Abstract
DNA polymerase (Pol) λ is a DNA repair enzyme involved in base excision repair, non-homologous end joining and translesion synthesis. Recently, we identified Pol λ as an interaction partner of cyclin-dependent kinase 2 (CDK2) that is central to the cell cycle G1/S transition and S-phase progression. This interaction leads to in vitro phosphorylation of Pol λ, and its in vivo phosphorylation pattern during cell cycle progression mimics the modulation of CDK2/cyclin A. Here, we identify several phosphorylation sites of Pol λ. Experiments with phosphorylation-defective mutants suggest that phosphorylation of Thr 553 is important for maintaining Pol λ stability, as it is targeted to the proteasomal degradation pathway through ubiquitination unless this residue is phosphorylated. In particular, Pol λ is stabilized during cell cycle progression in the late S and G2 phases. This most likely allows Pol λ to correctly conduct repair of damaged DNA during and after S phase.
Keywords: cell cycle, DNA polymerase λ, phosphorylation, protein stability, ubiquitination
Introduction
DNA polymerase (Pol) λ is a DNA repair enzyme with template-dependent and -independent pol activities, as well as a 5′-deoxyribose-5-phosphate (dRP) lyase function (Ramadan et al, 2004), and has been implicated in base excision repair (Garcia-Diaz et al, 2001; Braithwaite et al, 2005b), non-homologous end joining (Fan & Wu, 2004; Lee et al, 2004; Nick McElhinny et al, 2005) and translesion synthesis (TLS; Maga et al, 2002, 2007; Picher & Blanco, 2007). Although the biochemical properties of Pol λ are well characterized, its in vivo functions are far from being understood. Targeted deletion of Pol λ has no influence on the viability and fertility of mice (Bertocci et al, 2002), and cells lacking Pol λ show no increased sensitivity towards DNA-damaging agents such as ultraviolet light, ionizing radiation or DNA-alkylating agents (Kobayashi et al, 2002; Bertocci et al, 2006). Only a striking sensitivity to oxidative stress has been reported for Pol λ-deficient fibroblasts (Braithwaite et al, 2005a; Bertocci et al, 2006), which is probably due to the specific role of Pol λ in faithful bypass of 8-oxo-G lesions, a common form of oxidative damage (Maga et al, 2007; Picher & Blanco, 2007).
Recently, we discovered that human Pol λ can interact in vitro and in vivo with cyclin-dependent kinase 2 (CDK2; Frouin et al, 2005), which is crucial for G1/S transition (CDK2/cyclin E) and S-phase progression (CDK2/cyclin A) of the cell cycle (Malumbres & Barbacid, 2005). We found that Pol λ is phosphorylated by various CDK/cyclin complexes in vitro. Furthermore, the phosphorylation pattern of Pol λ in vivo during cell cycle progression mimics the modulation of CDK2/cyclin A (Frouin et al, 2005), indicating that this complex is responsible for the modification. Here, we tried to gain further insight into the post-translational regulation of Pol λ and its in vivo function. The phosphorylation sites of Pol λ were identified, and experiments with phosphorylation-defective mutants in human cells showed that the modification of Thr 553 is important for protein stability: Pol λ is targeted to the proteasomal degradation pathway through ubiquitination, but phosphorylation at this particular residue can prevent this process. Most notably, Pol λ seems to be stabilized during cell cycle progression in the late S and G2 phases, thus providing time for the enzyme to repair lesions during these stages.
