellow asterisks), MTJ boundaries are readily apparent (yellow arrowheads). Fast-twitch fibers often cross the myotome boundary denoted by slow fibers (E4, H4, white arrowheads). This uncoupling of fast-twitch and slow-twitch morphogenesis is readily apparent when the images are overlaid (E6, H6, white arrowheads).
F, I: Embryos injected with 6 ng fn 1+3 MOs display more severe disruptions of MTJ boundaries, fiber morphology, and uncoupling of MTJ boundaries in the fast-twitch and slow-twitch muscle domains.
J: Graph showing fast-twitch fiber and slow-twitch segment perimeters through time. In control embryos, fast-twitch and slow-twitch segment domains are approximately the same size (n = 165 myotomes). In contrast, fast and slow-twitch segment domains in fn1+3 morphant embryo are more variable (n = 126 3 ng fn morphant myotomes and n = 152 6 ng fn morphant myotomes). The trend towards a larger fast-twitch domain in fn1+3 morphant embryos reflects the uncoupling of fast-twitch and slow-twitch muscle morphogenesis.
K: Graph depicting ratio of fast-twitch to slow-twitch muscle domain segment perimeter through time. The ratio was obtained by dividing the average fast-twitch myotome perimeter by the average slow-twitch myotome perimeter. There is approximately a 1:1 ratio of fast to slow muscle segment perimeters in control embryos. This ratio is increased in fn1+3 morphant embryos, indicating that fast-twitch muscle domains are larger than the corresponding slow-twitch muscle domains.
The above data suggest that posterior somite formation is relatively normal in 3ng fn1+3 morphant embryos, but anterior somite formation is disrupted. In order to address potential roles for Fn during muscle morphogenesis, we did not analyze anterior muscle morphogenesis. Instead, we analyzed subsequent muscle morphogenesis in the region of the embryo where somites had formed, i.e. in the posterior of fn1+3 morphant embryos (Fig. 2 C). Thus, all subsequent analyses were performed in the posterior trunk and tail.
Both slow-twitch (green) and fast-twitch (red) fibers in control embryos are organized at 32 and 48 hpf (Fig. 2 D, G). Slow-twitch fibers are regularly spaced and are of constant length (Fig. 2 D1, G1). Higher magnification views show the myotome boundary, the nascent MTJ, very clearly (yellow arrowheads in Fig. 2 D2, G2). Fast-twitch muscle is medial to slow-twitch fibers at this stage. A projection of the fast-twitch muscle fibers shows the organized nature of control fast fibers (Fig. 2 D3, G3). The MTJ boundary in the fast-twitch muscle domain corresponds to the MTJ boundary in the slow-twitch muscle domain (compare Fig. 2 D2 with D4, and Fig. 2 G2 with G4). Thus, the MTJ persists throughout the slow-twitch and fast-twitch (medial-lateral) domains in control embryos.
There are two disruptions of muscle morphogenesis in 3ng fn1+3 morphant embryos. One is that slow fibers are not as organized as in control embryos. Some slow-twitch fibers are unusually long (Fig. 2 H1, yellow arrow) and others are unusually short (Fig. 2 H1, white arrow). Gaps in slow-twitch fibers are also observed (Fig. 2 E1, H1, yellow asterisks). The second major disruption in muscle development is that slow-twitch and fast-twitch muscle lengths are uncoupled in 3ng fn1+3 morphant embryos. Fast-twitch fibers cross MTJ boundaries denoted by slow-twitch fibers (Fig. 2 E2, E4, the arrowhead points to a boundary in the slow-twitch fibers, E2, that is crossed by medial fast-twitch fibers, E4, also see H2 and H4). Note that a 3-dimensional projection of fast-twitch fibers is shown, indicating that the disruption is present throughout the fast-twitch musculature. Thus, the MTJ does not persist throughout the medial-lateral extend of the musculature as it does in control embryos.
