Abstract
Five major products (adducts A1a, A1b, A2, A3,and B) from the reaction of guanosine (Guo) with 4-oxo-2(E)-nonenal (ONE) were detected by liquid chromatography-mass spectrometry (LC-MS). Tandem MS (MS/MS) analysis of these compounds suggested that modifications to the nucleoside had been introduced. Adducts A1a, A1b, A2, A3 were heptanone-ethano-2′-Guo adducts that all decomposed to adduct B. Adducts A1a and A1b were isomeric hemi-ketal forms. Adducts A2 and A3 were diastereomers of the open chain ketone form. The structure of adduct B was shown by LC-MS/MS and NMR spectroscopy to be the heptanone-etheno-Guo (HεGuo) adduct, 3-(D-erythropentafuranosyl)imidazo-7-(heptane-2″-one)-9-hydroxyl[ 1,2-α]purine. The overall reaction of Guo with ONE was very similar to its reaction with 2′-deoxyguanosine. Reaction of ONE with yeast transfer RNA also resulted in the formation of HεGuo. Finally, HεGuo was detected and quantified in the RNA from rat intestinal epithelial cells that stably express cyclooxygenase-2. These data show that RNA is modified by the same bifunctional reactive electrophiles derived from lipid peroxidation that covalently modify DNA.
Keywords: lipid peroxidation, RNA-adducts, heptanone-etheno-adducts, 4-oxo-2(E)-nonenal, LC-MS, stable isotope dilution
Introduction
Lipid peroxidation can occur when cells are subjected to oxidative stress (1,2). Homolytic decomposition of lipid hydroperoxides leads to the formation of reactive bifunctional electrophiles, including 4-hydroperoxy-2(E)-nonenal (HPNE) (1,2), 4-oxo-2(E)-nonenal (ONE) (1,2) 4-hydroxy-2(E)-nonenal (HNE) (2,3) 4,5-epoxy-2(E)-decenal (EDE) (4). HPNE is the precursor to both ONE and HNE (2,5,6). COX-2, which is upregulated in colon cancer (7–9) can also form lipid hydroperoxides from arachidonic acid (10). ONE forms both heptanone-ethano and heptanone-etheno-adducts with 2′-deoxyguanosine (dGuo) (11) 2′-deoxyadenosine (12,13) 2′-deoxycytidine (14) as well as heptanone-etheno-adducts with DNA in vitro (6,11,12,15). The heptanone-ethano adducts formed with 2′-deoxynucleosides readily decompose to heptanone-etheno adducts (11–14). Heptanone-etheno DNA-adducts have been detected in the DNA of rat intestinal epithelial cells that stably express COX-2 (RIES cells) (10). They were also more abundant in intestinal DNA of Min mice (a model of colon cancer) when compared with controls (15). Furthermore, we recently showed that the heptanone-etheno-2′-deoxycytidine adduct was highly mutagenic in both bacteria and human cells.1 These results implicate that oxidative stress and lipid peroxidation can play an important role in carcinogenesis and other age-related diseases.
While the formation of DNA-adducts has been studied extensively, relatively fewer studies have been conducted with RNA. Since RNA has a similar structure to DNA, RNA was expected to show similar reactivity to lipid hydroperoxide-derived bifunctional electrophiles. Previous studies have shown that RNA can react with many carcinogens to form RNA-adducts. For example, Cardona et al. (16) reported that adducts can form between RNA and arylamines such as arylacethydroxamic acids, N-hydroxy-N-2-acetylaminofluorene, N-hydroxy-N-4-acetylaminobiphenyl and their corresponding acetamides under radiation. Subsequently, King et al. showed that these adducts can be formed in isolated parenchymal rat liver cells (17). Studies with aminostilbene, N-nitroso-di-n-propylamine and 7-sulfooxymethyl-12-methyl-benz[a]anthracene have shown that RNA-adducts can be formed in vivo (18–20). Murphy et al. have shown that RNA-adducts can arise from the reactive eicosanoid leukotriene A4 in calcium ionophore-treated human neutrophils (21,22). Finally, Sotomayor et al. identified RNA-adducts that were derived from the environmental carcinogen aflatoxin B1 (23). These studies suggest that RNA is a target of electrophiles in a similar manner to DNA, although further studies are required in order to address the biological significance of these observations.
Unlike DNA, modifications to RNA do not lead to mutations. However, the biological properties of RNA provide unique opportunities for using it as a biomarker. Compared with DNA, RNA is not usually as densely folded, so it may be more susceptible to reaction with lipid peroxidation products. Also, RNA exists in both the nucleus and cytoplasm, which makes RNA a potentially more sensitive biomarker for oxidative stress than DNA. For example, the lipid peroxidation products produced by the COX-2 enzyme, which resides mostly on the inside of the nuclear membrane, could covalently modify both DNA and RNA. However, the lipid peroxidation products produced by lipoxygenases or reactive oxygen species in the cytoplasm would be primarily captured by RNA. In addition, there are many species of RNA, e.g., mRNA, tRNA, and ribozymes, and they all have their unique cellular functions. Therefore, RNA-adduct formation could interfere with the normal function of mRNA and induce epigenetic responses. For example, it was reported that various RNA-binding proteins are involved in many important biological pathways (24–26). Adduct formation could interfere with the binding between RNA and its binding proteins. Also, the catalytic effects of ribozymes might be abolished by adduct formation. Moreover, the RNA mononucleoside adducts themselves could be toxic to cells and induce apoptosis.
In the present study, the reaction between guanosine (Guo) and ONE was carried out, and the products were characterized using LC-MSn and NMR spectroscopy. This reaction produced four ethano-adducts and one etheno-adduct of guanosine (HεGuo). These Guo adducts were very similar to their dGuo counterparts. The reaction between ONE and yeast tRNA also produced HεGuo. Finally, we were able to detect and quantify HεGuo in RIES cells.
Materials and Methods
Materials
Guanosine (Guo), 3-(N-morpholino)propanesulfonic acid (MOPS), ammonium acetate, dimethyl sulfoxide-d6 (DMSO-d6), deuterium oxide (D2O) and trimethylsilane (TMS) were purchased from Sigma Chemical Co. (St. Louis, MO). [15N]-Guo was purchased from Medical Isotopes, Inc. (Pelham, NH). Yeast tRNA was purchased from Invitrogen Inc. (Carlsbad, California). RNase (DNase free) and shrimp alkaline phosphatase (SAP) were obtained from Roche Applied Science, Inc. (Indianapolis, IN). Phosphodiesterase was purchased from Worthington Biochemical Corporation. Supelclean LC-18 SPE columns were acquired from Supelco Inc. (Bellefonte, PA). HPLC-grade water, hexane and acetonitrile were obtained from Fisher Scientific Co. (Fair Lawn, NJ). Gases were supplied by BOC Gases (Lebanon, NJ).
NMR
The 1H NMR spectra were measured at 25 °C using a Bruker DMX-400 (wide bore) instrument equipped with a 5 mm inverse proton probe. The sample of adduct B was dissolved in 600 μL of DMSO-d6, and the residual proton signal of DMSO was used as a reference in the proton spectrum. Acquisition conditions were as follows: spectral width (SW) of 6775 Hz, 30 ° pulse flip angle (PW), 64K data points (SI), and 64 scans. The 1H-1H two-dimensional correlation spectroscopy (COSY) spectrum was obtained with an SW1 and SW2 of 5200 Hz, 2K SI2, 512 experiments zerofilled to 1K, and 64 scans. Data were processed with a sinebell window.