Results
Identification of phosphorylation sites of Pol λ
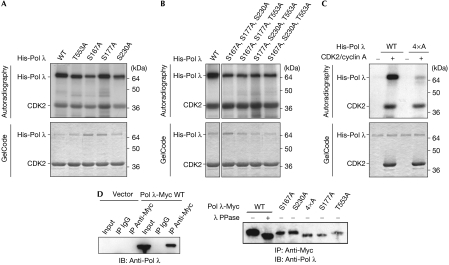
Four sites providing the minimal consensus motif Ser/Thr-Pro for CDK phosphorylation were found in the human Pol λ sequence, namely Ser 167, Ser 177, Ser 230 and Thr 553. These four residues were individually converted to alanine by site-directed mutagenesis, and His-tagged Pol λ (His-Pol λ) wild-type and mutant proteins were expressed in Escherichia coli and purified (supplementary Fig S1 online). The four single mutants showed only a slight reduction in the level of phosphorylation in an in vitro kinase assay with purified recombinant human CDK2/cyclin A (Fig 1A), and therefore the mutations were combined. Phosphorylation was still clearly detectable in the case of triple mutants (Fig 1B); however, a decrease of more than 95% was achieved if all four sites were converted to alanine (4 × A; Fig 1C), suggesting that all four residues can be phosphorylated by CDK2/cyclin A in vitro. To analyse in vivo phosphorylation, Myc-tagged Pol λ (Pol λ-Myc) wild-type, quadruple (4 × A) or single mutants were expressed in 293T cells by transient transfection and analysed by immunoprecipitation (Fig 1D). In line with our previous findings (Frouin et al, 2005), an increase in electrophoretic mobility, when compared with untreated wild type, was observed as a result of dephosphorylation of wild-type protein by λ phosphatase. A similar migration to phosphatase-treated Pol λ wild type was detected for untreated 4 × A and the S177A mutant, indicating a lack of phosphorylation for these mutants. The other single mutants showed no increase in mobility, and thus the shift seems to be a result of Ser 177 phosphorylation only. Indeed, in vivo phosphorylation of the other sites can occur without a change in electrophoretic mobility (Solan et al, 2003; Yu et al, 2005).
Figure 1.
DNA polymerase λ is phosphorylated in vitro and in vivo. (A) Human His-Pol λ WT or single mutant proteins were incubated with human CDK2/cyclin A in the presence of [γ-32P]ATP. Phosphorylated products were separated by SDS–PAGE, stained for total protein with GelCode (lower panel) and the level of phosphorylation was visualized by autoradiography (top panel). (B,C) Assay as described in (A), but comparing His-Pol λ WT with triple mutant (B) and 4 × A protein (C). (D) 293T cells were transfected with pcDNA3 vector or constructs encoding human Pol λ-Myc WT, 4 × A or single mutants. At 2 days after transfection, Pol λ proteins were immunoprecipitated (IP) with Myc antibody and rabbit IgG was used as a control. A sample of WT immune complexes was treated in addition with λ PPase. Immunoblots (IB) were probed with Pol λ antibody. 4 × A, quadruple mutant; CDK2, cyclin-dependent kinase 2; PAGE, polyacrylamide gel electrophoresis; Pol, polymerase; λ PPase, bacteriophage λ protein phosphatase; WT, wild type.
In vitro phosphorylation of Pol λ by CDK2/cyclin A does not affect its DNA Pol, its terminal deoxynucleotidyl transferase (Frouin et al, 2005) or dRP lyase activity (supplementary Fig S2 online).
Phosphorylation modulates Pol λ protein stability
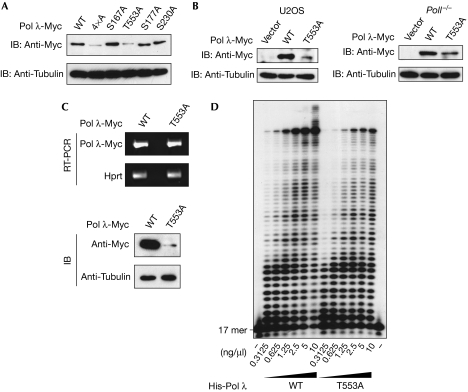
Most interestingly, expression analysis of Pol λ-Myc proteins in 293T cells showed a marked reduction in the level of protein for 4 × A, as well as the T553A mutant, when compared with wild type (Fig 2A). A similar decrease in protein amount was observed in U2OS cells and stably transduced mouse embryonic fibroblasts (MEFs) lacking Pol λ (Poll−/−; Fig 2B; supplementary Fig S5 online). Transfection of 293T cells with constructs encoding haemagglutinin (HA)-tagged (Pol λ-HA) and untagged Pol λ wild type or T553A further confirmed these data (supplementary Fig S3 online). In summary, the T553A mutant was less stable than wild type in three different cell lines: 293T, U2OS and MEFs. To ascertain that transcription of the phosphorylation-defective T553A mutant was not impaired, the level of transcripts encoding Pol λ-Myc proteins was analysed by semiquantitative reverse transcription (RT)–PCR (Fig 2C). No obvious difference in the levels of transcript was observed, although the same cells showed a clear reduction in the level of T553A mutant protein. These data indicate that phosphorylation at Thr 553 of Pol λ stabilizes the protein. However, the mutant protein might be degraded due to improper folding and elimination by cellular protein quality control mechanisms (Goldberg, 2003). The residue Thr 553 is situated in the thumb region of the Pol λ polymerization domain (Garcia-Diaz et al, 2004) and a T553A substitution resulted in a reduction of DNA pol activity to around 60% of wild type (Fig 2D; supplementary Fig S4 online). This might be due to slight alterations in, for example, dNTP binding, positioning of the primer/templating base or the stability of the closed conformation (Garcia-Diaz et al, 2004, 2005). Nevertheless, polymerization of the T553A mutant was still clearly detectable with an identical pattern to wild-type protein, and elimination by cellular quality control seems rather unlikely. Taken together, these data suggest that phosphorylation at Thr 553 modulates protein stability of Pol λ.
