Abstract
Transforming growth factor-β (TGFβ) induces the expression of the pro-apoptotic protein BIM, and mediates apoptosis in hepatocytes and B lymphocytes. BIM is regulated through a post-translational mechanism involving ERK-dependent phosphorylation and ubiquitin-mediated proteasomal degradation. Here, we show that TGFβ induces BIM through its rapid inhibition of ERK, thereby preventing the phosphorylation and degradation of BIM. TGFβ, through a SMAD3-dependent mechanism, transcriptionally induces the mitogen-activated protein kinase (MAPK) phosphatase MKP2, encoded by an immediate early gene, to attenuate ERK and promote the accumulation of BIM protein. Overexpression of MKP2 in hepatocytes modulates ERK-mediated phosphorylation of BIM and apoptosis in the absence of TGFβ, whereas its ablation in pro-B cells, derived from MKP2-deficient mice, protects cells from TGFβ-mediated apoptosis, and blocks TGFβ-induced ERK inhibition and BIM induction. Furthermore, in pro-B cells derived from SMAD3-deficient mice, induction of MKP2 by TGFβ, inhibition of ERK, induction of BIM and apoptosis do not occur. Our results indicate that MKP2 mediates TGFβ-dependent apoptosis by linking SMAD3 to the modulation of ERK activity and mitochondrial-mediated pro-apoptotic events.
Keywords: apoptosis, BIM, ERK, MKP2, TGFβ
Introduction
Transforming growth factor-β (TGFβ) regulates essential cellular functions ranging from cellular proliferation and differentiation to apoptosis (Siegel & Massague, 2003). Signalling by TGFβ is initiated by a receptor complex consisting of two subunits, the type I (TβRI) and type II (TβRII) receptors, each having Ser/Thr kinase activity (Siegel & Massague, 2003). Activation of the receptor complex propagates the signal to downstream signalling cascades, which include the SMADs, MAPK (mitogen-activated protein kinase) and PI(3)K (phosphatidylinositol 3-kinase; Siegel & Massague, 2003). SMAD signalling is activated by receptor-mediated phosphorylation of SMAD2 and SMAD3, which then form heteromeric complexes with SMAD4 and translocate to the nucleus to regulate the transcriptional response to TGFβ (Schmierer & Hill, 2007).
The apoptotic response to TGFβ is well documented in many cell types and several apoptotic mediators have been implicated, including the BCL2 (B-cell lymphoma 2) family of proteins (Siegel & Massague, 2003; Wildey et al, 2003; Yang et al, 2006). Previously, we have shown that TGFβ induces the expression of the pro-apoptotic protein BIM to induce cell death in B lymphocytes (Wildey et al, 2003). BIM induction has more recently been shown in gastric epithelial cells undergoing TGFβ-induced apoptosis (Ohgushi et al, 2005) and confirmed in AML-12 (acute myeloid leukaemia) hepatocytes (Ramjaun et al, 2007). BIM is a member of the BH3-only family of pro-apoptotic proteins (Huang & Strasser, 2000). It is expressed in a wide variety of tissues but most prominently by cells of haematopoietic origin (O'Reilly et al, 2000), and has been shown to be important for apoptosis in B and T lymphocytes, macrophages and granulocytes (Bouillet et al, 1999). Studies in BIM knockout mice indicate that it is required for B-cell antigen receptor (BCR)-induced apoptosis in immature and mature B cells, and for the negative selection of autoreactive B cells (Enders et al, 2003). Under cell growth conditions, BIM is bound to dynein light chain (LC8), of the microtubular motor complex, and is sequestered away from other BCL2 family members (Puthalakath et al, 1999). Following a pro-apoptotic stimulus, BIM localizes to the mitochondria, where it initiates the mitochondrial cell death pathway either by directly activating BAX-like proteins or by binding to pro-survival BCL2 family members and thereby releasing BAX-like proteins (Marani et al, 2002; Kim et al, 2006).