Mass Spectrometry
Data were acquired on a Finnigan LCQ ion trap mass spectrometer (Thermo Electron Corp., San Jose, CA) equipped with an atmospheric pressure chemical ionization (APCI) or electrospray ionization (ESI) source in positive ion mode. Instrument conditions for APCI mode were as follows: vaporizer temperature at 450 °C, heated capillary temperature at 150 °C, with a discharge current of 5 μA applied to the corona needle. Nitrogen was used as the sheath (80 psi) and auxiliary (5 arbitrary units) gas to assist with nebulization. Instrument conditions for the ESI mode were as follows: capillary temperature at 200 °C, spray voltage of 4.5 kV applied to the electrospray needle. Nitrogen was used as the sheath (80 psi) and auxiliary (20 psi) gas to assist with nebulization. Full scanning analyses were performed in the range of m/z 100 – 600. Collision-induced dissociation (CID) experiments coupled with multiple tandem mass Spectrometry (MSn) employed argon as the collision gas. The relative collision energy was set at 20–40 % of the maximum.
The quantification data were acquired on an Applied Biosystems API4000 triple quadrupole mass spectrometer (Foster City, CA) equipped with an ESI turboionspray source in positive ion mode. The operating conditions were as follows: source temperature at 550 °C, spray voltage at 4.5 kV, collision cell exit potential at 10 V, collision energy at 23 V, the collision gas pressure at 6 psi and curtain gas at 20 psi, and Gas1 and Gas2 at 30 psi and 10 psi, respectively.
Liquid Chromatography
Chromatography was performed using a Waters Alliance 2690 HPLC system (Waters Corp., Milford, MA). Gradient elutions were all performed in the linear mode. Gradient systems 1, 2, 3, and 5 employed a Phenomenex Luna C8 column (250 mm × 4.6 mm i.d., 5 μm; Phenomenex, Inc., Torrance, CA) at a mobile phase flow rate of 1 mL/min. Gradient system 4 employed a Phenomenex Luna C18(2) column (150 mm × 2 mm i.d., 3 μm; Phenomenex, Inc., Torrance, CA) at a flow rate of 0.2 mL/min. Gradient system 6 employed the Phenomenex Luna C8 column with the mobile phase flow rates detailed below. LC-UV/MS experiments were performed using a Hitachi L-4200 UV detector at 236 nm for adducts analysis and 260 nm for RNA base analysis. For systems 1-3, solvent A was 5 mM ammonium acetate in water and solvent B was 5 mM ammonium acetate in acetonitrile. For system 1, the gradient conditions were as follows: 16% B at 0 min, 16% B at 40 min, 60% B at 41 min, 60% B at 55 min, and 16% B at 56 min, followed by a 10 min equilibration period. For system 2, the gradient conditions were as follows: 20% B at 0 min, 20% B at 25 min, 60% B at 26 min, 60% B at 36 min, and 20% B at 37 min, followed by a 10 min equilibration period. For system 3, the gradient conditions were as follows: 6% B at 0 min, 6% B at 3 min, 15% B at 10 min, 15% B at 55 min, 80% B at 57 min, 80% B at 65 min, and 6% B at 67 min, followed by a 10 min equilibration period. For system 4, solvent A was 0.02% formic acid in water and solvent B was acetonitrile. The gradient conditions were as follows: 15% B as 0 min, 50% B at 15 min, 90% B at 16 min, 90% B at 22 min, 15% B at 23 min, followed by an 8 min equilibration. For system 5, solvent A was water and solvent B was acetonitrile. The gradient conditions were as follows: 0% B at 0 min, 0% B at 3 min, 13% B at 15 min, 100% B at 20 min, 100% B at 22 min, and 0% B at 25 min, followed by equilibration for 5 min. For system 6, solvent A was 0.02% formic acid in water and solvent B was acetonitrile. The gradient conditions were as follows: 30% B with a flow rate 0.5 mL/min at 0 min, 60% B with a flow rate of 0.5 mL/min at 13 min, 100% B with a flow rate of 1.0 mL/min at 16 min, 100% B with flow rate 1.0 mL/min at 25 min, 30% B with a flow rate of 1.0 mL/min at 28 min, 30% B with a flow rate of 1.0 mL/min at 35 min, followed by equilibration for 2 min.
Synthesis of 4-Oxo-2(E)-nonenal
4-Hydroxy-2(E)-nonenal diethyl acetal was oxidized with activated MnO2 as described by Esterbauer and Weger (27). The resulting 4-oxo-2(E)-nonenal diethyl acetal was then hydrolyzed by citric acid/HCl as described previously (11) and purified by flash chromatography on silica. It was judged to be > 99 % pure based on its NMR spectrum and HPLC analysis.
Reaction of 4-Oxo-2(E)-nonenal with guanosine
A solution of ONE (770 μg, 5.0 μmol) in ethanol (20 μL) was added to Guo (267 μg, 1.0 μmol) in water (250 μL), which had a pH of 5.5. The final concentrations of ONE and Guo were 18.5 mM and 3.7 mM respectively. The reaction mixture was sonicated for 15 min at room temperature and incubated at 37 °C or 60 °C for 24 h, after which it was extracted twice with 200 μL hexane to remove excess ONE. Finally the sample was centrifuged at 5000 rpm for 10 min, and the supernatants were analyzed by LC-MS using gradient system 3. The reaction was conducted in MOPS buffer (100 mM, pH 7.4) and in phosphate buffer (100 mM, pH 7.4). Very similar results were obtained but the yields were slightly lower.
Preparation of Adducts A1a and A1b for LC-MS Analysis
A solution of ONE (7.7 mg, 0.05 mmol) in 200 μL of ethanol was added to guanosine (2.83 mg, 0.01 mmol) in 2.0 mL of water. After sonication for 20 min, the reaction mixture was incubated at 37 °C for 24 h. The reaction product was isolated by preparative HPLC. Chromatography was conducted using gradient system 1. Reaction products were collected between 20.0 and 40.0 min by monitoring the UV response at 236 nm. Fractions containing primarily A1a or A1b were concentrated under nitrogen at room temperature. Adduct A1a had a retention time of 23.3 min under these conditions, and adduct A1b had a retention time of 25.3 min.
Dehydration of Adducts A1a and A1b
HPLC-purified adducts A1a or A1b were dissolved in water/acetonitrile (100 μL, 84:16 v/v) and then mixed with 100 mM Chelex-treated MOPS buffer (100 μL) containing 150 mM NaCl at pH 3.0, 7.0, or 9.0. The solutions were vortex-mixed for 2 min then maintained at 37°C or 60 °C. Samples were incubated for different times and a portion of each sample (50 μL) was then analyzed by LC-UV using gradient system 2. One sample (at 60 °C, pH 7.4 for 4 h) was analyzed by LC-MS using gradient system 3 on the Finnigan LCQ.
Preparation of Adduct B
A solution of ONE (7.7 mg, 0.05 mmol) in 200 μL of ethanol was added to Guo (2.83 mg, 0.01 mmol) in 2.0 mL of water. The reaction mixture was sonicated for 20 min, incubated at 37 °C for 24 h, and then 60 °C for 6 h. Adduct B was isolated using gradient system 2, by monitoring the UV absorbance at 236 nm. The fraction, which eluted between 20.0 and 21.2 min, was concentrated under nitrogen at room temperature. The purity of the collected fraction was checked by LC-UV/MS. Fractions containing pure adduct B were combined, concentrated under reduced pressure, and dried under vacuum. To prepare the [15N]-labeled adduct B, the same procedure was carried out using [15N5]-Guo and ONE as starting materials.