Figure 2.
Influence of DNA polymerase λ phosphorylation on the levels of protein and RNA. (A) 293T cells were transfected with constructs encoding Pol λ-Myc WT, 4 × A or single mutants, and the levels of protein were determined 2 days after transfection by immunoblot (IB) with the indicated antibodies. (B) U2OS cells were transfected with pcDNA3 vector or constructs encoding human Pol λ-Myc WT or T553A, and the levels of protein were determined 2 days after transfection by immunoblot. Poll−/− MEFs were stably transduced with pLPCX vector or constructs encoding Pol λ-Myc WT or T553A. (C) 293T cells were transfected and analysed by immunoblotting as described in (B). Semiquantitative RT–PCR was performed in a linear range of detection by using primers specifically amplifying mRNA encoding Pol λ-Myc. Hprt mRNA was amplified as a control. (D) Various amounts of His-Pol λ WT or T553A were incubated with a 5′-[32P] 17-mer primer annealed to a 73-mer template in the presence of 0.5 mM MnCl2 and 10 μM dNTPs. The resulting products were analysed by autoradiography after separation on a sequencing gel. 4 × A, quadruple mutant; Hprt, hypoxanthin-guanine-phosphoribosyltransferase; MEF, mouse embryonic fibroblast; Pol, polymerase; RT, reverse transcription; WT, wild type.
Pol λ phosphorylation prevents degradation
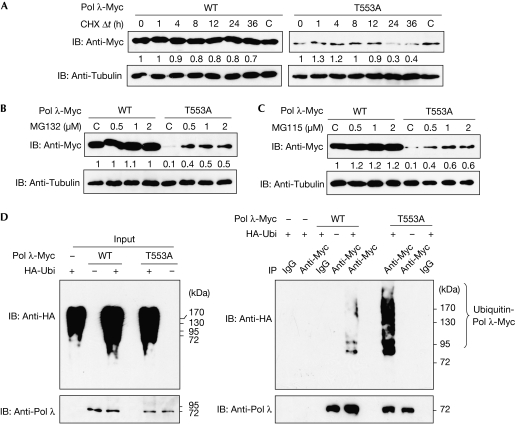
To assess further whether a lack of Thr 553 phosphorylation reduces Pol λ stability, the protein half-life of the T553A mutant was compared with wild type following treatment of transfected 293T cells with the translation inhibitor cycloheximide (Fig 3A). Indeed, the half-life of the wild-type protein was significantly longer than that of the phosphorylation-defective mutant. To investigate whether the stability of Pol λ was linked to the ubiquitin–proteasome pathway (Pickart, 2004; Elsasser & Finley, 2005), the proteasome inhibitor MG132 or MG115 was added to 293T cells transfected with constructs encoding Pol λ-Myc wild type and T553A (Fig 3B,C). The level of mutant protein was recovered with increasing amounts of both inhibitors, suggesting that Pol λ lacking Thr 553 phosphorylation is targeted for proteasomal degradation. As the proteasome recognizes and degrades proteins conjugated to polyubiquitin chains, it was tested whether Pol λ featured this covalent modification. 293T cells were transfected with plasmids encoding HA-tagged ubiquitin (HA-Ubi), as well as Pol λ-Myc proteins, and treated with MG132 (Fig 3D). Immunoprecipitation with Myc antibody under denaturing conditions showed an accumulation of polyubiquitinated Pol λ-Myc wild type and T553A. The accumulation was more pronounced in the case of the mutant protein, indicating that Pol λ is subject to ubiquitination and that phosphorylation at Thr 553 can prevent this process.