Muscle development in 6ng fn1+3 morphant embryos exhibits the two main disruptions mentioned above, but with greater severity. Slow-twitch fibers are much less organized and abnormally long or short and large gaps (Fig. 2 F1, I1, yellow asterisks) are observed. Fast-twitch fibers also cross MTJ boundaries denoted by slow-twitch fibers (Fig. 2 F2, F4, and I2, I4). Taken together, these data suggest that Fn plays a role in both slow-twitch fiber organization and in the coupling of MTJ boundaries in the slow-twitch and fast-twitch domains.
The uncoupling of MTJ formation between the slow-twitch and fast-twitch domains in fn morphant embryos is reflected when the perimeter of myotomes in slow-twitch and fast-twitch domains is analyzed. MTJs in slow-twitch and fast-twitch muscle domains were interactively traced such that the myotome flanked by MTJs was outlined. Myotome perimeters were measured with the Zeiss Axiovision Automeasure software module at 20, 24, 32, and 48 hpf. In control embryos, MTJs persist throughout the medial-lateral axis, for slow-twitch and fast-twitch domains. Thus, the perimeter of fast-twitch and slow-twitch myotomes is fairly consistent (Fig. 2 J, dark and light blue bars) and the ratio of fast to slow perimeter is approximately 1 (Fig. 2 K, blue bars). In contrast, the perimeter of fast-twitch myotomes is larger than the perimeter of slow-twitch myotomes in fn morphant embryos (Fig. 2 J, gray and black bars) and the ratio of fast to slow twitch perimeter is greater than 1 (Fig. 2 K). Thus, quantification of myotome perimeter supports the qualitative assessment that fast- and slow-twitch muscle morphogenesis is uncoupled in fn morphant embryos.
The muscle phenotype of fn1+3 morphants is distinctly different than a Notch pathway mutant
The above data indicate that Fn is required for normal muscle morphogenesis. Muscle development is disrupted in the posterior of 3ng fn1+3 morphants, where initial somite boundaries formed. This fact suggests that Fn is required subsequent to somite boundary formation. However, the possibility that Fn in initial somite boundaries regulates muscle development cannot be excluded. In order to address this issue, we analyzed muscle development in a mutant where initial somites do not form. aei/deltaD mutant embryos have disrupted somite formation posterior to somite 8 (Holley et al., 2000), but Fn protein is expressed in these embryos (Henry et al., 2005). If Fn is indeed required for muscle morphogenesis outside of its role in somite formation, then we would expect that muscle morphogenesis in 3ng fn1+3 morphant embryos would be more disrupted than muscle morphogenesis in aei/deltaD mutant embryos. We thus assayed 3 aspects of muscle morphogenesis in the posterior (posterior to somite 8): (1) uncoupling of fast-twitch and slow-twitch fiber lengths, (2) the relative lengths of fast-twitch and slow-twitch fibers, and (3) anisotropy of fast-twitch and slow-twitch muscle fibers.
Although the MTJ is derived from the initial epithelial somite boundary, somite boundary formation is not required for MTJ formation. This is evidenced by the fact that in many zebrafish Notch pathway mutants, initial somite boundary formation is disrupted; however MTJs do form by 26 hpf (van Eeden et al., 1998; Henry et al., 2005; Julich et al., 2005). MTJs boundaries are not regularly spaced or V-shaped, but they persist throughout both the slow-twitch and fast-twitch muscle domains (Henry et al., 2005). Significantly, fast-twitch and slow-twitch lengths are coupled in 92% of boundaries in aei/deltaD mutant embryos (203/220), but only 69% of boundaries in 3ng fn1+3 morphant embryos (166/240, Fig. 3). Fast-twitch fibers are longer than slow-twitch fibers in 31% of boundaries in 3ng fn1+3 morphant embryos (Fig. 3 D, D1, F, F1, 74/240 boundaries assessed). Slow-twitch fibers in 31 (13%) of the boundaries showed minor disruptions and fast-twitch fibers were longer than slow-twitch fibers in all of these boundaries (Fig. 3 D, D1). Interestingly, F59 antibody staining for slow-twitch muscle did not reveal any slow-twitch fiber defects, but fast-twitch fibers were longer than slow-twitch fibers at 18% (43/240) of the boundaries (Fig. 3 F, F1). Thus, fast-twitch and slow-twitch muscle fiber morphogenesis are more frequently uncoupled in 3ng fn1+3 morphant embryos than in aei/dlD mutants where initial somite boundaries did not form.