Reaction of 4-Oxo-2(E)-nonenal with yeast tRNA
A solution of ONE (1.0 mg, 6.5 (μmol) in 68 μL ethanol was added to 200 μg yeast tRNA in 250 μL water at a pH of 5.5. The reaction mixture was incubated at 37 °C for 24 h, after which it was extracted twice with 200 μL hexane to remove excess ONE. The tRNA was then precipitated and hydrolyzed as described below. [15N]-labeled adduct B internal standard was added, and solid phase extraction (SPE) was performed as described below in order to enrich for adduct B. After SPE extraction, the sample was analyzed with LC-MS/MS using gradient system 4 and multiple reaction monitoring (MRM) (described below).
Cell culture
RIES cells were obtained from Dr. R.N. DuBois (Vanderbilt University). Cells were grown on plastic dishes in Dulbecco’s modified Eagle’s medium with 10% FBS (Summit, Fort Collins, CO), 4,500 mg/L D-glucose, and 100,000 units/liter streptomycin until almost confluent. The cells were not synchronized so potential effects of cell cycle on RNA expression would be random. Cells were collected as described previously when they reached 90 % confluency (10). There was a total of 1 × 106 cell on each plate.
RNA Extraction, Hydrolysis, and Solid Phase Extraction of Adduct B
RNA from 3 plates of RIES cells (3 × 106 cells) was extracted using Ambion RNAqueous Kit (Austin, TX). RNA was precipitated and dissolved in 200 μL of 2 mM Tris/20 mM MgCl2, pH 7.4. DNase-free RNase (2.5 μg) was added, the sample was incubated at 60 °C for 1 h and then at 37 °C for an additional 1 h. At the end of the incubation, 30 μL 0.2 M glycine buffer (pH 10) and phosphodiesterase (0.5 units) were added, and the sample was incubated at 37 °C for 2 h. Next, SAP (15 units), 30 μL 10x SAP buffer, and 30 μL of 50 mM Tris-HCL pH 7.4 were added, and the sample was incubated for 2 h. Finally, the sample was filtered through a 0.2 μm Costar cartridge before further analysis. At this time, an aliquot of hydrolyzed RNA was removed for LC-UV RNA base analysis (described below). After adding internal standard, the remaining sample was applied to Supelclean LC-18 SPE columns that had been pre-conditioned by pre-washing with acetonitrile (18 mL) followed by water (18 mL). After sample loading, the column was washed with water (4 mL) and methanol/water mixture (1 mL, 5:95 v/v). Finally, the adduct was eluted with acetonitrile/water mixture (6 mL, 1:1 v/v) and dried under nitrogen
RNA Base Analysis
RNA hydrolysis products were analyzed with HPLC gradient system 5 and at a UV wavelength 260 nm. Quantitation of RNA bases was achieved through constructing standard curves of known amounts of adenosine, cytidine, guanosine, and uridine in the range of 0.005 mg/mL to 0.25 mg/mL.
MS Analysis of Adduct B
The sample was re-dissolved in an acetonitrile/water mixture (200 μL, 10:90 v/v) and analyzed by LC-MRM/MS using gradient system 4 for the nucleoside and tRNA adducts and gradient system 6 for the RIES cell RNA-adducts. For the MRM analysis, parent ions were monitored for the protonated molecule (MH+) of adduct B and for its [15N5]-internal standard. Product ions were monitored that corresponded to the loss of the ribose moiety from adduct B (BH2+) and from its corresponding [15N5]-internal standard. The following transitions were monitored: m/z 420 → m/z 288 (endogenous adduct B) and m/z 425 → m/z 293 ([15N5]-internal standard). Quantitation was performed from standard curves constructed by the ratio of known amounts of authentic standards and internal standards. Adduct levels were normalized to the amount of RNA as detected in the RNA base analysis described above.
Results
Reaction of 4-Oxo-2(E)-nonenal with Guanosine
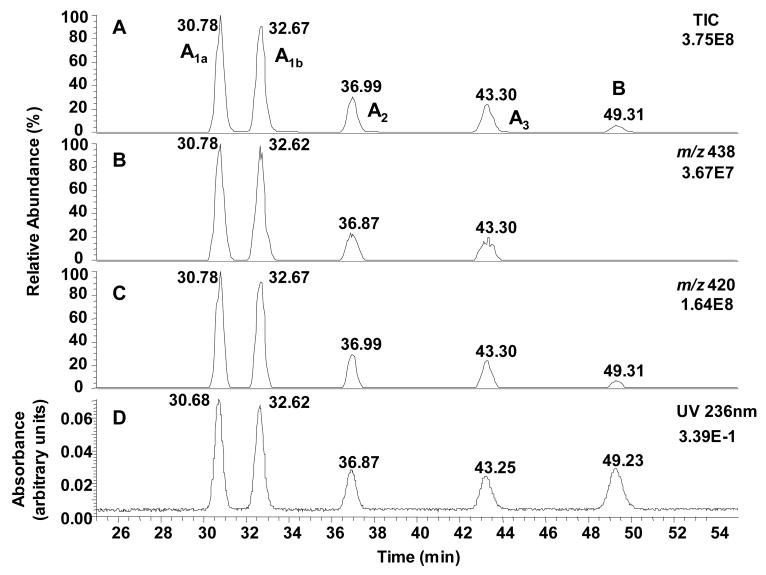
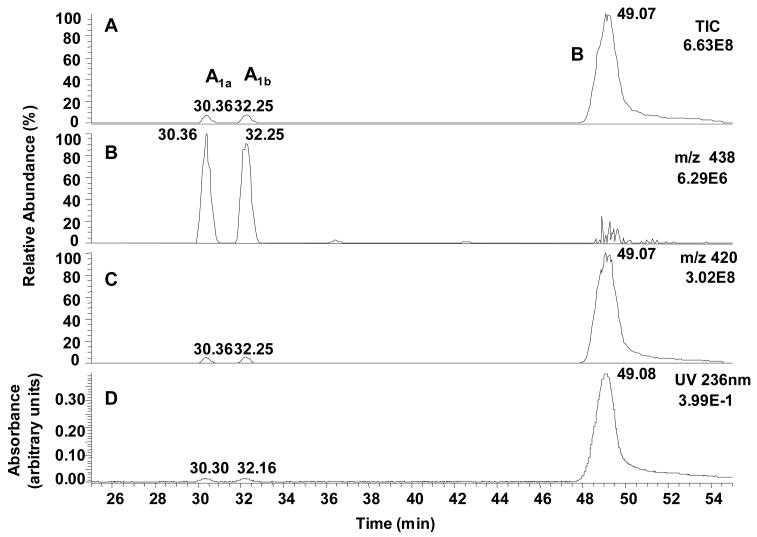
Using gradient system 3, LC-UV/MS analysis of the products from the reaction between Guo and ONE at 37 °C for 24 h revealed five major compounds with MH+ ions at m/z 438 (adducts A1a, A1b, A2 and A3) and m/z 420 (adduct B) (Figure 1). The UV chromatogram also confirmed the formation of five adducts together with residual Guo. When the reaction was heated at 60 °C for 24 h, the magnitudes ofthe UV signals for adducts A1a, A1b, A2 and A3 were reduced considerably with a concomitant increase in the magnitude of the UV signal for adduct B (Figure 2).