Figure 3.
Phosphorylation of DNA polymerase λ is required to maintain its stability. (A) 293T cells were transfected with constructs encoding human Pol λ-Myc WT or T553A. At 40 h after transfection, the cells were incubated in medium containing 50 μM CHX for the indicated time. As a control, the cells were incubated with DMSO alone for 36 h. The levels of protein were determined by immunoblot (IB) with the indicated antibodies. The amount of Pol λ protein was normalized to the amount of the corresponding tubulin protein, and the relative level of each sample was determined using the level of Pol λ at time point 0 h. (B,C) 293T cells were transfected as described in (A). At 1 day after transfection, the cells were exposed for 16 h to various concentrations of MG132 (B) or MG115 (C) and to DMSO as a control. The levels of protein were analysed by immunoblot; the amount of Pol λ protein was normalized to the amount of the corresponding tubulin protein and the relative level of each sample was determined using the level of Pol λ in the DMSO-treated sample. (D) 293T cells were co-transfected with constructs encoding HA-Ubi, Pol λ-Myc WT or T553A, and 1 day after transfection, the cells were treated for 16 h with 5 μM MG132. Pol λ proteins were immunoprecipitated (IP) from denatured extracts with Myc antibody, and rabbit IgG was used as a control. Input (left panel) and immunoprecipitation experiment (right panel) are shown. C, control; CHX, cycloheximide; DMSO, dimethyl sulphoxide; HA, haemagglutinin; HA-Ubi, HA-ubiquitin; Pol, polymerase; Δt, time; WT, wild type.
Phosphorylation stabilizes Pol λ in the late S and G2 phases
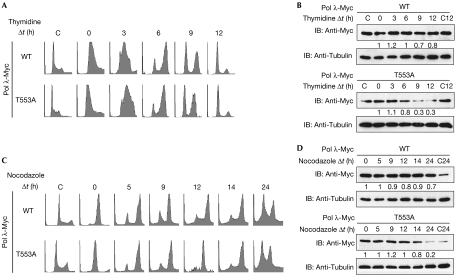
Pol λ is a substrate and interaction partner of the cell cycle regulator CDK2 (Frouin et al, 2005). Thus, we investigated whether Pol λ phosphorylation modulates its stability during cell cycle progression. 293T cells transfected with constructs encoding Pol λ-Myc wild type or T553A were synchronized at the G1/S-phase boundary by double thymidine block and examined at different time points after release (Fig 4A,B). Pol λ wild type was present at all time points examined, with a slight decline in the level of protein between 3 and 9 h—that is, during progression through S and G2 phases (Fig 4B, top panel). However, the level of the phosphorylation-defective mutant was significantly decreased 9 h after release (Fig 4B, bottom panel). These data were confirmed by analysis of transfected 293T cells synchronized at G2/M phase by treatment with nocodazole (Fig 4C,D). Taken together, these results indicate that phosphorylation of Pol λ at Thr 553 stabilizes the protein in the late S and G2 phases of the cell cycle.
Figure 4.
DNA polymerase λ is stabilized by phosphorylation during the late S and G2 phases of the cell cycle. 293T cells were transfected with constructs encoding human Pol λ-Myc WT or T553A. The transfected cells were synchronized 1 day after transfection at the G1/S phase boundary by double blocks with 2 mM thymidine (A,B) or at G2/M phase by incubation with 300 ng/ml nocodazole (C,D) and collected immediately or released in fresh medium for the indicated time. Unsynchronized control cells were collected at time points 0, 12 or 24 h as indicated. (A,C) Synchronization and release was verified by flow cytometric analysis of DNA content. (B,D) The levels of protein were analysed by immunoblotting (IB) with the indicated antibodies. The amount of Pol λ protein was normalized to the amount of the corresponding tubulin protein, and the relative level of each sample was determined using the level of Pol λ at time point 0 h. C, control 0 h; C12, control 12 h; C24, control 24 h; Pol, polymerase; Δt, time; WT, wild type.