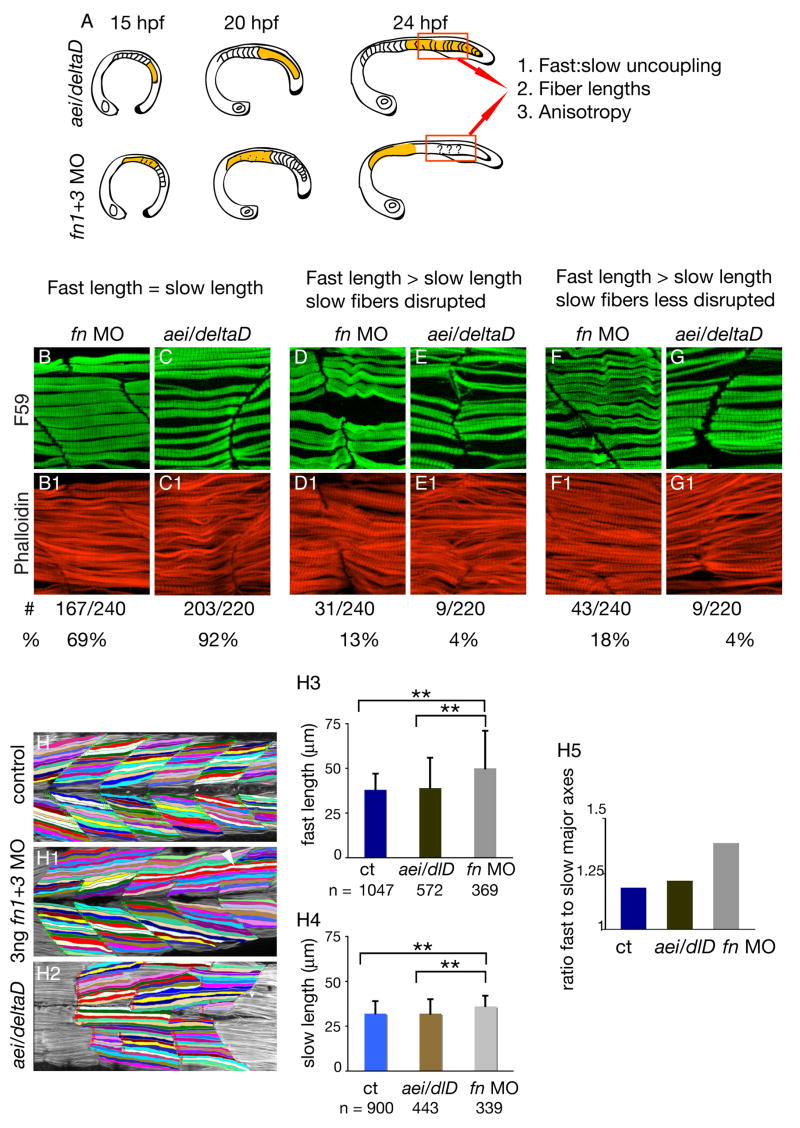
Figure 3. Muscle morphogenesis in fn morphant embryos is more disrupted than in aei/deltaD mutant embryos.
A: Summary of experimental design. Muscle development in the posterior of aei/deltaD mutant embryos, where initial somite boundaries did not form, was compared to muscle development in the posterior of 3 ng fn1+3 morphant embryos, where initial somite boundaries did form. Three phenotypes were assessed: uncoupling of fast and slow fiber lengths, absolute fast and slow fiber lengths, and anisotropy (a quantitative measure of organization, see Fig. 4).