Figure 1.
Analysis of the reaction between ONE and Guo for 24 h at 37 °C by concurrent LC-MS and UV detection using gradient system 3. (A) Total ion current (TIC) chromatogram. (B and C). Reconstructed selected ion chromatograms of the MH+ of adduct A1a, A1b, A2 and A3 (m/z 438), and adduct B (m/z 420). (D) UV absorbance at 236 nm.
Figure 2.
Analysis of the reaction between ONE and Guo for 24 h at 37 °C and then heated for 7 h at 60 °C by concurrent LC-MS and UV detection using gradient system 3. (A) Total ion current (TIC) chromatogram. (B and C). Reconstructed selected ion chromatograms of the MH+ of adduct A1a, AH1b,A2 and A3 (m/z 438), and adduct B (m/z 420). (D) UV absorbance at 236 nm.
LC-MSn Analysis of Adducts A1a, A1b, A2, A3 and B
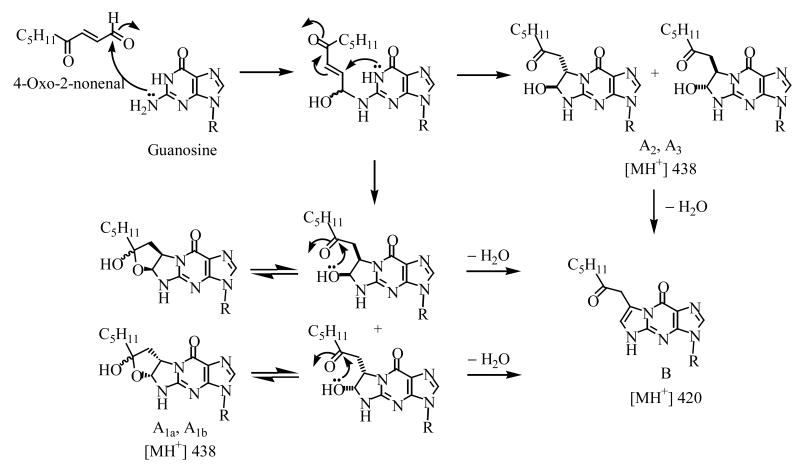
The ESI-MS spectrum for adduct A1a showed an intense MH+ at m/z 438 and a sodiated ion [MNa]+ at m/z 460. MS2 analysis of m/z 438 resulted in a BH2+ ion at m/z 306. BH2+ ion can lose a water to produce an ion at m/z 288. Further fragmentation of m/z 288 gives m/z 190 (Supplementary Figure 1). Adducts A1b, A2, and A3 produced the same fragmentation pattern. Adduct B showed a MH+ at m/z 420 in the ESI-MS spectrum. MS2 analysis of this ion generated the BH2+ ion at m/z 288. MS3 analysis of m/z 288 again produced an ion with m/z 190 (Supplementary Figure 2). These data were consistent with the proposed reaction scheme (Scheme I) that adduct B was a dehydration product of adducts A1a, A1b, A2, and A3.
Scheme I.
Structures of adducts A1a, AH1b A2, A3 and B and the mechanism for their formation.
UV Analysis of Adduct B
The UV spectrum of adduct B was very similar to that of the corresponding adduct formed between ONE and dGuo, with a λmax at 229 nm at pH 7. The λmax was shifted to 236 nm at pH 13 and to 226 nm at pH 1 (Supplementary Figure 3).
NMR Analysis of Adduct B
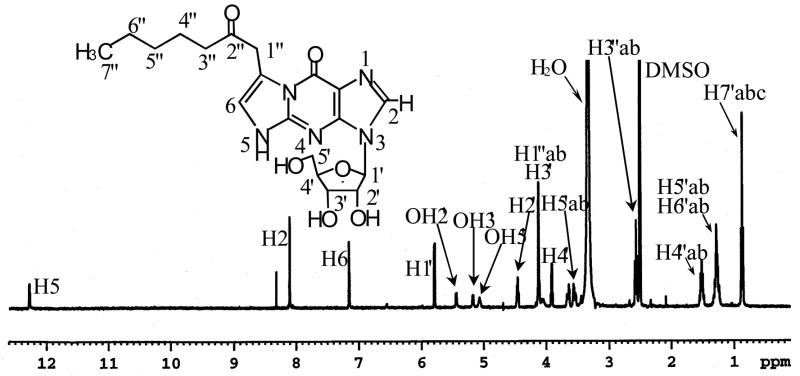
Assignments were made on the basis of chemical shifts and proton-proton couplings (Table 1 and Figure 3) together with COSY correlations (Figure 4). In the 1H NMR spectrum, besides the aromatic AB system of the Guo moiety, there was an additional aromatic proton singlet at 7.14 ppm (H-6). This proton showed COSY correlations with the methylene protons at H-1″ (4.12 ppm). These data were consistent with the presence of an olefinic bond between C-6 and C-7. The NH signal corresponding to H-5 appeared as a doublet at 12.25 ppm coupled to H-6 (J = 2.3 Hz). However the integration of this peak was less than one proton. There was an extra singlet at 8.32 ppm, which is most likely, the C-9 enol proton resulting from the shift of H-5 proton to the C-9 carbonyl group (Table 1). H-6 appe ared at 7.14 ppm as a broad singlet coupled to H-5. The triplet at 2.57 ppm was assigned to the methylene protons at H-3″ because of its chemical shift and its multiplicit y. The COSY spectrum also showed the correlations between the H-3″ protons with the H-4″ protons (1.51 ppm). Similarly, the multiplets at 1.28 ppm were assigned to the four methylene protons at H-4″ and H-5″. The triplet at 0.87 ppm was assigned to the methyl group at C-7″. The COSY spectrum also showed correlations between the H-1′ proton (5.79 ppm) and the proton at H-2′ (4.45 ppm), the H-2′ protons with the hydroxyl at C-2′ (5.46 ppm), the H-3′ protons with the hydroxyl at C-3′ (5.13 ppm) and with the H-4′ proton (3.92 ppm), the H-4′ proton with the H-5′ protons (3.55 and 3.65 ppm) and the H-5′ protons with the hydroxyl at C-5′ (5.04 ppm). A deuterium exchange experiment was done to further confirm the assignments of the exchangeable protons. After exchange with D2O, the NMR signals from the assigned H-5, C9-OH and 2′,3′,5′-OH protons all disappeared (Supplementary Figure 4). This confirmed the structure of adduct B as heptanone-etheno-Guo (HεGuo), 3-(D-erythropentafuranosyl)imidazo-7-(heptane-2″-one)-9-hydroxyl[l,2-α]purine (Figure 3, inset).
Table 1.