Discussion
In summary, we have shown that human Pol λ is degraded in vivo by the proteasome–ubiquitination pathway; however, it can be stabilized as a result of phosphorylation at Thr 553. This stabilization seems to occur in particular in the late S and G2 phases of the cell cycle, which probably allows Pol λ to fulfil its specific functions in DNA damage repair during these stages. This might be especially important for the repair of 8-oxo-G lesions, a common form of oxidative damage. During replication, dATP is wrongly incorporated opposite to 8-oxo-G by Pols δ and ɛ, and later removed by the MUTYH DNA glycosylase, leaving the lesion on the DNA. Subsequent incorporation of the correct dCTP opposite to 8-oxo-G on the gapped DNA is essential for the removal of the damaged base. We showed recently that Pol λ, together with proliferating cell nuclear antigen (PCNA) and replication protein A (RPA), incorporates the correct dCTP opposite to an 8-oxo-G template 1200-fold more efficiently than the incorrect dATP, suggesting a new mechanism to reduce the deleterious consequences of oxidative damage (Maga et al, 2007). Stabilization of phosphorylated Pol λ in the late S and G2 phases might allow Pol λ to perform the accurate dCTP incorporation before the cell enters mitosis, hence ensuring genome stability by preventing the propagation of a G:C to T:A transversion mutation. Future work aimed at identifying the key proteins involved in these processes will be necessary to completely understand this mechanism and its physiological impact on the cell.
Recently, it was shown that the base excision repair enzymes Pol β and XRCC1 are ubiquitinated by the E3 ubiquitin ligase CHIP (carboxyl terminus of Hsc70-interacting protein) and subsequently degraded by the proteasome, unless they are bound in repair complexes (Parsons et al, 2008). Both repair proteins are targets for various post-translational modifications such as methylation, acetylation and phosphorylation, and Sobol (2008) predicted that such modifications might prevent proteasome-dependent degradation by inhibiting ubiquitination. This corresponds exactly to our findings that phosphorylation protects the DNA repair protein Pol λ from degradation. It remains to be elucidated whether such a cross-talk between post-translational modifications and protein stability is a general regulatory process for repair enzymes.
Methods
Plasmids and proteins. Cloning of Pol λ constructs and purification of recombinant proteins are described in the supplementary information online.
In vitro kinase assay. His-Pol λ wild-type or mutant proteins (300–400 ng) were incubated for 30 min at 37°C with purified human CDK2/cyclin A in a final volume of 15 μl containing kinase buffer (40 mM HEPES pH 7.5, 8 mM MgCl2), 66.6 μM ATP and 3 μCi [γ-32P]ATP. The samples were separated by SDS–PAGE and stained with GelCode Blue Stain Reagent (Pierce, Rockford, IL, USA) according to the manufacturer's instructions.
Cell culture and transfection. 293T and U2OS cells, as well as immortalized Poll−/− MEFs (Bertocci et al, 2002; Maga et al, 2007), were grown at 37°C in a 5% CO2 incubator in DMEM containing GlutaMAX-I supplemented with 10% fetal bovine serum and 100 U/ml penicillin–streptomycin (all obtained from Gibco, Invitrogen, Carlsbad, CA, USA). All transfections were performed by calcium phosphate precipitation, and further details are described in the supplementary information online.
Cell treatment. Re-seeding was performed 16 h after transfection. To measure protein half-life, 1.0 × 106 cells were seeded in six-well plates, treated 24 h later with medium containing 50 μM cycloheximide (Sigma-Aldrich, St Louis, MO, USA) and collected at the time points indicated in the figure legends. For analysis of proteasomal degradation, 1.0 × 106 cells were seeded in six-well plates and various concentrations of MG132 or MG115 (both obtained from Sigma) were added 8 h later to the medium for 16 h. Double thymidine block was achieved by seeding 1.8 × 106 cells in 60-mm plates 8 h before incubation in medium with 2 mM thymidine for 16 h. The cells were washed and incubated in fresh medium for 8 h. A second 16 h incubation in 2 mM thymidine was carried out before releasing the block, and the cells were collected at the time points indicated in the figure legends. For synchronization at G2/M phase, the cells were treated 24 h after transfection with 300 ng/ml nocodazole (Sigma) for 16 h. The cells were washed, and 1.0 × 106 cells were seeded in six-well plates in a fresh medium for release and collected at the indicated time points. For cell cycle analysis, the cells were fixed in 70% ethanol, washed and incubated with 200 μg/ml of RNase A (Roche Diagnostics, Mannheim, Germany) in PBS for 30 min at 37°C. Propidium iodide (20 μg/ml; Sigma) was added and DNA content was analysed by flow cytometric analysis using a Cytomics FC 500 (Beckman Coulter, Fullerton, CA, USA).