B–G: Fast-twitch and slow-twitch fiber lengths are more frequently uncoupled in fn morphant embryos than in aei/deltaD mutant embryos. Side views, anterior left, F59 (green) denotes slow-twitch muscle and phalloidin (red) denotes fast-twitch muscle. B, C: Fast-twitch and slow-twitch fibers are approximately the same length at 69% of boundaries in fn1+3 morphant embryos, and at 92% of boundaries in aei/deltaD morphant embryos. D, E: When slow-twitch fibers exhibit disruptions such as gaps, fast-twitch fibers are longer than slow-twitch fibers. F, G: Fast-twitch fibers are sometimes longer than slow-twitch fibers when slow-twitch fibers either appear normal or show only minor disruptions.
H: Fast-twitch fibers are longer in fn1+3 morphant embryos than in aei/deltaD mutant embryos. H-H2: Fibers were interactively traced in Axiovision and the major axes calculated. Note that some fibers are unusually long (white arrowhead). H3, H4: Average fast-twitch and slow-twitch fiber lengths are significantly longer in fn morphant embryos than in either control or aei/deltaD mutant embryos (p < 0.01). H5: The ratio of fast-twitch to slow-twitch fiber lengths is higher in fn 1+3 morphant embryos. This increase is indicative of the more frequent uncoupling of fast-twitch and slow-twitch fiber lengths in fn morphant embryos.
The above data suggest that Fn regulates fast-twitch fiber length. Thus, we hypothesized that the average length of fast-twitch muscle fibers in fn1+3 morphant embryos would be longer than in control embryos. We quantified fiber length by interactively tracing individual slow and fast twitch muscle fibers using the Zeiss Axiovision Automeasure software module, which calculated the major axis (Fig 3 H–H5). The major axis is the longest length of the best fitting ellipse. Fast-twitch fibers in fn1+3 morphant embryos are not only significantly longer than control fast-twitch fibers, but also than aei/deltaD fast-twitch fibers (Fig. 3 H3). Interestingly, slow-twitch fibers are significantly longer in fn1+3 morphant embryos than in control and aei/deltaD mutant embryos (Fig. 3 H4).
In control embryos, fast-twitch fibers are angled slightly towards the midline, but slow-twitch fibers are oriented more parallel to the anterior-posterior axis. Thus, fast-twitch fibers are slightly longer than slow-twitch fibers in control embryos. The ratio of fast fiber to slow fiber length in control embryos is slightly above 1 (1.19). The fast:slow fiber length ratio in aei/deltaD mutants is similar to controls (1.22). Significantly, the fast:slow fiber length ratio in 3 ng fn1+3 morphant embryos is larger than in control and aei/deltaD mutant embryos (1.39) (Fig. 3 H5). Thus, quantification of muscle fiber length indicates that muscle fiber morphogenesis is more disrupted in fn1+3 morphant embryos that in aei/deltaD mutant embryos.
Finally, in order to rigorously assess the structural organization of muscle fibers in fn1+3 morphant and aei/deltaD mutant embryos, the 2D wavelet transform modulus maxima method (Arneodo et al., 2000; Arneodo et al., 2003; Khalil et al., 2006) was used to characterize the anisotropic signature found in the muscle fiber images. Isotropy is uniformity in all directions, and in this context can be thought of as a randomly organized structure. In contrast, anisotropy is the property of having directional dependence and within this context can be thought of as a measure of cellular organization. As described in the Materials & Methods section, the 2D WTMM method is perfectly suited to quantify structure by performing a statistical analysis of the angles of specific image points in the analyzed images where the intensity variation was locally maximal (i.e. the WTMMM, see Materials and Methods). The anisotropy factor Fa is then calculated from the probability density function (pdf) of the angles of these WTMMM vectors.