1H NMR assignments for adduct B. Spectra were obtained in DMSO-d6.
| Assigned H | δ(ppm) | multiplet | H-coupled [J(Hz)] | type |
|---|---|---|---|---|
| H-5 | 12.25 | d | H-6 (2.3 Hz) | C=NH |
| 9-OH | 8.32 | s | =C-OH | |
| H-2 | 8.09 | s | N=CH | |
| H-6 | 7.14 | bs | H-1″a,b | C=CH |
| H-1′ | 5.79 | d | H-2′ a,b (5.5) | -NCHO |
| 2′-OH | 5.46 | d | H-2′ (6.0) | -CHOH |
| 3′-OH | 5.13 | d | H-3′ (5.0) | -CHOH |
| 5 ′-OH | 5.04 | t | H-5′ a,b (5.5) | -CH2OH |
| H-2′ | 4.45 | t | H-3′, H-1′ (5.25) OH-2′ (5.0) | -OCH- |
| H-1″a,b H-3′ | 4.12 | m | H-6 (5.5) H-2′, H-4′, OH-3′ (5.0) | -CH2-C-OCH- |
| H-4′ | 3.92 | q | H-3′, H-5′ a,b (4.0) | -CHO |
| H-5′ a | 3.65 | abxy | H-4′ (4.0) OH-5′ (5.5) | -CH2O |
| H-5′b | 3.55 | abxy | H-4′ (4.0) OH-5 ′(5.5) | -CH2O |
| H-3″a,b | 2.57 | t | H-4″a,b (7.5) | -CH2-C |
| H-4″a,b | 1.51 | q | H-5′a,b, H-3″a,b (7.5) | -CH2-C |
| H-5″a,b | 1.28 | m | H-4″a,b, H-6″a,b | -CH2-C |
| H-6″a,b | 1.28 | m | H-5″a,b, H-7″a,b | -CH2-C |
| H-7″a,b,c | 0.87 | t | H-6″a,b (7.0) | -CH3 |
Figure 3.
1H NMR spectrum of adduct B in DMSO-d6. The inset shows the structure of adduct B as HεGuo based on NMR data.
Figure 4.
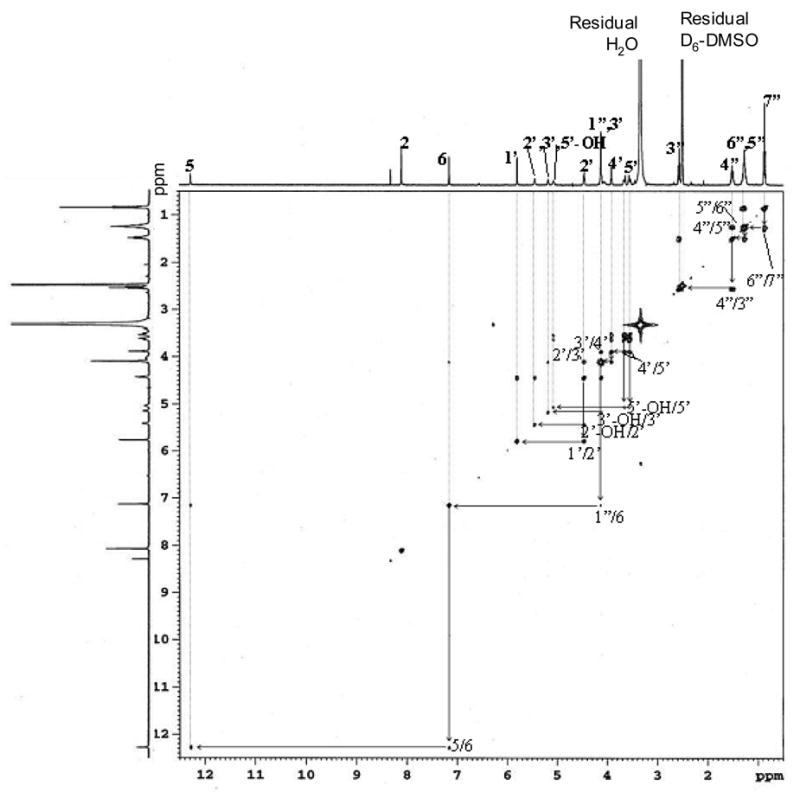
1H-1H two-dimensional COSY NMR spectrum of adduct B (HεGuo) in DMSO-d6.
Dehydration of Adducts A1a and A1b
Time course experiments were performed at 37 °C and 60 °C and at pH 3.0, 7.4, and 9.0 for adducts A1a and A1b by measuring the peak areas in the LC-UV spectrum. Both adduct A1a (Figure 5) and adduct A1b (data not shown) were converted to adduct B similarly in a temperature- and pH-dependent manner. At higher temperatures, the transformation was faster. Lower pH also increased the transformation rate. The dehydration of A1a to B (Supplementary Figure 5) and A1b to B (data not shown) was confirmed by LC-MS. Adducts A2 and A3 much more readily dehydrated to adduct B than adducts A1a and A1b, which made it impossible to prepare pure samples of the individual isomers. This suggested that adducts A1a and A1b were the more stable isomeric hemi-ketal adducts (Scheme I) similar to the more stable hemi-ketal adduct identified in the reaction of ONE with dGuo (11).
Figure 5.
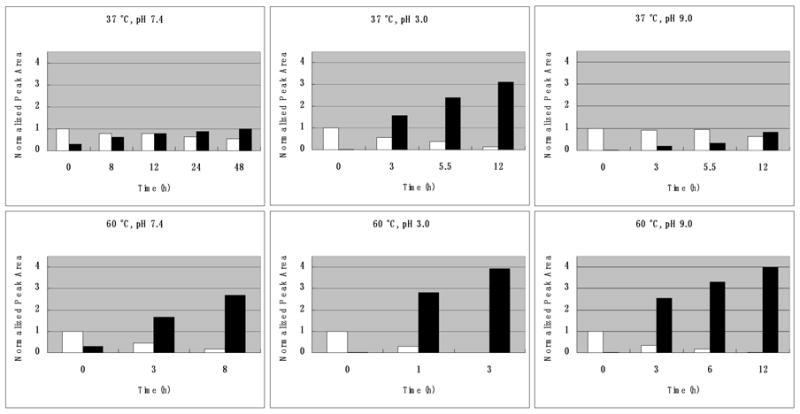
Time courses for the dehydration of adduct A1a to adduct B at 37 °C vs. 60 °C and at pH 3.0, 7.4 and 9.0 as determined by LC/UV monitoring 236 nm. White bars represent adduct A1a. Black bars represent HεGuo. The normalized peak area for peak B was > 1 because the λmax for adduct B under acidic conditions was 226 nM (Supplementary Figure 3) whereas, the λmax for adduct Ala was 252 nM. Results for dehydration of adduct A1b were essentially identical.
