Abstract
Histone H3K9 methylation is required for DNA methylation and silencing of repetitive elements in plants and filamentous fungi. In mammalian cells however, deletion of the H3K9 histone methyltransferases (HMTases) Suv39h1 and Suv39h2 does not affect DNA methylation of the endogenous retrovirus murine leukaemia virus, indicating that H3K9 methylation is dispensable for DNA methylation of retrotransposons, or that a different HMTase is involved. We demonstrate that embryonic stem (ES) cells lacking the H3K9 HMTase G9a show a significant reduction in DNA methylation of retrotransposons, major satellite repeats and densely methylated CpG-rich promoters. Surprisingly, demethylated retrotransposons remain transcriptionally silent in G9a−/− cells, and show only a modest decrease in H3K9me2 and no decrease in H3K9me3 or HP1α binding, indicating that H3K9 methylation per se is not the relevant trigger for DNA methylation. Indeed, introduction of catalytically inactive G9a transgenes partially ‘rescues' the DNA methylation defect observed in G9a−/− cells. Taken together, these observations reveal that H3K9me3 and HP1α recruitment to retrotransposons occurs independent of DNA methylation in ES cells and that G9a promotes DNA methylation independent of its HMTase activity.
Keywords: chromatin, DNA methylation, ERV, H3K9 methylation, HP1α
Introduction
Retrotransposons, including long terminal repeat (LTR) and non-LTR elements, are widely dispersed in the euchromatic compartment in higher mammals (Kuff and Lueders, 1988; Medstrand et al, 2002), constituting ∼37% of the mouse genome (Mouse Genome Sequencing Consortium, 2002). A subset of these elements are transcriptionally competent, placing a significant mutational load on their hosts (Maksakova et al, 2006). To minimize the likelihood of retrotransposition, a number of pathways that function at the transcriptional or post-transcriptional stages of the replicative cycle have evolved to inhibit the expression of these parasitic elements. DNA methylation for example, has an important function in transcriptional silencing of retrotransposons in mammalian cells (Li et al, 1992; Yoder et al, 1997; Walsh et al, 1998), as illustrated by the high level of expression of the intracisternal A particle (IAP) endogenous retrovirus (ERV) in mouse embryos deficient in the DNA methyltransferase (DNMT), Dnmt1 (Walsh et al, 1998). DNA methylation also has a critical function in transcriptional silencing of repetitive elements and their relics in filamentous fungi and plants (Goyon et al, 1996; Lindroth et al, 2001; Zhou et al, 2001), substantiating the importance of this epigenetic mark in suppressing transposable elements in distantly related eukaryotes.
Repetitive elements in eukaryotes are also marked by specific covalent histone modifications (Bernstein et al, 2007). Methylation of lysine 9 of the histone H3 tail (H3K9) in particular, has an important function in silencing of these elements in yeast (Nakayama et al, 2001), filamentous fungi (Tamaru and Selker, 2001), plants (Jackson et al, 2002) and animals (Martens et al, 2005). Recent genome-wide studies reveal that ERVs are marked by H3K9 dimethylation (H3K9me2) and/or H3K9 trimethylation (H3K9me3) in murine cells (Peters et al, 2003; Martens et al, 2005; Mikkelsen et al, 2007); however, the specific histone methyltransferases (HMTases) responsible have not been identified.
Intriguingly, the H3K9 HMTase DIM-5 is required for CpG methylation in Neurospora (Tamaru and Selker, 2001) and the H3K9 HMTase KRYPTONITE is required for CpNpG methylation in Arabidopsis (Jackson et al, 2002), suggesting the existence of an evolutionarily conserved silencing pathway in which H3K9 methylation promotes de novo DNA methylation of repetitive elements (Freitag and Selker, 2005; Stancheva, 2005). However, the role, if any, that H3K9 methylation has in DNA methylation of retrotransposons in mammalian cells has not been systematically addressed.