The expression of BIM is regulated at the transcriptional level (Dijkers et al, 2000), and post-translationally by phosphorylation and ubiquitination (Luciano et al, 2003; Ley et al, 2004; Qi et al, 2006). Phosphorylated forms of BIM were initially reported in Ba/F3 cells grown in the presence, but not the absence, of interleukin-3 (IL-3; Shinjyo et al, 2001) and were blocked by the inhibitors of the MEK/ERK (mitogen activated protein kinase kinase/extracellular signal-regulated kinase kinase) pathway, PD98059 or U0126. In vitro studies showed that purified ERK could phosphorylate BIM (Ley et al, 2004; Qi et al, 2006). Phosphorylation of BIM is associated with a time-dependent loss in total BIM protein that was blocked by lactacystin and MG-132 (Luciano et al, 2003; Ley et al, 2004), implicating the proteasomal pathway in BIM degradation. Further analysis showed that ERK-mediated phosphorylation of Ser 65 in mouse or Ser 69 in human BIM promotes BIM ubiquitination and degradation (Luciano et al, 2003; Ley et al, 2004).
Here, we investigated the effect of TGFβ on the regulation of BIM in two cellular models of TGFβ-mediated apoptosis: the Ba/F3 B lymphocyte and the AML-12 hepatocyte cell lines. We show that while TGFβ transcriptionally induces BIM following a sustained treatment (24 h), it also results in a rapid and time-dependent attenuation of ERK phosphorylation concomitant with its inhibition of BIM phosphorylation. TGFβ mediates these inhibitory effects on ERK, and subsequently BIM, through the induced expression of MKP2. Transcriptional induction of MKP2 by TGFβ is rapid and SMAD3 dependent. TGFβ induction of MKP2 is not observed in SMAD3−/−-derived pro-B lymphocytes and inhibition of ERK activity, induction of BIM expression and induction of apoptosis in response to TGFβ is not observed in either SMAD3−/−- or MKP2−/−-derived pro-B cells. Our findings delineate a TGFβ apoptotic pathway proceeding from SMAD3 to MKP2 and ERK, leading to the induction of BIM by TGFβ and apoptosis.
Results And Discussion
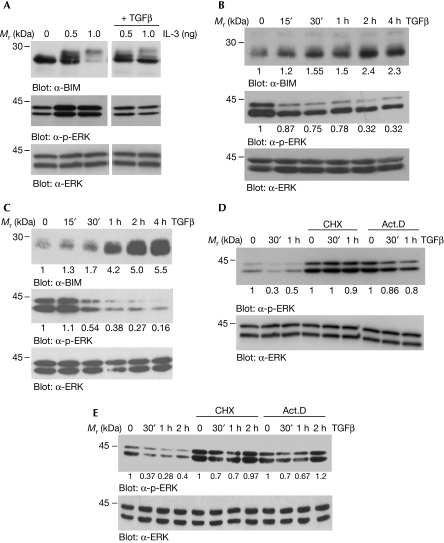
As phosphorylation of BIM has been shown to regulate its expression (Shinjyo et al, 2001; Ley et al, 2004) and as we previously observed that TGFβ promoted the accumulation of BIM (Wildey et al, 2003), we investigated the effect of TGFβ on BIM phosphorylation. Ba/F3 lymphocytes require IL-3 to survive and the withdrawal of IL-3 leads to increased BIM expression (Qi et al, 2006). As shown in Fig 1A, Ba/F3 cells were starved of IL-3 for 16 h to accumulate BIM levels (0) and treated with IL-3 in the presence or absence of TGFβ for 1 h. Treatment with IL-3 (0.5 and 1.0 ng/ml) resulted in the appearance of several phosphorylated BIM bands and a loss of immunoreactivity at the higher dose of IL-3. In the presence of TGFβ, however, the IL-3-dependent phosphorylation (band shift) and degradation of BIM were not as significant (Fig 1A, top panel). The addition of TGFβ to exponentially growing Ba/F3 cells (non-IL-3 starved) resulted in a rapid and time-dependent accumulation of BIM, with maximal effects observed at about 2 h (Fig 1B, top panel). The rapid effect of TGFβ on the accumulation of BIM is also observed in the mouse hepatocyte AML-12 cell line (Fig 1C, top panel).
Figure 1.