MS Analysis of the Reaction of 4-Oxo-2(E)-nonenal with Yeast tRNA
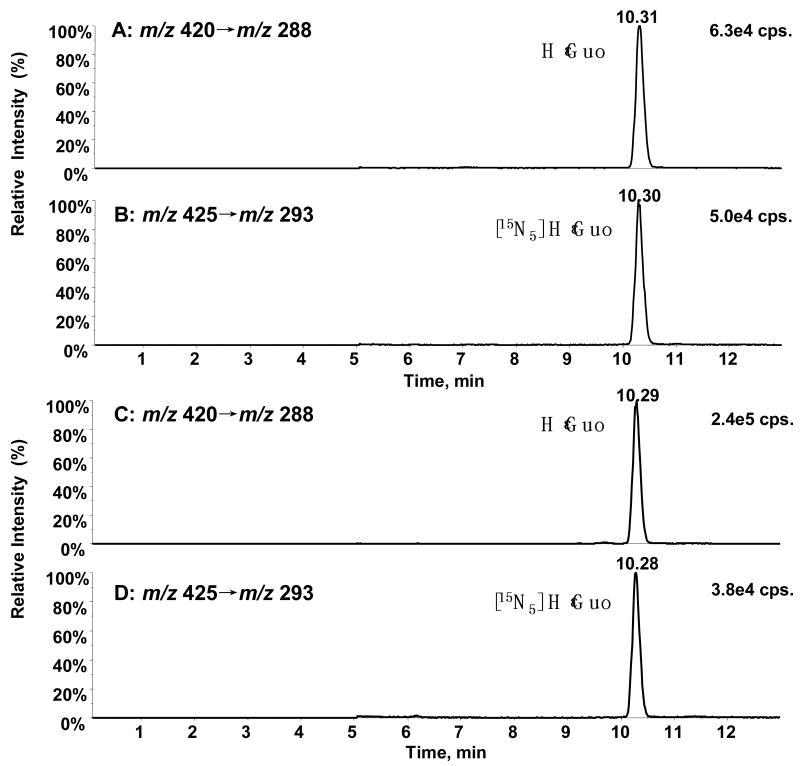
Typical chromatograms from LC-ESI/MRM/MS analysis of 0.2 ng/mL authentic adduct B (HεGuo) and its corresponding [l5N5] internal standard are shown in Figure 6 (A and B). HεGuo was quantified using calibration curves obtained with a linear regression analysis of peak area ratios of HεGuo to its [15N5]-labeled internal standard (Figure 6, C and D); concentrations of HεGuo were calculated by interpolation from the calculated regression line y = 0.7004x + 0.0264 (r2=0.998). HεGuo levels were normalized to the amount of tRNA in the sample. The final quantification of HεGuo corresponded to 24.4 adducts per 103 normal bases.
Figure 6.
LC-ESFMRM/MS analysis of HεGuo and its corresponding [15N5] internal standard. (A) Authentic HεGuo standard (200 pg/mL). (B) Internal standard. (C) HεGuo (24.4 adducts per 103 normal Guo bases) from yeast tRNA (200 mg) treated with ONE (1.0 mg, 6.5 (μmol). (D). Internal standard for yeast tRNA. Ion chromatograms for HεGuo (m/z 420 → m/z 288), [15N5]HεGuo (m/z 425 → m/z 293).
MS Analysis of RNA from RIES Cells
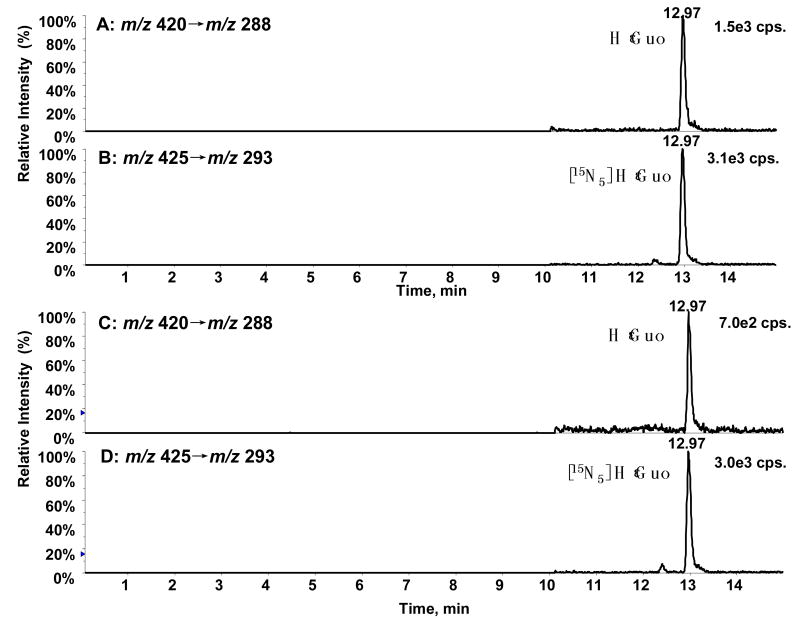
RIES cells were cultured to confluence. RNA was extracted from the cells, hydrolyzed, and analyzed using the same method as described above for tRNA. Typical chromatograms from LC-ESI/MRM/MS analysis of 400 pg/mL of authentic adduct B (HeGuo) and its corresponding [15N5] internal standard are shown in Figure 7 (A and B). Typical chromatograms obtained from RIES cell RNA for HεGuo and its internal are shown in Figures 7C and 7D, respectively. Concentrations of HεGuo in the RIES cell RNA were calculated by interpolation of the peak area ratios of analyte to internal standard using the calculated regression line y = 0.1334x + 0.0169 (r2=0.998). HεGuo levels of 7.0 adducts per 106 normal bases (± 1.1; SD) were found (n=4) after normalization for the amount of RNA that was isolated from the RIES cells.
Figure 7.
LC-ESI/MRM/MS analysis of HεGuo and its corresponding [15N5] internal standard. (A) Authentic HεGuo standard (400 pg/mL). (B) Internal standard. (C) HεGuo (6.6 adducts per 106 normal Guo bases) from RIES cell RNA. (D) Internal standard for RIES cell RNA. Ion chromatograms for HεGuo (m/z 420 → m/z 288), [15N5]HεGuo (m/z 425 → m/z 293).
Discussion
Cellular oxidative stress of polyunsaturated fatty acids leads to the formation of lipid hydroperoxides. The lipid hydroperoxides can then undergo homolytic decomposition to the reactive bifunctional electrophile, ONE, which reacts with DNA bases (4,11–14), peptides (28–30), and proteins (31–33) to form adducts. The reaction between ONE and dGuo is thought to take place through a two-step mechanism involving initial Michael addition of N2 to the C-1 aldehyde. Four major adducts were formed in the reaction: three of them were ethano adducts; one was the dehydrated etheno adduct, HεdGuo (11). Although many studies have been conducted to characterize the reactions of bifunctional electrophiles, such as ONE with DNA and proteins, little attention has been paid to their potential reactions with RNA. The present investigation of the reaction between Guo and ONE revealed the formation of five adducts, each of them very similar to adducts formed between dGuo and ONE (11). Adducts A2 and A3 were identified as heptanone-ethano-guanosine adducts, and A1a, A1b as hemi-ketal isomers. Because no interconversion between A1a and A1b was observed, A1a and A1b are proposed to be the 6(R), 7(S) and the 6(S), 7(R) isomers (Scheme I). Adduct B was characterized as the heptanone-etheno-Guo (HεGuo) adduct, 3-(D-erythropentafuranosyl)imidazo-7-(heptane-2″-one)-9-hydroxyl[1,2-α]purine adduct by LC-MSn (Supplementary Figures 1 and 2) and NMR spectroscopy (Figures 3 and 4). Adduct A1a was unstable and readily dehydrated to HεGuo as shown for adduct A1a in Figure 5. The dehydration process was catalyzed by acid. Under alkaline conditions, the rate of dehydration decreased significantly (Figure 5). Overall the Guo adducts with ONE have similar structures and chemical properties to the dGuo adducts. This suggests that the mechanism of formation is very similar (Scheme I) (11–14).