Five HMTases in the ‘Suv39' subfamily of SET (Suv39, Enhancer of Zeste, Trithorax) domain-containing proteins with H3K9 catalytic activity, including Suv39h1 and the closely related Suv39h2, G9a and the closely related GLP/EuHMTase1 and SETDB1/Eset, have been characterized in mammalian cells. On the basis of its sequence similarity to SETDB1, the sixth Suv39 family member, SETDB2/CLLD8, is also likely to have specificity for H3K9 (Mabuchi et al, 2001; Kouzarides, 2007). Although Suv39h1 and Suv39h2 double-negative (Suv39h1/2−/−) embryonic stem (ES) cells show a dramatic reduction in H3K9me3 and DNA methylation at major satellite repeats, IAP elements show no reduction in H3K9 methylation (Peters et al, 2003; Martens et al, 2005; Mikkelsen et al, 2007) and murine leukaemia virus (MLV) ERVs show no reduction in DNA methylation (Lehnertz et al, 2003) in these cells. Taken together, these results indicate that Suv39h1 and Suv39h2 do not have a major function in H3K9 methylation or DNA methylation of LTR retrotransposons in mammalian cells.
In contrast to the Suv39h HMTases, G9a and GLP/Eu-HMTase1, which form a heteromeric complex in vivo, are widely dispersed in the euchromatic compartment and deletion of either leads to a dramatic decrease in H3K9me1 and H3K9me2 in ES cells (Tachibana et al, 2002, 2005). A recent analysis revealed that ∼300–400 genes show altered expression in G9a−/− cells (Sampath et al, 2007) and several studies have shown that G9a regulates the expression and/or DNA methylation status of specific genes (Feldman et al, 2006; Ikegami et al, 2007). However, experiments aimed at determining whether G9a influences the expression and/or DNA methylation states of interspersed repetitive elements have not been reported.
Here, we investigated the function that G9a and GLP have in DNA methylation and silencing of potentially active ERVs and non-LTR retrotransposons. We show that DNA methylation of these elements, and a subset of non-repetitive sequences including CpG-rich promoters, is reduced in G9a−/− cells and that Dnmt3a recruitment to retrotransposons is decreased in these cells. However, H3K9me3 enrichment and HP1α binding are unaltered, demonstrating that an alternative H3K9 HMTase marks ERVs and that H3K9 methylation per se is not sufficient to promote DNA methylation of these elements. In support of this model, we show that the introduction of two different G9a transgenes that lack catalytic activity into G9a−/− cells partially ‘rescues' the observed DNA methylation defect, indicating that G9a promotes DNA methylation of retrotransposons independent of its catalytic activity.
Results
G9a is required for DNA methylation of retrotransposons
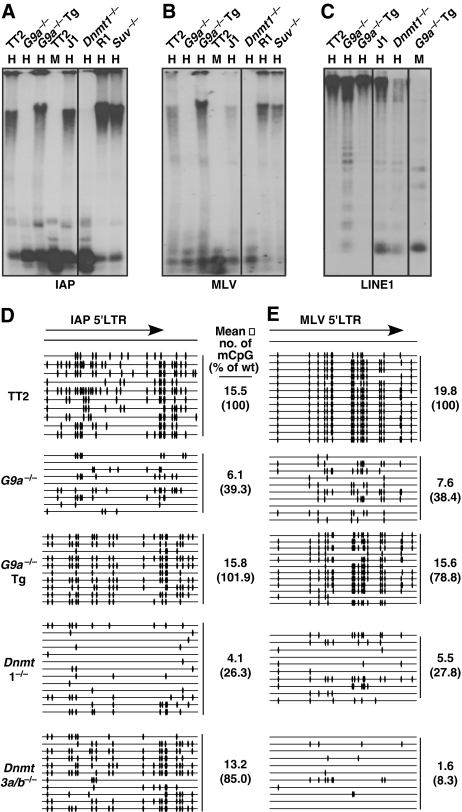
To establish whether G9a is required for DNA methylation of ERVs, genomic DNA isolated from TT2 wild-type (wt) and G9a−/− ES cells (Tachibana et al, 2002) (Supplementary Figure S1) was analysed by Southern blotting using the methylation-sensitive restriction enzyme HpaII and probes specific for IAP and MLV ERVs, of which there are ∼1200 and ∼60 copies in the mouse genome, respectively (Figure 1A and B). Genomic DNA samples isolated from Dnmt1−/− (Lei et al, 1996), Suv39h1/2−/− (Peters et al, 2001) and the wt parent ES cell lines from which they were derived were analysed in parallel. A dramatic reduction in DNA methylation of both ERVs was detected in the G9a−/− line relative to the wt control. At the resolution of Southern blot analysis, this reduction in methylation is not distinguishable from that observed for the Dnmt1−/− ES line, or TT2 genomic DNA digested with the methylation-insensitive isoschizomer MspI.
