The development of multicellular organisms is controlled by gene-regulatory cascades. Generally, these cascades are headed by a master regulator that determines the development of a precursor cell lineage towards a given tissue or organ during embryogenesis. A notable feature is that the genes at the top of the cascade are more highly conserved than the downstream-acting factors. An outstanding example of this phenomenon is the homeobox-containing paired box 6 (PAX6) gene in its function as the master regulator of eye development. When, for example, the human gene is transgenically expressed in a Drosophila leg, it is able to induce the development of an eye at this ectopic and xenogenic position (Halder et al, 1995). What develops is not a vertebrate-type eye but a fly eye because the genes downstream in the eye-development cascade are not conserved and are those that specify the development of the insect visual organ, which consists of the complex and very specific ommatidia.
However, the opposite is true for the genes of the sex-determining cascade. In this case, the genes at the top of the cascade are not conserved, whereas the downstream genes have homologues in a much broader spectrum of species (Graham et al, 2003). The SRY gene—the master male sex-determining gene of mammals—has not been detected outside the eutherians. However, the other known members of the gene network involved in the development of the bipotential gonad anlage of the embryo towards either an ovary or testis—for example, the transcription factors Wilms tumour 1 (WT1) and SOX9 (sex-determining region Y-type high-mobility group box 9), and the bone morphogenetic protein (BMP) growth factor family member anti-Mullerian hormone (AMH; for a review, see Wilhelm et al, 2007)—are present in all vertebrates, including fish (Devlin & Nagahama, 2002). The most downstream component of this cascade, DMRT1 (doublesex and male abnormal 3-related transcription factor 1), has homologues even in Drosophila (doublesex; dsx) and Caenorhabditis elegans (male abnormal 3; Mab-3) (Raymond et al, 1998), whereas the top sex-regulatory genes of the fly and the worm are not conserved. This diversity at the molecular level seems to reflect the range of mechanisms by which sex is determined, including the X-chromosome-to-autosome ratio in Drosophila and C. elegans, and the presence of the Y chromosome in mammals. Notably, these are the only three groups of organisms in the animal kingdom for which we have a reasonably good knowledge about the molecular factors involved in sex determination and how they function. So far, we still do not know how sex determination is initiated in the WZ system of birds (although some candidate genes have been named), or how the incubation temperature of the eggs of crocodiles and turtles is perceived and translated into the action of a—probably the same or highly similar—transcription factor cascade that initiates male or female gonad development in birds and mammals.
One of the few exceptions to our lack of knowledge about the master regulators of sex is the honeybee. Hasselmann and colleagues have recently reported exciting insights about how such a molecular diversity of regulatory pathways could have evolved (Hasselmann et al, 2008). In the honeybee—as well as the more than 200,000 species of the insect order Hymenoptera (which includes bees, wasps and ants), many other insect groups and rotifers (which amount to an estimated 20% of all animal species)—sex determination follows a haplodiploid mode in which males are derived from unfertilized haploid eggs, whereas the fertilized diploid eggs generally develop into females. Sex determination in such haplodiploid species does not rely on a difference in heteromorphic sex-chromosome composition. The underlying genetic basis of haplodiploid sex determination was first elucidated in the Hymenoptera by the discovery that certain inbred crosses of wasps resulted in 50% of the fertilized eggs becoming diploid male offspring of biparental origin (Whiting, 1933). This finding suggested that the process of fertilization is not itself the signal of sexual regulation but, conversely, that the development into one sex or the other is controlled by an initial signal coming from either one allele or two different alleles of a single gene, the so called complementary sex determiner (csd). The biochemical nature of the initial signal of complementary sex determination remained unknown until the groups of Martin Beye and Robert Page jointly used fine-scale mapping and positional cloning approaches to identify and isolate the genomic region of the csd of the honeybee (Beye et al, 2003). Sequence comparisons indicated that csd is a member of an Arg/Ser-rich (RS) protein family, and the highest similarity was found between its carboxy-terminal part and the Drosophila protein transformer (Tra), which is a downstream component of the genetic sex-determination cascade in the fruitfly (Beye et al, 2003). No differences in the transcription or splicing of csd were detected between males and females that could act as sex-specific information. Thus, it is assumed that the Csd proteins act as dimers, and that heterodimers or homodimers differ in their ability to regulate the downstream factors of the cascade (Beye, 2004).
In August 2008, Hasselmann and co-workers from the Beye laboratory published the identification of a second gene involved in the sexual development of the honeybee that also localizes to the sex-determination locus (Hasselmann et al, 2008); this gene was called feminizer (fem). Interestingly, the fem gene encodes a protein that has a C-terminal RS-rich and Pro-rich domain with a high degree of sequence identity (>70%) to the Csd protein. Therefore, Fem is also a member of the Ser/Arg (SR)-type proteins, which are commonly thought to be involved in regulation of RNA splicing. Molecular phylogenetic analyses showed that csd is a duplicate of fem that arose between 10 and 70 million years ago during the evolution of the honeybee lineage. Furthermore, RNA interference (RNAi)-induced knockdown of fem resulted in a developmental switch that changed females into a male phenotype, whereas it had no effect in males. As identical results were observed for csd RNAi-knockdown experiments (Beye et al, 2003), these findings convincingly define fem as a second binary switch gene of the honeybee sex-determination pathway, in addition to csd.