The reaction between yeast tRNA and ONE efficiently produced the same adducts as in nucleoside reactions (Figure 6) showing that adduction of RNA by ONE was feasible. This raised the possibility that ONE adduction would occur in cells. RIES cells that stably express COX-2 were selected as a model cell system because COX-2 is highly expressed in colon cancer tissue and other cancers, but not in normal tissue (35–37). In addition to its role in prostaglandin formation, COX-2 mediates the formation of lipid hydroperoxide-derived DNA-adducts (10). In order to assess the formation of RNA-adduct formation in RIES cells, a high sensitivity LC-MS/MS method was developed in order to quantify low pg amounts of H εGuo. Using this method, the adduct abundance in RIES cells was 7.0 adducts per 106 normal Guo bases (Figure 7). The similar HεdGuo-adduct was present at 2.0 adducts per 107 normal bases in the RIES cells under the same conditions (10). Thus, the heptanone-etheno adducts in RNA were approximately 35 times more abundant than the corresponding DNA-adducts.
The increased formation of RNA-adducts is most likely the consequence of a number of factors. First, RNA is present in both the cytoplasm and nucleus, while DNA is only found within the nucleus. Thus, RNA is more accessible to lipid peroxidation products released in both the nucleus and in the cytoplasm. Second, RNA exists in a variety of forms (tRNA, mRNA, ribosomes, ribozymes, microRNAs) with extended structures; in contrast, DNA is typically more densely folded and wrapped around histone proteins. So the various RNA forms would be more susceptible to reaction with bifunctional electrophiles than DNA. Third, RNA-adducts may be less readily repaired than the corresponding DNA-adducts. Besides serving as biomarkers, lipid hydroperoxide-derived RNA-adducts could have a variety of potential biological and toxicological effects, given that RNA is involved in so many diverse cellular functions. For example, mRNA-adduct formation could potentially modulate transcription and translation resulting in altered protein expression.
In summary, we have identified five RNA-adducts formed in the reaction between Guo and ONE. These adducts are very similar to those formed between dGuo and ONE, which suggests that lipid peroxidation can damage RNA in a similar manner to DNA. Detection and quantification of the HεGuo adduct in RIES cells confirmed this hypothesis. These findings suggest that heptanone-etheno-RNA-adducts in cell culture models and in tissue samples will provide biomarkers of oxidative stress. However, the biological role of these lipid hydroperoxide-derived RNA-adducts awaits further studies.
Supplementary Material
Acknowledgments
We gratefully acknowledge the support of NIH grant RO1 CA91016 and the editorial assistance of Dr. Michael Pollack.
Abbreviations
- APCI
atmospheric pressure chemical ionization
- bs
broad singlet
- CID
collision induced dissociation
- COSY
1H-1H 2D correlation spectroscopy
- COX
cyclooxygenase
- D2O
deuterium oxide
- d
doublet
- DMSO-d6
dimethyl sulfoxide-d6
- EDE
4,5-epoxy-2(E)-decenal
- ESI
electrospray ionization
- Guo
guanosine
- HεGuo
heptanone-etheno-Guo
- HεdGuo
heptanone-etheno-2′-dGuo
- HNE
4-hydroxy-2(E)-nonenal
- HPNE
4-hydroperoxy-2(E)-nonenal
- LC-MS
liquid chromatography-mass spectrometry
- m
multiplet
- MH+
protonated molecule
- MSn
multiple tandem mass spectrometry
- MOPS
morpholinopropanesulfonic acid
- MRM
multiple reaction monitoring
- NMR
nuclear magnetic resonance
- ONE
4-oxo-2(E)-nonenal
- q
quintet
- RIES
rat intestinal epithelial cells that stably express COX-2
- s
singlet
- SAP
shrimp alkaline phosphatase
- SPE
solid phase extraction
- t
triplet
- TIC
total ion current
- TMS
trimethylsilane
Footnotes
Pollack, M., Yang I.-Y., Kim, H.-Y., Blair, I.A., and Moriya, M. Submitted to Nucleic Acids Res.
References
- 1.Lee SH, Blair IA. Characterization of 4-oxo-2-nonenal as a novel product of lipid peroxidation. Chem Res Toxicol. 2000;13:698–702. doi: 10.1021/tx000101a. [DOI] [PubMed] [Google Scholar]
- 2.Lee SH, Oe T, Blair IA. Vitamin C-induced decomposition of lipid hydroperoxides to endogenous genotoxins. Science. 2001;292:2083–2086. doi: 10.1126/science.1059501. [DOI] [PubMed] [Google Scholar]
- 3.Benedetti A, Comporti M, Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochim Biophys Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- 4.Lee SH, Oe T, Blair IA. 4,5-Epoxy-2(E)-decenal-induced formation of 1,N(6)-etheno-2′-deoxyadenosine and 1,N(2)-etheno-2′-deoxyguanosine adducts. Chem Res Toxicol. 2002;15:300–304. doi: 10.1021/tx010147j. [DOI] [PubMed] [Google Scholar]
- 5.Lee SH, Oe T, Arora JS, Blair IA. Analysis of Fell-mediated decomposition of a linoleic acid-derived lipid hydroperoxide by liquid chromatography/mass spectrometry. J Mass Spectrom. 2005;40:661–668. doi: 10.1002/jms.838. [DOI] [PubMed] [Google Scholar]
- 6.Williams MV, Lee SH, Blair IA. Liquid chromatography/mass spectrometry analysis of bifunctional electrophiles and DNA adducts from vitamin C mediated decomposition of 15-hydroperoxyeicosatetraenoic acid. Rapid Commun Mass Spectrom. 2005;19:849–858. doi: 10.1002/rcm.1854. [DOI] [PubMed] [Google Scholar]
- 7.Birkenkamp-Demtroder K, Olesen SH, Sorensen FB, Laurberg S, Laiho P, Aaltonen LA, Orntoft TF. Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut. 2005;54:374–384. doi: 10.1136/gut.2003.036848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Lu W, Chen YG, Zhou XM, Gu JR. Comparison of gene expression between normal colon mucosa and colon carcinoma by means of messenger RNA differential display. World J Gastroenterol. 1999;5:533–534. doi: 10.3748/wjg.v5.i6.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backert S, Gelos M, Kobalz U, Hanski ML, Bohm C, Mann B, Lovin N, Gratchev A, Mansmann U, Moyer MP, Riecken EO, Hanski C. Differential gene expression in colon carcinoma cells and tissues detected with a cDNA array. Int J Cancer. 1999;82:868–874. doi: 10.1002/(sici)1097-0215(19990909)82:6<868::aid-ijc16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 10.Lee SH, Williams MV, Dubois RN, Blair IA. Cyclooxygenase-2-mediated DNA damage. J Biol Chem. 2005;280:28337–28346. doi: 10.1074/jbc.M504178200. [DOI] [PubMed] [Google Scholar]