Figure 1.
DNA methylation of MLV, IAP and LINE1 retrotransposons is reduced in G9a−/− cells. Genomic DNA isolated from G9a−/−, G9a−/−Tg, Dnmt1−/−, Suv39h1/2−/− (Suv−/−) and the wt parent lines TT2, J1 and R1, respectively, was digested with MspI (M) or the methylation-sensitive restriction enzyme HpaII (H) and subject to Southern blotting using probes specific for (A) IAP, (B) MLV or (C) LINE1 retrotransposons. The G9a−/− line shows a dramatic decrease in DNA methylation at each of these repetitive elements that is reversed in the G9a−/−Tg line. In contrast, the Suv39h1/2−/− line shows no DNA methylation defect at IAP or MLV repeats. (D, E) Bisulphite analysis of the 5′LTR regions of IAP and MLV elements was conducted on TT2, G9a−/−, G9a−/−Tg (15-3), Dnmt1−/− and Dnmt3a/b−/− cells. For each molecule sequenced (horizontal bar), filled ovals represent the presence of an mCpG. The mean number of mCpGs per molecule sequenced is shown to the right of each set of sequenced samples. The mean % of mCpGs relative to the wild-type line is also shown (in parentheses).
To obtain a more accurate measure of the methylation status of these elements, high-resolution bisulphite sequencing analysis was conducted using primers specific for the CpG-rich 5′LTR regions of IAP and MLV elements. A >2.5-fold decrease in the mean number of mCpGs per molecule sequenced was detected in the G9a−/− line relative to the wt parent line (Figure 1D and E), with a subset of sequenced molecules in the G9a−/− line showing almost complete loss of methylation in these regions. A decrease in DNA methylation across the LTR and downstream regions of the potentially active class II ERV MusD (Mager and Freeman, 2000), of which there are ∼90 full-length copies in the mouse genome, was also detected in the G9a−/− line (Supplementary Figure S2). All three of these LTR retrotransposons show an even more severe DNA methylation defect in Dnmt1−/− cells (Figure 1D and E; Supplementary Figure S2). Consistent with the observations of Chen et al (2003), early passage Dnmt3a/b−/− cells (Okano et al, 1999) show a significantly more severe DNA methylation defect for MLV elements than IAP elements. Interestingly, whereas IAP elements show a less severe defect in Dnmt3a/b−/− cells than in G9a−/− cells, the reverse is true of MLV repeats. Thus, although it is clear that G9a is required for DNA methylation of distantly related ERVs in murine ES cells, the degree of demethylation is distinct from that observed for Dnmt1−/− or Dnmt3a/b−/− cells. In contrast, no reduction in DNA methylation of MLV or IAP elements was detected in Suv39h1/2−/− cells (Figure 1A and B), consistent with a previous report showing that Suv39h1 and Suv39h2 are not required for DNA methylation of MLV (Lehnertz et al, 2003).
DNA methylation of LINE1 (L1) elements, non-LTR retrotransposons that comprise ∼20% of the mouse genome (Mouse Genome Sequencing Consortium, 2002), also depends on the presence of both Dnmt1 and Dnmt3a and/or Dnmt3b in ES cells (Liang et al, 2002). To determine whether G9a has a function in DNA methylation of this class of interspersed repeats, Southern blot analysis was conducted with a probe that spans the promoter region of the L1Md-A2 subfamily of L1 elements. A significant decrease in DNA methylation of L1 elements is also apparent in G9a−/− cells, although this defect is not as severe as that detected in the DNMT mutant lines (Figure 1C). Taken together, these observations indicate that G9a influences DNA methylation of both LTR and non-LTR retrotransposons in ES cells.