The formalism was used to quantify the strength of the anisotropic signature found in the slow fibers from the aei/deltaD mutants in order to compare it to the slow fibers from the fn1+3 morphants. A visual inspection of Figure 4 B1, B2 clearly shows that the WTMMM vector arrows are almost all aligned vertically in the aei/deltaD mutant image (green arrows). Correspondingly, the angle pdf associated to the aei/deltaD mutants shown in Figure 4 C (blue curve) is very strongly peaked at ± π/2. This means that the arrows are either pointing up (+90 degrees = +π/2 radians with respect to an imaginary horizontal line) or down (−90 degrees = −π/2 radians). The majority of WTMMM vector arrows from the fn1+3 morphants (green arrows in Figure 4 A1, A2) are also aligned either up or down. However, a visual inspection clearly shows that several are aligned at different angles. Correspondingly, the peak of the angle pdf is not as high (Fig. 4 C, red curve). This lower peak indicates a wider variety of WTMMM vector arrows in fn morphants than in aei/deltaD mutants. The calculated anisotropy factors unambiguously quantify this information (Fig. 4 D, D1). Both slow-twitch and fast-twitch fibers in 3ng fn1+3 morphants have a significantly lower anisotropy factor, indicating a more unstructured, random organization. This analysis was performed on 20 images of 256 × 256 pixels each for both sets of data.
For calibration purposes the angle pdf of the WTMMM vectors was also obtained from the analysis of 20 isotropic fBm images of 256 × 256 pixels. The angle pdf (black curve shown in Figure 4, panel C) is clearly fluctuating very closely to the theoretical ½π pointed black flat curve shown in Figure 4C. The calculation of the corresponding anisotropy factor Fa =0.12 +/− 0.02 is very close to 0 and thus clearly demonstrates the robustness of the formalism.
Our data indicate that Fn is not only required for normal somite formation, but also for subsequent muscle development. Specifically, either reduction or abrogation of Fn protein results in uncoupling of fast-twitch and slow-twitch fiber lengths. Two lines of evidence suggest that Fn may be required subsequent to somite formation. One, posterior somites form when Fn levels are reduced but not inhibited. In these embryos, muscle development is disrupted in the posterior segments, where initial somites formed. The fact that somites formed but muscle development was disrupted suggests that Fn plays a role in muscle development. The second line of evidence is that the muscle defects in fn1+3 morphant embryos are distinct from muscle defects in a mutant where initial somites did not form, but Fn protein is present. Taken together, these data suggest the hypothesis that Fn is not only critical for initial somite formation, but is also required for normal slow-twitch muscle morphogenesis and coupling of fast-twitch and slow-twitch fiber lengths. Development of tools to inhibit Fn function later in development will facilitate testing of this hypothesis.
Acknowledgments
The authors would like to thank Scott Holley for helpful discussions and Xuesong Feng, Chunyue Yin, and Mary Simon for critical reading of the manuscript. The authors also thank Ian M. McNulty for initial pilot experiments that led to this work. Sincere thanks to Dr. Alain Arneodo for critical reading of the manuscript and technical discussions. CJS was supported in part by an undergraduate research award from the University of Maine as well as NIH grant P20 RR-016463 from the INBRE program of the National Center for Research Resources. This research was supported by the Muscular Dystrophy Association and also in part by NIH grant RO1 HD052934-01A1.