TGFβ inhibits the phosphorylation of ERK and BIM. (A) Ba/F3 lymphocytes were starved of IL-3 for 16 h to accumulate BIM (0), followed by stimulation in medium containing 0.5 or 1.0 ng/ml IL-3 in the presence or absence of TGFβ for 1 h. WCLs were prepared and analysed by immunoblotting with α-BIM, α-p-ERK and α-ERK2 antibodies. (B) Asynchronously growing Ba/F3 cells were treated with TGFβ for the indicated times, and WCLs were prepared and analysed by immunoblotting. (C) AML-12 hepatocytes were treated with TGFβ for the indicated times and WCLs were analysed by immunoblotting. Ba/F3 lymphocytes (D) and AML-12 hepatocytes (E) were treated for 4 h with 10 μg/ml of cycloheximide (CHX) or 1 μg/ml of actinomycin D (Act.D) before stimulation with TGFβ for 30 min, 1 h and/or 2 h. WCLs were prepared and analysed by immunoblotting. B and intensities for the immunoblots of (B–E) were quantified as described in the Methods and the fold change over control untreated cells is shown below each corresponding band. IL-3, interleukin-3; TGFβ, transforming growth factor-β; WCL, whole cell lysate.
ERK-dependent phosphorylation of BIM leads to its proteasomal-mediated degradation (Luciano et al, 2003; Ley et al, 2004; Qi et al, 2006). Thus, we examined whether the inhibition of BIM phosphorylation by TGFβ was due to its inhibition of ERK, as measured by α-phospho-ERK (α-p-ERK) immunoblot analysis. In both Ba/F3 (Fig 1A,B, middle panels) and in AML-12 cells (Fig 1C, middle panel), TGFβ inhibited ERK phosphorylation. In Ba/F3 cells, TGFβ attenuated the levels of p-ERK when added to overnight starved and IL-3 re-stimulated cells (Fig 1A) or to exponentially growing cells (Fig 1B). Kinetically, TGFβ inhibited p-ERK as rapidly as 30 min and sustained inhibitory effects until around 4 h (Fig 1B). Similarly, in AML-12 cells, treatment with TGFβ resulted in a rapid and sustained attenuation of p-ERK (Fig 1C). Levels of total ERK (α-ERK; lower panels) were not altered by the addition of TGFβ in either cell type. These data show the inverse correlation between the expression of p-ERK and BIM, and suggest that TGFβ inhibition of ERK results in the inhibition of BIM phosphorylation and accumulation of BIM in both B lymphocytes and hepatocytes. Next, we assessed the effect of the transcription inhibitor actinomycin D (Act.D) and of the protein synthesis inhibitor cycloheximide (CHX) on the regulation of phosphorylation of ERK by TGFβ. In both Ba/F3 (Fig 1D) and AML-12 cells (Fig 1E), CHX and Act.D blocked the inhibition of p-ERK by TGFβ. Both CHX and Act.D caused the induction of ERK, compared with non-treated control cells, perhaps through the inhibition of an ERK-negative regulator (Cook et al, 1997), and TGFβ was without an inhibitory effect on p-ERK in the presence of these inhibitors. These results suggest that the inhibitory effects of TGFβ on ERK require de novo transcription and protein synthesis.
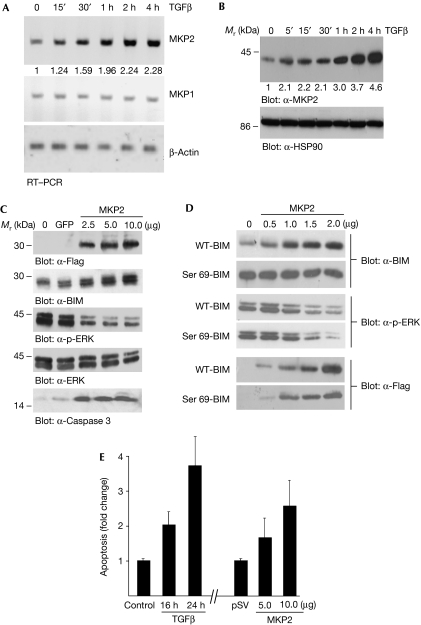
A complementary DNA microarray approach was performed to identify genes for which levels of messenger RNA were rapidly induced by TGFβ using the Illumina BeadChip system. The original data are available from the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress; accession number E-TABM-503). Our analysis focused on genes that were (1) rapidly induced by TGFβ, as the inhibitory effect of TGFβ on the phosphorylation of ERK and BIM is observed within 1 h; and (2) potential modulators of ERK. We observed that the mitogen-activated protein kinase phosphatase MKP2/DUSP4 (dual specificity protein phosphatase 4) met both these criteria. MKP2 has been shown to be a regulator of the MAPK pathway (Lang et al, 2006) and it was rapidly induced by TGFβ (1.7±0.23-fold at 30 min and 2.2±0.33-fold at 1 h). A list of the 12 DUSP genes on the array and their regulation following a 1 h treatment with TGFβ is shown in supplementary Fig S1 online. The rapid induction of MKP2 by TGFβ in AML-12 cells was confirmed at the mRNA level by reverse transcription (RT)–PCR analysis (Fig 2A, top panel) showing maximal induction at about 1 h; TGFβ had no effect on MKP1/DUSP1 (Fig 2A). Immunoblot analysis with an α-MKP2 antibody shows that the treatment of AML-12 cells with TGFβ leads to a time-dependent increase in the levels of MKP2 (Fig 2B). Thus, we conclude that both mRNA and protein levels of MKP2 are upregulated by TGFβ.