RT–PCR. Total RNA was isolated with TRIZOL® Reagent (Invitrogen, Carlsbad, CA, USA) and RT–PCRs were performed with QIAGEN® OneStep RT–PCR Kit (Qiagen, Hiden, Germany) using 150 ng RNA. Further details are described in the supplementary information online.
Immunoblotting and immunoprecipitation. Cellular proteins were extracted using extraction buffer (50 mM Tris–Cl pH 7.5, 400 mM NaCl, 2.5 mM MgCl2, 0.5% (v/v) NP-40, 2 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml bestatin, 2 μg/ml leupeptin, 10 mM glycerophosphate, 10 mM Na4P2O7 and 1 mM NaF). Here, 10–30 μg of the respective cell extracts was used for immunoblotting with the following antibodies: anti-Myc A-14 (Santa Cruz, Santa Cruz, CA, USA), anti-HA.11 (16B12; Covance, Berkeley, CA, USA) or anti-α-Tubulin DM1A (Sigma). Densitometric analysis of immunoblots was performed with ImageJ (Rasband, 1997–2007). For immunoprecipitation, the cell extracts were diluted with hypotonic buffer—that is, extraction buffer without NaCl—to a final concentration of 100 mM NaCl. Pol λ-Myc was immunoprecipitated from 0.5 to 1 mg cell extract for 2 h at 4°C using 3 μg of Myc antibody. Immune complexes were absorbed for 1 h at 4°C on Protein A Sepharose™ CL-4B beads (GE Healthcare, Amersham, Buckinghamshire, UK) coated with BSA, and the beads were washed later 3 × with wash buffer (50 mM Tris–Cl pH 8, 0.1% (v/v) NP-40, 50 mM NaCl, 75 mM KCl, 2 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml bestatin, 2 μg/ml leupeptin, 10 mM glycerophosphate, 10 mM Na4P2O7 and 1 mM NaF). As indicated in the figures, some samples were also washed twice with λ protein phosphatase buffer (New England Biolabs, Ipswich, MA, USA) and incubated for 30 min at 30°C with 600 U λ protein phosphatase (New England Biolabs) in a final volume of 100 μl λ protein phosphatase buffer. All immune complexes were eluted from beads by heating for 10 min at 95°C in 2 × SDS sample buffer and analysed by immunoblotting. Detection of ubiquitination was performed as described previously (El-Shemerly et al, 2005).
DNA Pol assay (primer extension). The reaction mixture (10 μl) included Pol buffer (50 mM Tris-CL pH 7.5, 0.25 mg/ml BSA, 1 mM DTT), 0.5 mM MnCl2, 10 μM dNTPs and 20 fmol 5′-[32P]17-mer primer annealed to a 73-mer template (Frouin et al, 2005). The reactions were incubated for 15 min at 37°C, stopped by the addition of 10 μl 2 × loading buffer (96% (v/v) formamide, 20 mM EDTA pH 8.0) and heated for 3 min at 95°C. 10 μl were loaded on a 10% polyacrylamide, 7 M urea sequencing gel and examined by autoradiography.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
This paper is dedicated to Arthur Kornberg, who discovered DNA polymerase. We thank H.P. Nasheuer for providing pGEX2T-suc1 and baculoviruses for CDK2/cyclin A, D. Bohmann for HA-ubiquitin plasmid, O. Georgiev for competent E. coli and M. El-Shemerly for help with the ubiquitination analysis. This study was supported by the Swiss Cancer League (Oncosuisse; 01996-02-2007) and the Swiss National Science Foundation (3100A0-109312).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bertocci B, De Smet A, Flatter E, Dahan A, Bories JC, Landreau C, Weill JC, Reynaud CA (2002) Cutting edge: DNA polymerases μ and λ are dispensable for Ig gene hypermutation. J Immunol 168: 3702–3706 [DOI] [PubMed] [Google Scholar]
- Bertocci B, De Smet A, Weill JC, Reynaud CA (2006) Nonoverlapping functions of DNA polymerases μ, λ, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity 25: 31–41 [DOI] [PubMed] [Google Scholar]
- Braithwaite EK et al. (2005a) DNA polymerase λ protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem 280: 31641–31647 [DOI] [PubMed] [Google Scholar]
- Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH (2005b) DNA polymerase λ mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem 280: 18469–18475 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Finley D (2005) Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol 7: 742–749 [DOI] [PubMed] [Google Scholar]