To determine whether G9a is also required for DNA methylation of tandem repeats in ES cells, Southern blot analysis using a probe specific for major satellite repeats (present at approximately 700 000 copies per cell) was conducted using the methylation-sensitive restriction enzyme HpyCH4IV (Supplementary Figure S3). Consistent with a previous report showing that Suv39h1 and Suv39h2 are required for methylation of major satellite repeats (Lehnertz et al, 2003), a dramatic reduction in DNA methylation of major satellite repeats was detected in Suv39h1/2−/− cells. Unexpectedly, a dramatic reduction in DNA methylation of this class of repeats was also detected in the G9a−/− line, revealing that G9a is required for DNA methylation of pericentromeric heterochromatin as well.
Introduction of a G9a transgene rescues the DNA methylation defect observed in G9a−/− cells
Reintroduction of Dnmt3a, Dnmt3a2 (the predominant isoform of Dnmt3a in ES cells; Chen et al (2002)) or Dnmt3b1 into Dnmt3a/b−/− ES cells restores DNA methylation of MLV and IAP elements (Chen et al, 2003), indicating that the de novo DNMTs are capable of reestablishing DNA methylation patterns in these cells. To determine whether reintroduction of G9a is capable of reversing the DNA methylation defect observed in G9a−/− cells, the methylation status of these elements was also analysed in a G9a−/− line stably expressing a wt G9a transgene (G9a−/−Tg) (Tachibana et al, 2002) at a level similar to that of the endogenous protein (Supplementary Figure S1). Strikingly, the DNA methylation state of MLV, IAP, L1 (Figure 1) and MusD (Supplementary Figure S2) retrotransposons and major satellite repeats (Supplementary Figure S3) in the G9a−/−Tg resembles that of the original wt parent line (TT2) rather than the G9a−/− line from which they were directly derived. These observations indicate that loss of DNA methylation in G9a−/− ES cells is not an irreversible process and that reintroduction of G9a is sufficient for the reestablishment of DNA methylation in G9a-deficient ES cells.
GLP is required for DNA methylation of retrotransposons
As G9a forms a complex with the closely related HMTase GLP, and both are required for the deposition of the H3K9me2 mark (Tachibana et al, 2005), we next determined whether GLP is also required for DNA methylation of retrotransposons. Genomic DNA isolated from wt TT2 and GLP−/− ES cells (Tachibana et al, 2005) was analysed by Southern blotting as above, using probes specific for IAP, MLV (Supplementary Figure S4) and L1 elements (data not shown). A significant DNA methylation defect is apparent for all three elements in the GLP−/− line as well, with IAP elements showing the most dramatic decrease. Furthermore, introduction of a wt GLP transgene into the GLP−/− line (generating the GLP−/−Tg line (see Supplementary Figure S1; Tachibana et al, 2005) rescues the IAP DNA methylation defect and partially rescues the MLV methylation defect, revealing that DNA methylation can be reestablished on reintroduction of this HMTase as well. Consistent with these results, bisulphite sequencing analysis of polytrophic MLV elements reveals an ∼40% reduction in DNA methylation across the 5′LTR in the GLP−/− line relative to the wt control, and a partial rescue of this methylation defect in the GLP−/−Tg line (Supplementary Figure S4). Thus, both G9a and GLP have a function in DNA methylation of retrotransposons in ES cells.
DNA methylation at non-repetitive genomic regions is reduced in G9a−/− cells
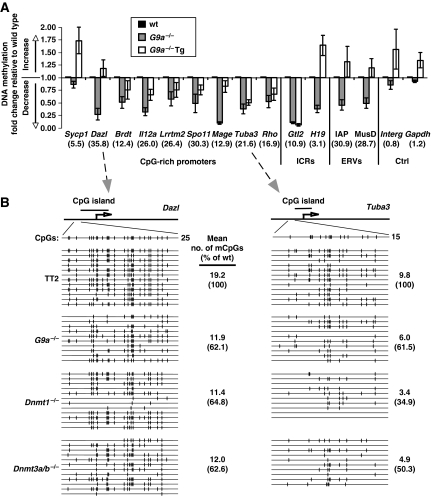
To determine whether this DNA methylation defect extends to non-repetitive elements in the genome, we carried out MeDIP (Weber et al, 2005) on genomic DNA isolated from TT2, G9a−/− and G9a−/−Tg ES cells and analysed the methylation status of 11 single-copy genomic regions, including 9 CpG-rich promoters shown previously to be methylated in ES cells (Mohn et al, 2008) (Figure 2A). Strikingly, all of the regions that are highly methylated in the TT2 line show a significant decrease in the G9a−/− line, including the germline-specific gene Mage-a2, shown previously to be aberrantly expressed in G9a−/− cells (Tachibana et al, 2002). As for the repetitive elements, DNA methylation is increased at most of these regions in the G9a−/−Tg line. The DNA methylation defect was confirmed through bisulphite sequencing of the Dazl and Tuba3 promoter regions, both of which show an ∼40% reduction in DNA methylation in the G9a−/− line (Figure 2B). The degree of demethylation across the Dazl promoter is similar to that observed in Dnmt1−/− and Dnmt3a/b−/− ES cells. In contrast, the degree of demethylation across the Tuba3 promoter in G9a−/− cells more closely resembles that observed in the Dnmt3a/b−/− line. Thus, although DNA methylation of CpG-rich promoter regions is also dependent on G9a, the degree to which G9a influences DNA methylation state depends on the genomic context.