References
- Arneodo A, Decoster N, Kestener P, Roux S. A wavelet-based method for multifractal image analysis: from theoretical concepts to experimental applications. In: Hawkes PW, editor. Advances in imaging and electron physics. Academic Press; 2003. p. 1. [Google Scholar]
- Arneodo A, Decoster N, Roux S. A wavelet-based method for multifractal image analysis. I. Methodology and test applications on isotropic and anisotropic random rough surfaces. European Journal of Physics B. 2000;15:567–600. [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM. Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes Dev. 1997;11:2163–2175. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent AE, Tabin CJ. Developmental regulation of somite derivatives: muscle, cartilage and tendon. Curr Opin Genet Dev. 2002;12:548–557. doi: 10.1016/s0959-437x(02)00339-8. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes F, Daggett D, Bryson-Richardson RJ, Neyt C, Maule J, Gautier P, Hollway GE, Keenan D, Currie PD. Cadherin-mediated differential cell adhesion controls slow muscle cell migration in the developing zebrafish myotome. Dev Cell. 2003;5:865–876. doi: 10.1016/s1534-5807(03)00362-9. [DOI] [PubMed] [Google Scholar]
- Crawford BD, Henry CA, Clason TA, Becker AL, Hille MB. Activity and distribution of paxillin, focal adhesion kinase, and cadherin indicate cooperative roles during zebrafish morphogenesis. Mol Biol Cell. 2003;14:3065–3081. doi: 10.1091/mbc.E02-08-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MT, Stockdale FE. Myosin expression and specialization among the earliest muscle fibers of the developing avian limb. Dev Biol. 1986;113:238–254. doi: 10.1016/0012-1606(86)90126-0. [DOI] [PubMed] [Google Scholar]
- Dahm R, Geisler R. Learning from small fry: the zebrafish as a genetic model organism for aquaculture fish species. Mar Biotechnol (NY) 2006;8:329–345. doi: 10.1007/s10126-006-5139-0. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Du SJ, Devoto SH, Westerfield M, Moon RT. Positive and negative regulation of muscle cell identity by members of the hedgehog and TGF-beta gene families. J Cell Biol. 1997;139:145–156. doi: 10.1083/jcb.139.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. Stretching fibronectin. J Muscle Res Cell Motil. 2002;23:575–580. doi: 10.1023/a:1023427026818. [DOI] [PubMed] [Google Scholar]
- Gemballa S, Vogel F. Spatial arrangement of white muscle fibers and myoseptal tendons in fishes. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:1013–1037. doi: 10.1016/s1095-6433(02)00186-1. [DOI] [PubMed] [Google Scholar]
- George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- Georges-Labouesse EN, George EL, Rayburn H, Hynes RO. Mesodermal development in mouse embryos mutant for fibronectin. Dev Dyn. 1996;207:145–156. doi: 10.1002/(SICI)1097-0177(199610)207:2<145::AID-AJA3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Heasman J. Morpholino oligos: making sense of antisense? Dev Biol. 2002;243:209–214. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- Henry CA, Amacher SL. Zebrafish slow muscle cell migration induces a wave of fast muscle morphogenesis. Dev Cell. 2004;7:917–923. doi: 10.1016/j.devcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Henry CA, McNulty IM, Durst WA, Munchel SE, Amacher SL. Interactions between muscle fibers and segment boundaries in zebrafish. Dev Biol. 2005;287:346–360. doi: 10.1016/j.ydbio.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Holley SA, Geisler R, Nusslein-Volhard C. Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wave-front activity. Genes Dev. 2000;14:1678–1690. [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham KC, Brew SA, Huff S, Litvinovich SV. Cryptic self-association sites in type III modules of fibronectin. J Biol Chem. 1997;272:1718–1724. doi: 10.1074/jbc.272.3.1718. [DOI] [PubMed] [Google Scholar]
- Jowett T. Analysis of protein and gene expression. Methods Cell Biol. 1999;59:63–85. doi: 10.1016/s0091-679x(08)61821-x. [DOI] [PubMed] [Google Scholar]