- El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari S (2005) Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res 65: 3604–3609 [DOI] [PubMed] [Google Scholar]
- Fan W, Wu X (2004) DNA polymerase λ can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4–ligase IV complex. Biochem Biophys Res Commun 323: 1328–1333 [DOI] [PubMed] [Google Scholar]
- Frouin I, Toueille M, Ferrari E, Shevelev I, Hübscher U (2005) Phosphorylation of human DNA polymerase λ by the cyclin-dependent kinase Cdk2/cyclin A complex is modulated by its association with proliferating cell nuclear antigen. Nucleic Acids Res 33: 5354–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L (2001) Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase λ: a possible role in base excision repair. J Biol Chem 276: 34659–34663 [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Krahn JM, Blanco L, Kunkel TA, Pedersen LC (2004) A structural solution for the DNA polymerase λ-dependent repair of DNA gaps with minimal homology. Mol Cell 13: 561–572 [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC (2005) A closed conformation for the Pol λ catalytic cycle. Nat Struct Mol Biol 12: 97–98 [DOI] [PubMed] [Google Scholar]
- Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y et al. (2002) Hydrocephalus, situs inversus, chronic sinusitis, and male infertility in DNA polymerase λ-deficient mice: possible implication for the pathogenesis of immotile cilia syndrome. Mol Cell Biol 22: 2769–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF (2004) Implication of DNA polymerase λ in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem 279: 805–811 [DOI] [PubMed] [Google Scholar]
- Maga G, Villani G, Ramadan K, Shevelev I, Tanguy Le Gac N, Blanco L, Blanca G, Spadari S, Hübscher U (2002) Human DNA polymerase λ functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem 277: 48434–48440 [DOI] [PubMed] [Google Scholar]
- Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hübscher U (2007) 8-Oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 447: 606–608 [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M (2005) Mammalian cyclin-dependent kinases. Trends Biochem Sci 30: 630–641 [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA (2005) A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell 19: 357–366 [DOI] [PubMed] [Google Scholar]
- Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL (2008) CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol Cell 29: 477–487 [DOI] [PubMed] [Google Scholar]
- Picher AJ, Blanco L (2007) Human DNA polymerase λ is a proficient extender of primer ends paired to 7,8-dihydro-8-oxoguanine. DNA Repair 6: 1749–1756 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2004) Back to the future with ubiquitin. Cell 116: 181–190 [DOI] [PubMed] [Google Scholar]
- Ramadan K, Shevelev I, Hübscher U (2004) The DNA-polymerase-X family: controllers of DNA quality? Nat Rev Mol Cell Biol 5: 1038–1043 [DOI] [PubMed] [Google Scholar]
- Rasband WS (1997–2007) ImageJ. Bethesda, Maryland, USA: National Institutes of Health http://rsb.info.nih.gov/ij/ [Google Scholar]
- Sobol RW (2008) CHIPping away at base excision repair. Mol Cell 29: 413–415 [DOI] [PubMed] [Google Scholar]
- Solan JL, Fry MD, TenBroek EM, Lampe PD (2003) Connexin43 phosphorylation at S368 is acute during S and G2/M and in response to protein kinase C activation. J Cell Sci 116: 2203–2211 [DOI] [PubMed] [Google Scholar]
- Yu CT, Hsu JM, Lee YC, Tsou AP, Chou CK, Huang CY (2005) Phosphorylation and stabilization of HURP by Aurora-A: implication of HURP as a transforming target of Aurora-A. Mol Cell Biol 25: 5789–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information