Figure 2.
DNA methylation of promoter regions is reduced in G9a−/− cells. (A) MeDIP followed by quantitative PCR of nine CpG-rich promoter regions and two imprinting control loci (ICR) shown previously to be methylated in ES cells (Mohn et al, 2008) was conducted on wt, G9a−/− and G9a−/−Tg lines. IAP and MusD amplicons were included as positive controls. An active housekeeping gene (Gapdh) and a CpG-poor intergenic region (Interg) were included as negative controls. A bar graph illustrating DNA methylation changes in G9a−/− and G9a−/−Tg ES cells relative to wt ES cells (set to 1) is shown. The fold change is normalized to an unmethylated control gene (Hprt). Numbers in parentheses indicate the enrichment in MeDIP relative to Hprt. Error bars indicate the s.e.m. of at least three independent experiments. A lower level of methylation was detected in the G9a−/− line than the wt or rescued lines for all of the genes that show a high level of methylation in the TT2 parent line. (B) DNA methylation status of the germline-specific Dazl and Tuba3 genes in wt, G9a−/−, Dnmt1−/− and Dnmt3a/b−/− cells was confirmed by bisulphite sequencing. The mean number of mCpGs per molecule sequenced is shown, along with the mean % of mCpGs relative to the wild-type line (in parentheses). Both promoters show an ∼40% reduction in DNA methylation density in the G9a−/− line.
DNMT expression is not dramatically altered in G9a−/− cells
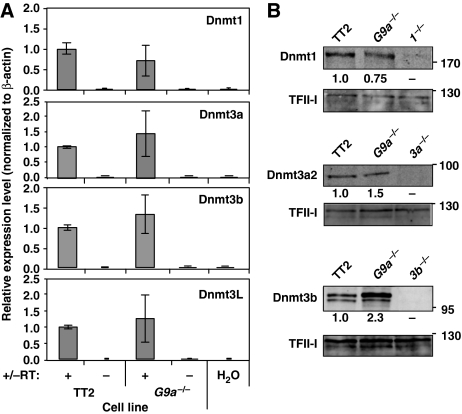
The observed DNA methylation defect prompted us to address whether Dnmt1, Dnmt3a, Dnmt3b or DNMT-like (Dnmt3L) are downregulated in G9a−/− ES cells. Quantitative RT–PCR analysis did not reveal a significant difference in mRNA levels of any of the DNMT family members in these lines (Figure 3A). Similarly, quantitative western blot analyses revealed a <2-fold difference in Dnmt1 or Dnmt3a2 expression levels and an ∼2-fold higher level of Dnmt3b expression in the G9a−/− line than the wt parent line (Figure 3B). These data indicate that the DNA methylation defect observed in G9a−/− cells is unlikely to be a consequence of decreased DNMT expression.
Figure 3.
Expression of DNMTs in G9a−/− cells. (A) RNA was isolated from TT2 wt and G9a−/− cells and expression levels of Dnmt1, Dnmt3a, Dnmt3b and Dnmt3L were determined by real-time quantitative RT–PCR, normalized to β-actin (RT–reverse transcriptase). Values represent the mean (±s.d.) expression level relative to the wild-type line, from three independent experiments. (B) Western blot analyses using quantitative two-colour fluorescence imaging was performed on nuclear extracts isolated from TT2 and G9a−/− cells, using antibodies specific for Dnmt1, Dnmt3a and DNMT3b and TFII-I as an internal control. Extract isolated from DNMT-deficient cells was used as a control for antibody specificity. Relative protein expression levels, normalized to TFII-I, are shown beneath each blot.