- Julich D, Geisler R, Holley SA. Integrinalpha5 and delta/notch signaling have complementary spatiotemporal requirements during zebrafish somitogenesis. Dev Cell. 2005;8:575–586. doi: 10.1016/j.devcel.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Kalcheim C, Ben-Yair R. Cell rearrangements during development of the somite and its derivatives. Curr Opin Genet Dev. 2005;15:371–380. doi: 10.1016/j.gde.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Kanagawa M, Toda T. The genetic and molecular basis of muscular dystrophy: roles of cell-matrix linkage in the pathogenesis. J Hum Genet. 2006 doi: 10.1007/s10038-006-0056-7. [DOI] [PubMed] [Google Scholar]
- Khalil A, Joncas G, Nekka F, Kestener P, Arneodo A. Morphological analysis of HI features. II. Wavelet-based multifractal formalism. Astrophysical Journal Supplement Series. 2006;165:512–550. [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koshida S, Kishimoto Y, Ustumi H, Shimizu T, Furutani-Seiki M, Kondoh H, Takada S. Integrinalpha5-dependent fibronectin accumulation for maintenance of somite boundaries in zebrafish embryos. Dev Cell. 2005;8:587–598. doi: 10.1016/j.devcel.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Kragtorp KA, Miller JR. Regulation of somitogenesis by Ena/VASP proteins and FAK during Xenopus development. Development. 2006;133:685–695. doi: 10.1242/dev.02230. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Currie PD, Roy S, Schauerte H, Haffter P, Ingham PW. Control of muscle cell-type specification in the zebrafish embryo by Hedgehog signalling. Dev Biol. 1999;216:469–480. doi: 10.1006/dbio.1999.9519. [DOI] [PubMed] [Google Scholar]
- Long JH, Adcock B, Root RG. Force transmission via axial tendons in undulating fish: a dynamic analysis. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:911–929. doi: 10.1016/s1095-6433(02)00211-8. [DOI] [PubMed] [Google Scholar]
- Mallat S, Hwang WL. IEEE Trans on Information Theory. 1992;38:617. [Google Scholar]
- Mallat S, Zhong S. IEEE Trans on Patern Analysis and Machine Intelligence. 1992;14:710. [Google Scholar]
- Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389–399. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Morla A, Ruoslahti E. A fibronectin self-assembly site involved in fibronectin matrix assembly: reconstruction in a synthetic peptide. J Cell Biol. 1992;118:421–429. doi: 10.1083/jcb.118.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitgen H-O, Saupe D, Barnsley MF. The Science of fractal images. xiii. New York: Springer-Verlag; 1988. p. 312. [Google Scholar]
- Riedel-Kruse IH, Muller C, Oates AC. Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science. 2007;317:1911–1915. doi: 10.1126/science.1142538. [DOI] [PubMed] [Google Scholar]
- Rifes P, Carvalho L, Lopes C, Andrade RP, Rodrigues G, Palmeirim I, Thorsteinsdottir S. Redefining the role of ectoderm in somitogenesis: a player in the formation of the fibronectin matrix of presomitic mesoderm. Development. 2007;134:3155–3165. doi: 10.1242/dev.003665. [DOI] [PubMed] [Google Scholar]
- Schessl J, Zou Y, Bonnemann CG. Congenital Muscular Dystrophies and the Extracellular Matrix. Semin Pediatr Neurol. 2006;13:80–89. doi: 10.1016/j.spen.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Summers AP, Koob TJ. The evolution of tendon--morphology and material properties. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:1159–1170. doi: 10.1016/s1095-6433(02)00241-6. [DOI] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6:371–382. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Vakonakis I, Campbell ID. Extracellular matrix: from atomic resolution to ultrastructure. Curr Opin Cell Biol. 2007 doi: 10.1016/j.ceb.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden FJ, Holley SA, Haffter P, Nusslein-Volhard C. Zebrafish segmentation and pair-rule patterning. Dev Genet. 1998;23:65–76. doi: 10.1002/(SICI)1520-6408(1998)23:1<65::AID-DVG7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Winklbauer R, Keller RE. Fibronectin, mesoderm migration, and gastrulation in Xenopus. Dev Biol. 1996;177:413–426. doi: 10.1006/dbio.1996.0174. [DOI] [PubMed] [Google Scholar]