Figure 2.
Induction of the MAPK phosphatase MKP2 by TGFβ. (A) AML-12 cells were treated for the indicated times with TGFβ. RNA was isolated and RT–PCR analysis was carried out with primers specific for MKP2, MKP1 and β-Actin; β-Actin acts as a loading control. (B) WCLs were prepared and analysed by immunoblotting; α-HSP90 blot acts as a loading control. RT–PCR gels (A) and immunoblots (B) were quantified as described in the Methods and the fold change over control untreated cells is shown below each corresponding band. (C) Flag-tagged MKP2 (2.5, 5 and 10 μg DNA) was transfected into COS7 cells (GFP was used as a control transfected DNA) and after 24 h incubation WCLs were analysed by immunoblotting for Flag-MKP2, BIM, p-ERK, total ERK and cleaved caspase 3. (D) COS7 cells were co-transfected with 2.0 μg of either WT BIM or the phosphorylation mutant, Ser 69-BIM, and increasing concentrations (0.5, 1, 1.5 and 2 μg) of MKP2. WCLs were prepared and analysed by immunoblotting. (E) AML-12 hepatocytes were either treated with TGFβ for 16 or 24 h, or transfected with MKP2 (5 and 10 μg) for 24 h and apoptosis was quantified by ELISA. The mean±s.d. of three independent experiments each performed in duplicate (n=3) is shown. AML, acute myeloid leukaemia; ELISA, enzyme-linked immunosorbent assay; GFP, green fluorescent protein; HSP90, heat shock protein 90; IL-3, interleukin-3; MAPK, mitogen-activated protein kinase; RT–PCR, reverse transcription–PCR; TGFβ, transforming growth factor-β; WCL, whole cell lysate; WT, wild type.
Next, we investigated whether ectopic expression of MKP2 could lead to the modulation of p-ERK, levels of BIM and apoptosis in the absence of TGFβ. A Flag-tagged MKP2 expression construct was generated and COS7 cells were transfected with increasing concentrations of DNA. Overexpression of MKP2 resulted in increased expression of BIM (Fig 2C; α-BIM panel), inhibition of p-ERK (α-p-ERK panel) and an increased level in the active, cleaved product of caspase 3 (α-caspase 3 panel). Levels of total ERK (α-ERK panel) were unaltered by MKP2. ERK phosphorylation of human BIM has been shown to occur on Ser 69 (Luciano et al, 2003). If MKP2 inactivates ERK, then its overexpression should lead to the modulation of BIM stability through Ser 69. We generated a wild-type and a Ser 69 mutant expression construct (Ser/Ala mutation) and co-transfected these into COS7 cells in the presence or absence of increasing concentrations of MKP2. The data show that the expression of MKP2 leads to an MKP2-dependent induction of wild-type BIM but does not modulate the levels of the Ser 69 mutant form of BIM (Fig 2D; α-BIM panels). Confirming our previous data (Qi et al, 2006), expression levels of the wild-type BIM construct are much lower than the Ser 69 mutant (comparing control levels of overexpression), suggesting that Ser 69 mediates ERK-dependent phosphorylation and degradation of BIM. Controls show levels of MKP2 overexpression (α-Flag panels) and confirm that its overexpression leads to a decrease in p-ERK (α-p-ERK panels). In addition, as analysed by enzyme-linked immunosorbent assay (Fig 2E) in AML-12 hepatocytes, there was an increase in apoptosis as a result of MKP2 overexpression. These data suggest that the regulation of ERK by MKP2 mediates the expression of BIM and ultimately leads to apoptosis.