Dnmt3a recruitment is reduced in G9a−/− ES cells
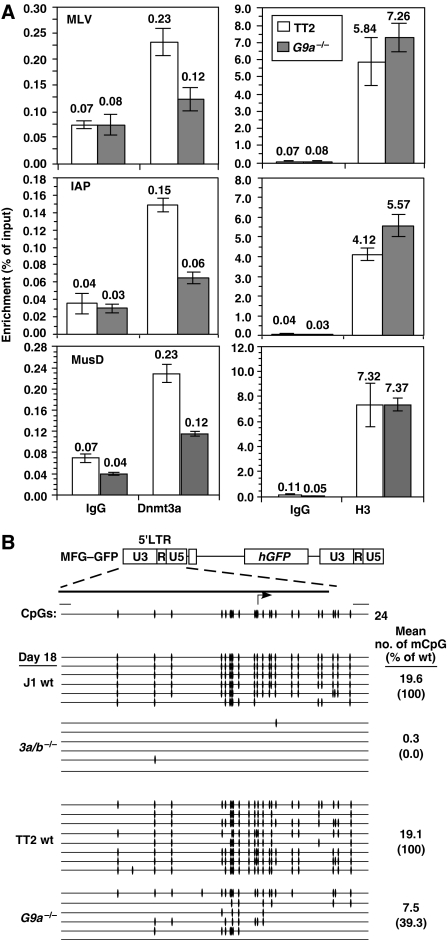
Introduction of Dnmt3a, Dnmt3a2 or Dnmt3b1 is sufficient to restore DNA methylation of retrotransposons in highly demethylated Dnmt3a/b−/− ES cells (Chen et al, 2003), indicating that de novo DNMT activity is required to maintain retrotransposons in a densely methylated state. To determine whether DNMT recruitment to such elements is perturbed in G9a−/− cells, chromatin immunoprecipitation (ChIP) was conducted using Dnmt1-, Dnmt3a/Dnmt3a2- or Dnmt3b-specific antibodies and unmodified histone H3 as an internal control. For Dnmt1 and Dnmt3b, ChIP experiments with two different antibodies specific for each did not yield conclusive results (data not shown). In contrast, a significant reduction in enrichment of Dnmt3a was detected in the LTR regions of MLV, IAP and MusD retrotransposons in the G9a−/− line (Figure 4A). Thus, the decrease in DNA methylation observed in G9a−/− cells can be attributed at least in part to a decrease in the efficiency of recruitment of de novo DNMT activity.
Figure 4.
G9a−/− ES cells show defects in recruitment of Dnmt3a to ERVs and de novo methylation of introduced retroviruses. (A) Formaldehyde-fixed chromatin was isolated from TT2 and G9a−/− lines and ChIP was conducted using nonspecific IgG or antisera raised against Dnmt3a or unmodified histone H3. Real-time PCR using primers specific for the LTR regions of MLV, IAP and MusD ERVs was carried out and values are presented as percentage of input precipitated (±s.d.) relative to the input in the representative experiment shown. A significant reduction in Dnmt3a enrichment in the G9a−/− line relative to the wt control is clearly apparent. (B) TT2 wt, G9a−/−, J1 wt and Dnmt3a/b−/− (3a/b−/−) lines were infected with the retroviral vector MFG–GFP and passaged in the absence of selection. Genomic DNA was isolated on day 18 post-infection and analysed by bisulphite genomic sequencing.
To independently assess whether deletion of G9a not only impairs maintenance of DNA methylation on already methylated loci but also influences the efficiency of de novo DNA methylation on previously unmethylated DNA, TT2 wt, G9a−/−, J1 wt and Dnmt3a/b−/− ES cells were infected with the MLV-based retroviral vector MFG–GFP and the DNA methylation status of the proviral LTR was analysed at day 18 post-infection (Figure 4B). As expected, infected Dnmt3a/b−/− cells show virtually no DNA methylation at this time point. Strikingly, infected G9a−/− cells also show a significantly lower level of DNA methylation (>2.5-fold) than the parent line from which they were derived. Although not as dramatic as the methylation defect observed in the Dnmt3a/b−/−-deficient cell line, this observation indicates that G9a is required for efficient de novo methylation in ES cells.