Hasselmann and colleagues also analysed fem splicing. They could identify sex-specific fem transcripts with the same 5′ untranslated region (UTR) sequence, but with differences in their downstream exon composition that resulted in prematurely terminated male-specific fem transcripts. Notably, the occurrence of sex-specific splice variants is an important regulatory mechanism already described for the worm and fly sex-determination pathways (Fig 1). Finally, by showing csd-induced sex-specific splicing of fem, the authors could conclude that the binary switch of the sex-determination pathway is implemented by alternative splicing of the fem transcript in response to heterozygosity at csd.
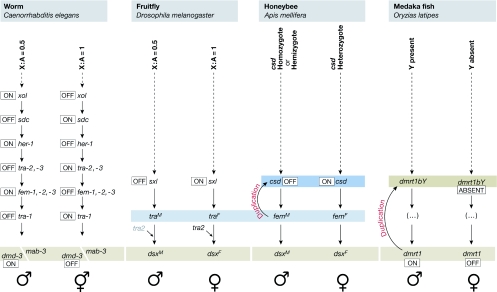
Figure 1.
Sex-determining cascades in the worm, insects and medaka fish. Molecular pathways leading to the formation of the gonad in Caenorhabditis elegans, Drosophila melanogaster, Apis mellifera and Oryzias latipes. Conserved doublesex/male abnormal 3/dsx and mab-3 related transcription factor 1 (dmrt1) or Drosophila transformer (tra)-like homologues are indicated with light-brown and light-blue boxes, respectively; duplicated factors in the cascades are also highlighted (brown and blue boxes, respectively). Note that tra of C. elegans is not related to tra of Drosophila. csd, complementary sex determiner; dmd3, doublesex/mab-3 domain family member 3; dsx, doublesex; fem, feminizer; her-1, hermaphroditization of XO-1; sdc, sex determination and dosage compensation defective; sx1, sex lethal; xol, XO lethal.
Interestingly, csd itself is not alternatively spliced. As it is a duplicated version of fem, this poses the question of whether the ability to be sex-specifically alternatively spliced has been lost by csd or gained by fem. As tra, the fem homologue in Drosophila (Fig 1), is analogously spliced, one can reasonably assume that sex-specific splicing was indeed lost for the duplicated copy that is now csd. Understandably, as it is at the top of the cascade, there is no possibility of splicing regulation for csd.
A homologue of dsx/mab-3, which is the most downstream factor of the fly and worm sex-determination cascades, has also been identified in the honeybee (Cho et al, 2007). In Drosophila, the Fem-like factor Tra regulates the differential splicing of dsx homologues. Therefore, it is reasonable to assume the same regulation from fem towards the dsx homologue in the honeybee, although this has not yet been confirmed experimentally (Beye, 2004).
The evolution of gene interactions in the sex-determining cascades characterized by conservation at the bottom and diversity at the top could be explained convincingly by an evolutionary scenario in which these hierarchies evolve from a common downstream component that acquires new upstream regulators—which will naturally vary in different evolutionary lineages (Wilkins, 1995). The case uncovered by Hanselmann and colleagues in the honeybee might seem to be an exception; here, in an extension of the Wilkins ‘bottom-up hypothesis', the master regulator is not elected and recruited from the outside to perform a new sex-determining function, but rather a member of the existing cascade usurps the position at the top. However, on closer inspection, this situation is not so exceptional. A similar situation exists in the medaka fish, which is the only non-mammalian vertebrate in which the master sex-determination gene is known (Matsuda et al, 2002; Nanda et al, 2002). In this case, the Dmrt1 gene—the most downstream gene in all sex-determining cascades known so far—underwent gene duplication approximately 5–10 million years ago. The new copy was inserted into another chromosome, which became the Y chromosome (Schartl, 2004). There, the duplicated Dmrt1 gene became the primary determinant of male sexual development and testes determination. In conclusion, it seems as if gene duplication of downstream components—as described for the medaka fish and now also for the honeybee—is more widespread than previously thought and provides an efficient evolutionary mechanism to create new sex-determination mechanisms.


Acknowledgments
Research in our laboratory is supported by grants from the Deutsche Forschungsgemeinschaft and the European Commission.
References
- Beye M (2004) The dice of fate: the csd gene and how its allelic composition regulates sexual development in the honey bee, Apis mellifera. Bioessays 26: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Beye M, Hasselmann M, Fondrk MK, Page RE, Omholt SW (2003) The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114: 419–429 [DOI] [PubMed] [Google Scholar]
- Cho S, Huang ZY, Zhang J (2007) Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177: 1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y (2002) Sex determination and sex differentiation in fish. Aquaculture 208: 191–364 [Google Scholar]
- Graham P, Penn JK, Schedl P (2003) Masters change, slaves remain. Bioessays 25: 1–4 [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ (1995) Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267: 1788–1792 [DOI] [PubMed] [Google Scholar]
- Hasselmann M, Gempe T, Schiott M, Nunes-Silva CG, Otte M, Beye M (2008) Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454: 519–522 [DOI] [PubMed] [Google Scholar]
- Matsuda M et al. (2002) DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563 [DOI] [PubMed] [Google Scholar]
- Nanda I et al. (2002) A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci USA 99: 11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D (1998) Evidence for evolutionary conservation of sex-determining genes. Nature 391: 691–695 [DOI] [PubMed] [Google Scholar]
- Schartl M (2004) A comparative view on sex determination in medaka. Mech Dev 121: 639–645 [DOI] [PubMed] [Google Scholar]
- Whiting PW (1933) Selective fertilization and sex determination in hymenoptera. Science 78: 537–538 [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Palmer S, Koopman P (2007) Sex determination and gonadal development in mammals. Physiol Rev 87: 1–28 [DOI] [PubMed] [Google Scholar]
- Wilkins AS (1995) Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17: 71–77 [DOI] [PubMed] [Google Scholar]