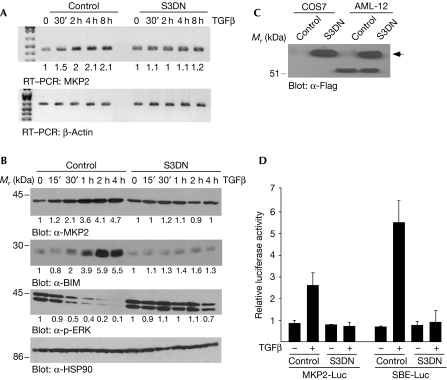
Next, we determined whether the effect of TGFβ on the induction of MKP2 is mediated by SMADs. We generated stable AML-12 cells overexpressing Flag-tagged, dominant-negative (DN) forms of SMAD2 and SMAD3 (S3DN), and determined that only S3DN-expressing cells showed any effects on TGFβ-mediated apoptosis (data not shown). In control cells (vector transfected), TGFβ induced MKP2 mRNA (Fig 3A, top panel) and protein (Fig 3B; α-MKP2 panel) in a time-dependent manner; this induction was not observed in AML-12 hepatocytes overexpressing S3DN. In S3DN-overexpressing cells, TGFβ did not induce BIM (α-BIM panel) or inhibit p-ERK (α-p-ERK panel), as it did in control cells (Fig 3B). The expression of Flag-tagged S3DN is confirmed in Fig 3C. To verify that SMAD3 functions in the induction of MKP2 by TGFβ, we performed transient transfection of luciferase/reporter assays with an MKP2 promoter construct (Zhang et al, 2001). Transactivation of this construct was induced 2.5-fold as a result of treatment of AML-12 cells with TGFβ (Fig 3D). Promoter transactivation was not observed in AML-12 cells expressing S3DN, showing the dependence of SMAD3 on TGFβ-mediated MKP2 induction. Although these levels of MKP2 induction are not as robust as those observed with a SMAD-binding element concatemerize promoter (around sixfold), they are similar to the levels of MKP2 induction by TGFβ that we observed by using the microarray analysis (supplementary Fig S1 online).
Figure 3.
TGFβ-mediated induction of MKP2 is SMAD3 dependent. Control AML-12 (vector alone transfected) and AML-12 cells stably expressing a dominant-negative form of SMAD3 (S3DN) were treated with TGFβ for the indicated times. (A) RNA was analysed by RT–PCR with primers specific for MKP2 and β-Actin (loading control). (B) WCLs were analysed by immunoblotting with α-MKP2, α-BIM, α-p-ERK and α-HSP90. RT–PCR gels (A) and immunoblots (B) were quantified as described in the Methods and the fold change over control untreated cells is shown below each corresponding band. (C) Western blotting was performed to detect the Flag-tagged S3DN protein in transiently transfected COS7 cells and the AML-12 S3DN cells. (D) AML-12 and S3DN-AML-12 cells were transfected with MKP2-Luc and SBE-Luc promoter/reporter constructs and treated with TGFβ for 24 h. The cell lysates were prepared and luciferase activity was measured and quantified. The mean±s.d. of three independent experiments each performed in duplicate (n=3) is shown. AML, acute myeloid leukaemia; HSP90, heat shock protein 90; RT–PCR, reverse transcription–PCR; SBE, SMAD binding element; TGFβ, transforming growth factor-β; WCL, whole cell lysate.