- 11.Rindgen D, Nakajima M, Wehrli S, Xu K, Blair IA. Covalent modifications to 2′-deoxyguanosine by 4-oxo-2-nonenal, a novel product of lipid peroxidation. Chem Res Toxicol. 1999;12:1195–1204. doi: 10.1021/tx990034o. [DOI] [PubMed] [Google Scholar]
- 12.Lee SH, Rindgen D, Bible RH, Jr, Hajdu E, Blair IA. Characterization of 2′-deoxyadenosine adducts derived from 4-oxo-2-nonenal, a novel product of lipid peroxidation. Chem Res Toxicol. 2000;13:565–574. doi: 10.1021/tx000057z. [DOI] [PubMed] [Google Scholar]
- 13.Rindgen D, Lee SH, Nakajima M, Blair IA. Formation of a substituted 1,N(6)-etheno-2′-deoxyadenosine adduct by lipid hydroperoxide-mediated generation of 4-oxo-2-nonenal. Chem Res Toxicol. 2000;13:846–852. doi: 10.1021/tx0000771. [DOI] [PubMed] [Google Scholar]
- 14.Pollack M, Oe T, Lee SH, Silva Elipe MV, Arison BH, Blair IA. Characterization of 2′-deoxycytidine adducts derived from 4-oxo-2-nonenal, a novel lipid peroxidation product. Chem Res Toxicol. 2003;16:893–900. doi: 10.1021/tx030009p. [DOI] [PubMed] [Google Scholar]
- 15.Williams MV, Lee SH, Pollack M, Blair IA. Endogenous lipid hydroperoxide-mediated DNA-adduct formation in min mice. J Biol Chem. 2006 doi: 10.1074/jbc.M600178200. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Cardona RA, King CM, Redpath JL. Irradiation-induced adduct formation of RNA with carcinogenic arylamine derivatives. Cancer Res. 1975;35:2007–2014. [PubMed] [Google Scholar]
- 17.King CM, Traub NR, Cardona RA, Howard RB. Comparative adduct formation of 4-aminobiphenyl and 2-aminofluorene derivatives with macromolecules of isolated liver parenchymal cells. Cancer Res. 1976;36:2374–2381. [PubMed] [Google Scholar]
- 18.Gaugler BJ, Neumann HG. The binding of metabolites formed from aminostilbene derivatives to nucleic acids in the liver of rats. Chem Biol Interact. 1979;24:355–372. doi: 10.1016/0009-2797(79)90083-8. [DOI] [PubMed] [Google Scholar]
- 19.Park KK, Archer MC, Wishnok JS. Alkylation of nucleic acids by N-nitrosodi-n-propyl-amine: evidence that carbonium ions are not significantly involved. Chem Biol Interact. 1980;29:139–144. doi: 10.1016/0009-2797(80)90028-9. [DOI] [PubMed] [Google Scholar]
- 20.Surh YJ, Lai CC, Miller JA, Miller EC. Hepatic DNA and RNA adduct formation from the carcinogen 7-hydroxymethyl-12-methylbenz[a]anthracene and its electrophilic sulfuric acid ester metabolite in preweanling rats and mice. Biochem Biophys Res Commun. 1987;144:576–582. doi: 10.1016/s0006-291x(87)80005-0. [DOI] [PubMed] [Google Scholar]
- 21.Reiber DC, Murphy RC. Covalent binding of LTA(4) to nucleosides and nucleotides. Arch Biochem Biophys. 2000;79:119–126. doi: 10.1006/abbi.2000.1851. [DOI] [PubMed] [Google Scholar]
- 22.Hankin JA, Jones DN, Murphy RC. Covalent binding of leukotriene A4 to DNA and RNA. Chem Res Toxicol. 2003;16:551–561. doi: 10.1021/tx034018+. [DOI] [PubMed] [Google Scholar]
- 23.Sotomayor RE, Washington M, Nguyen L, Nyang’anyi R, Hinton DM, Chou M. Effects of intermittent exposure to aflatoxin B1 on DNA and RNA adduct formation in rat liver: dose-response and temporal patterns. Toxicol Sci. 2003;73:329–338. doi: 10.1093/toxsci/kfg076. [DOI] [PubMed] [Google Scholar]
- 24.Gautrey H, McConnell J, Hall J, Hesketh J. Polarised distribution of the RNA-binding protein Staufen in differentiated intestinal epithelial cells. FEBS Lett. 2005;579:2226–2230. doi: 10.1016/j.febslet.2005.02.074. [DOI] [PubMed] [Google Scholar]
- 25.Noureddine MA, Qin XJ, Oliveira SA, Skelly TJ, van der Walt J, Hauser MA, Pericak-Vance MA, Vance JM, Li YJ. Association between the neuron-specific RNA-binding protein ELAVL4 and Parkinson disease. Hum Genet. 2005;117:27–33. doi: 10.1007/s00439-005-1259-2. [DOI] [PubMed] [Google Scholar]
- 26.Pullmann R, Jr, Juhaszova M, Lopez de Silanes L, Kawai T, Mazan-Mamczarz K, Halushka MK, Gorospe M. Enhanced proliferation of cultured human vascular smooth muscle cells linked to increased function of RNA-binding protein HuR. J Biol Chem. 2005;280:22819–22826. doi: 10.1074/jbc.M501106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esterbauer H, Weger W. Uber die wirkungen von aldehyden auf gesunde und maligne zellen, 3 mitt: Synthese von homologen 4-hydroxy-alkenalen, II (Actions of aldehydes on normal and malignant cells III Synthesis of homologous 4-hydroxy-2-alkenals 2) Monatsh Chem. 1967;98:1994–2000. [Google Scholar]
- 28.Doom JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 29.Oe T, Lee SH, Silva Elipe MV, Arisen BH, Blair IA. A novel lipid hydroperoxide-derived modification to arginine. Chem Res Toxicol. 2003;16:1598–1605. doi: 10.1021/tx034178l. [DOI] [PubMed] [Google Scholar]
- 30.Jian W, Arora JS, Oe T, Shuvaev VV, Blair IA. Induction of endothelial cell apoptosis by lipid hydroperoxide-derived bifunctional electrophiles. Free Radic Biol Med. 2005;39:1162–1176. doi: 10.1016/j.freeradbiomed.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Oe T, Arora JS, Lee SH, Blair IA. A novel lipid hydroperoxide-derived cyclic covalent modification to histone H4. J Biol Chem. 2003;278:42098–42105. doi: 10.1074/jbc.M308167200. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Minkler PE, Sayre LM. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2-(E)-nonenal. Chem Res Toxicol. 2003;16:901–911. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- 33.Yocum AK, Oe T, Yergey AL, Blair IA. Novel lipid hydroperoxide-derived hemoglobin histidine adducts as biomarkers of oxidative stress. J Mass Spectrom. 2005;40:754–764. doi: 10.1002/jms.847. [DOI] [PubMed] [Google Scholar]
- 34.Lin D, Lee HG, Liu Q, Perry G, Smith MA, Sayre LM. 4-Oxo-2-nonenal is both more neurotoxic and more protein reactive than 4-hydroxy-2-nonenal. Chem Res Toxicol. 2005;18:1219–1231. doi: 10.1021/tx050080q. [DOI] [PubMed] [Google Scholar]
- 35.Fujita H, Koshida K, Keller ET, Takahashi Y, Yoshimito T, Namiki M, Mizokami A. Cyclooxygenase-2 promotes prostate cancer progression. Prostate. 2002;53:232–240. doi: 10.1002/pros.10152. [DOI] [PubMed] [Google Scholar]
- 36.Sano H, Kawahito Y, Wilder RL, Hashiramoto A, Mukai S, Asai K, Kimura S, Kato H, Kondo M, Hla T. Expression of cyclooxygenase-1 and -2 in human colorectal cancer. Cancer Res. 1995;55:3785–3789. [PubMed] [Google Scholar]
- 37.Ranger GS, Thomas V, Jewell A, Mokbel K. Elevated cyclooxygenase-2 expression correlates with distant metastases in breast cancer. Anticancer Res. 2004;24:2349–2351. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.