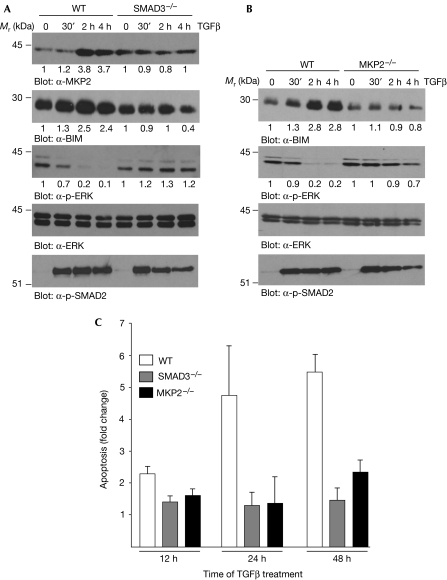
To confirm the role of SMAD3 and MKP2 in the TGFβ apoptotic pathway, we isolated primary bone marrow-derived, IL-7-dependent pro-B lymphocytes from wild-type, SMAD3-deficient (SMAD3−/−; Yang et al, 1999) and MKP2-deficient (MKP2−/−; obtained from Jeffery Molkentin) mice and examined TGFβ-induced responses. In pro-B cells derived from wild-type mice, TGFβ induced MKP2 in a time-dependent manner (Fig 4A; α-MKP2 panel), whereas this effect was not observed in pro-B cells derived from SMAD3−/− deficient mice. Furthermore, in SMAD3−/− pro-B cells, TGFβ did not induce BIM (α-BIM panel) or inhibit p-ERK (α-p-ERK panel), as it does in pro-B cells isolated from wild-type mice. In MKP2−/−-derived B cells (Fig 4B), stimulation of TGFβ did not lead to increased expression of BIM (α-BIM panel), nor did it inhibit p-ERK (α-p-ERK panel). In both SMAD3- and MKP2-deficient pro-B cells, TGFβ did induce the phosphorylation of SMAD2 (Fig 4A,B; α-p-SMAD2 panel) showing that not all TGFβ-mediated signalling in these cells is abrogated. In addition, in wild-type pro-B cells, stimulation of TGFβ caused increased apoptosis following a 24 and 48 h treatment (Fig 4C), whereas in SMAD3- and MKP2-deficient pro-B cells, TGFβ did not induce apoptosis. (The genotype of the wild-type, SMAD3- and MKP2-deficient mice, as well as the lack of SMAD3 phosphorylation in SMAD3-deficient mice is shown in supplementary Fig S2 online.) These results show that SMAD3 is required for TGFβ-induced MKP2 expression, and that both SMAD3 and MKP2 are necessary to mediate the TGFβ/BIM apoptotic pathway.
Figure 4.
TGFβ-mediated effects in wild-type, SMAD3- and MKP2-deficient pro-B lymphocytes. (A) Bone marrow-derived pro-B cells isolated from WT and SMAD3-deficient (SMAD3−/−) mice were treated with TGFβ for the indicated times, and WCLs were analysed by immunoblotting. (B) Pro-B cells isolated from WT and MKP2-deficient (MKP2−/−) mice were treated with TGFβ for the indicated times, and WCLs were analysed by immunoblotting. Immunoblots of (A,B) were quantified as described in the Methods and the fold change over control untreated cells is shown below each corresponding band. (C) WT and MKP2−/− pro-B cells were treated with TGFβ for 16 and 24 h, and apoptosis was quantified by ELISA. The mean±s.d. of three independent experiments each performed in duplicate (n=3) is shown. ELISA, enzyme-linked immunosorbent assay; TGFβ, transforming growth factor-β; WCL, whole cell lysates; WT, wild type.
Previously, we have shown that TGFβ-induced apoptosis is mediated through a SMAD3-dependent mechanism resulting in the accumulation of BIM (Wildey et al, 2003). The central role of BIM in TGFβ-induced apoptosis has been confirmed in gastric epithelial cells (Ohgushi et al, 2005) and in hepatocytes (Ramjaun et al, 2007). Here, we show that TGFβ induces BIM, thereby mediating apoptosis in hepatocytes and B lymphocytes, through its post-translational regulation of BIM. TGFβ induces MKP2 to rapidly target the levels of BIM by the inactivation of ERK, resulting in dephosphorylation and the escape of BIM from ubiquitin-mediated proteasomal decay. Induction of MKP2 is rapid, occurring within 30 min, and is SMAD3 dependent. Induction of MKP2 does not occur in hepatocytes expressing a dominant-interfering form of SMAD3 or in B lymphocytes derived from SMAD3-deficient mice.
The expression of BIM has also been shown to be regulated at the transcriptional level (Dijkers et al, 2000; Kuroda et al, 2006). We show in supplementary Fig S3 online using RT–PCR analysis in AML-12 hepatocytes that TGFβ induces the levels of BIM mRNA by around twofold following 24 h treatment and that this effect is SMAD3 dependent. The delayed kinetics (24 h) of this transcriptional induction of BIM by TGFβ is similar to the 16 h induction of TGFβ observed in hepatocytes by Ramjaun et al (2007). We postulate, therefore, that TGFβ targets BIM at both the transcriptional and post-translational levels. TGFβ induces MKP2 to rapidly target existing levels of BIM by the inactivation of ERK, resulting in dephosphorylation and the escape of BIM from ubiquitin-mediated decay; for the sustained modulation of BIM, TGFβ induces BIM mRNA. Thus, TGFβ not only induces BIM mRNA but also assures that the pathway that results in degradation of its product is inhibited, resulting in the accumulation of BIM protein and ultimately in cell death.
In summary, our results delineate an apoptotic signalling pathway emanating from TGFβ receptors and SMAD3 to MKP2 and ERK, leading to BIM induction and cell death. Thus, SMAD3 and MKP2 mediate the tumour suppressor function of TGFβ in hepatocytes and B lymphocytes by regulating TGFβ-induced apoptosis. Tumour cells often acquire resistance to the growth inhibitory and apoptotic effects of TGFβ. It might be expected that deregulation and/or defects in this TGFβ apoptotic pathway might underlie hepatocellular carcinoma and many haematological malignancies.
Methods
Cell culture. Ba/F3 cells were cultured in DMEM/F-12 medium supplemented with 10% fetal calf serum (FBS) and 30 μM 2-β-mercaptoethanol. IL-3 was added to a final concentration of 1 ng/ml. AML-12 cells were cultured in DMEM supplemented with 10% FBS, dexamethasone (40 μg/ml) and 1 U of recombinant insulin. COS7 cells were cultured in DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic. Primary pro-B lymphocytes were isolated from mouse femurs and maintained as previously described (Hardy et al, 1991). Briefly, bone marrow cells were flushed from the femurs of wild-type and knockout mice with Opti-MEM medium and cultured in Opti-MEM medium supplemented with 10% FBS, 1% antibiotic/antimycotic, 30 μM 2-β-mercaptoethanol and recombinant IL-7 (2 ng/ml). When required all cells were treated with 5 ng/ml of TGFβ2.
Western blot analysis. Whole cell lysates (WCLs) were prepared in TNMG lysis buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 5.0 mM MgCl2, 0.5% Nonidet P-40 and 10% glycerol) containing 1 mM sodium vanadate, protease inhibitor cocktail and phosphatase inhibitors. WCLs were sonicated and clarified by centrifugation at 12,000 r.p.m. for 20 min at 4°C. Typically 10–100 μg of WCL per well was separated on 12% acrylamide mini gels and transferred to Immobilon-P membrane (Millipore, Bedford, MA, USA). The membrane was blocked for 1 h in TBST buffer (Tris-buffered saline with 0.05% Tween 20) containing 5% non-fat dry milk. Primary antibodies were typically diluted between 1:500 and 1:10,000 in the same blocking buffer. Secondary antibodies were used at a dilution of 1:5000 and 1:10,000. Western blots were developed using ECL-Plus kit. The blots were stripped and reprobed with either total-ERK or HSP90 to check for consistent loading.
Typical immunoblots are shown in the individual figures, along with the corresponding fold change in the levels of protein. Protein intensity was quantified using ImageQuant software with the loading control, HSP90 or total ERK, to normalize. The normalized values were expressed as fold change over control untreated cells. Furthermore, each experiment was repeated three times independently and the results were quantified, and are shown graphically in supplementary Figs S5,S6 online. In supplementary Figs S5,S6 online, the mean±s.d. of three independent experiments is shown. A typical immunoblot for each of the proteins of interest is shown in supplementary Fig S4 online and shows migration in relation to molecular weight markers.
See supplementary information online for additional Methods.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
We thank Dr M. Roberson for generous provision of the MKP2 promoter construct and Dr S. Ledbetter at Genzyme Inc. for generous provision of TGFβ. This study was supported by grants CA55536 and CA80095 from the National Cancer Institute to P.H.H.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Köntgen F, Adams JM, Strasser A (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286: 1735–1738 [DOI] [PubMed] [Google Scholar]
- Cook SJ, Beltman J, Cadwallader KA, McMahon M, McCormick F (1997) Regulation of MKP-1 expression by Erk-dependent and Ca2+-dependent signal pathways in Rat-1 cells. J Biol Chem 272: 13309–13319 [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Lammers JJ, Koenderman L, Coffer PJ (2000) Expression of the pro-apoptotic bcl-2 family member bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol 10: 1201–1204 [DOI] [PubMed] [Google Scholar]
- Enders A, Bouillet P, Puthalakath H, Xu Y, Tarlinton DM, Strasser A (2003) Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim inhibits BCR stimulation-induced apoptosis and deletion of autoreactive B cells. J Exp Med 198: 1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayabawa K (1991) Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med 173: 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DC, Strasser A (2000) BH3-only proteins—essential initiators of apoptotic cell death. Cell 103: 839–842 [DOI] [PubMed] [Google Scholar]
- Kim H, Shah MR, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJD, Cheng EHY (2006) Hierarchical regulation of mitochondrion-dependent apoptosis by Bcl-2 subfamilies. Nat Cell Biol 8: 1348–1358 [DOI] [PubMed] [Google Scholar]
- Kuroda J et al. (2006) Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci USA 103: 14907–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R, Hammer M, Mages J (2006) DUSP meet immunology: dual specificty MAPK phosphatases in control of the inflammatory response. J Immunol 177: 7497–7504 [DOI] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Howes E, Balmanno K, Cook SJ (2004) Extracellular signal-regulated kinases 1/2 are serum-stimulated BimEL kinases that bind to the BH3-only protein Bim EL causing its phosphorylation and turnover. J Biol Chem 279: 8837–8847 [DOI] [PubMed] [Google Scholar]
- Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, Auberger P (2003) Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 22: 6785–6793 [DOI] [PubMed] [Google Scholar]
- Marani M, Tenev T, Hancock D, Downward J, Lemoine NR (2002) Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol Cell Biol 22: 3577–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi M, Kuroki S, Fukamachi H, O'Reilly LA, Kuida K, Strasser A, Yonehara S (2005) TGFβ-dependent sequential activation of Smad, Bim, and caspase-9 mediates physiological apoptosis in gastric epithelial cells. Mol Cell Biol 25: 10017–10028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly LA, Cullen L, Visvader H, Lindeman GJ, Print C, Bath ML, Huang DC, Strasser A (2000) The pro-apoptotic BH3-ony protein Bim is expressed in hematopoietic, epithelial, neuronal and germ cells. Am J Pathol 157: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Huang DC, O-Reilly LA, King SM, Strasser A (1999) The pro-apoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 3: 287–296 [DOI] [PubMed] [Google Scholar]
- Qi XJ, Wildey GM, Howe PH (2006) Evidence that Ser87 of BimEL is phosphorylated by Akt and regulates BimEL apoptotic function. J Biol Chem 281: 813–823 [DOI] [PubMed] [Google Scholar]
- Ramjaun AR, Tomlinson S, Eddaoudi A, Downward J (2007) Upregulation of two BH3-only proteins, Bim and Bmf, during TGFβ induced apoptosis. Oncogene 26: 970–981 [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS (2007) TGFβ-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol 8: 970–982 [DOI] [PubMed] [Google Scholar]
- Shinjyo T, Kuribara P, Inukai T, Hosoi H, Kinoshita T, Miyajima A, Houghton P, Look AT, Ozawa K, Inaba T (2001) Downregulation of Bim, a proapoptotic relative of Bcl-2 is a pivotal step in cytokine-initiated survival signaling in murine hematopoietic progenitors. Mol Cell Biol 21: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Massague J (2003) Cytostatic and apoptotic actions of TGFβ in homeostasis and cancer. Nat Rev Cancer 3: 807–821 [DOI] [PubMed] [Google Scholar]
- Wildey GM, Patil S, Howe PH (2003) Smad3 potentiates TGFβ induced apoptosis and expression of the BH3-only protein Bim in WEHI B-lymphocytes. J Biol Chem 278: 18069–18077 [DOI] [PubMed] [Google Scholar]
- Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, Roberts AB, Deng C (1999) Targeted disruption of Smad3 results in impaired mucosal immunity and diminished T cell responsiveness to TGFβ. EMBO J 18: 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang G, Feigenbaum L, Zhang YE (2006) Smad3 reduces susceptibility to hepatocarcinoma by sensitizing hepatocytes to apoptosis through downregulation of Bcl-2. Cancer Cell 9: 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Choy M, Jo M, Roberson MS (2001) Structural organization of the rat mitogen-activated protein kinase phophatase 2 gene. Gene 273: 71–79 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information