G9a is not required for transcriptional silencing of retrotransposons
As Dnmt1 was previously shown to be required for silencing of IAP elements in embryos (Walsh et al, 1998), we next determined whether the defect in DNA methylation of retrotransposons in G9a−/− cells is associated with aberrant expression of these potentially active endogenous elements. Expression of MLV and LINE1 elements was not detected above background levels in wt, G9a−/−, Dnmt1−/− or Dnmt3a/b−/− lines by northern blotting (Figure 5A and data not shown). In contrast, IAP elements of each subtype (I, IΔ1 and II) (Kuff and Lueders, 1988) (Figure 5B) and MusD elements (Supplementary Figure S5) are expressed at a significantly higher level in the Dnmt1−/− line than the G9a−/− line, relative to the parent lines from which they were derived. Although not as dramatic as that observed in the Dnmt1−/− line, aberrant IAP expression was also observed in the Dnmt3a/b−/− line by RT–PCR (Figure 5C). As the G9a and DNMT deletions were generated in ES cells of differing genetic backgrounds, it is not possible to attribute the differences in ERV expression exclusively to the genes deleted. Nevertheless, taken together with the DNA methylation data, these results indicate either that the level of residual DNA methylation is sufficient to maintain potentially active retroelements in a silent state in TT2 G9a−/− cells, or that an alternative repressive pathway exerts an effect on these elements independent of DNA methylation.
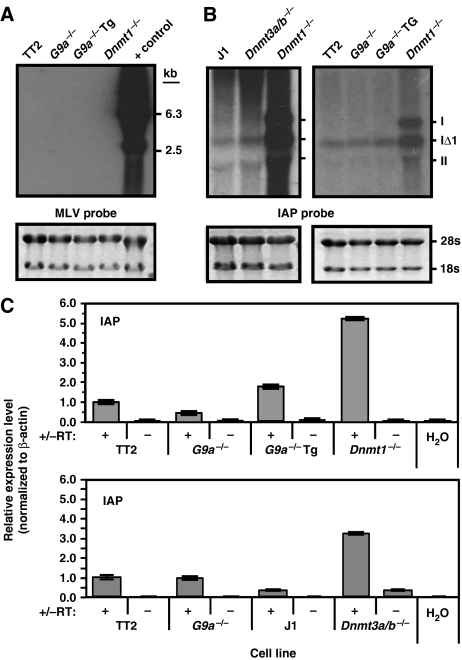
Figure 5.
ERVs are not aberrantly expressed in G9a−/− cells. RNA was isolated from TT2 wt, G9a−/−, G9a−/−Tg, J1 wt, Dnmt3a/b−/− and Dnmt1−/− cells and analysed by northern blotting. RNA isolated from a cell line harbouring an active MLV-based retroviral vector was used as a positive control. 18 and 28s RNA loading controls are shown for each blot. (A) No expression of MLV was detected in any of the lines tested, using a probe specific for the MLV LTR region. (B) A high level of aberrant expression of the three subtypes (I, IΔ1 and II) of IAP elements was detected in the Dnmt1−/− line, but not the G9a−/− line, using a probe specific for the IAP LTR region. (C) Quantitative RT–PCR (+/−RT) using primers specific for the Pol region of full-length IAP elements revealed no increase in expression in the parent or G9a−/− lines, but a significant increase in expression in the Dnmt1−/− and Dnmt3a/b−/− lines.
IAP and MusD ERVs show reduced H3K9 dimethylation in G9a−/− cells, whereas H3K9 trimethylation and HP1α binding are unaffected
Several groups have reported that ERVs and other repetitive sequences are marked by H3K9me2 and/or H3K9me3 in murine ES cells (Peters et al, 2003; Martens et al, 2005; Mikkelsen et al, 2007). As G9a is responsible for the majority of H3K9me2 in euchromatin (Tachibana et al, 2002), we next wished to determine whether ERVs show a decrease in either of these marks in G9a−/− cells. In addition, as methylation of H3K9 creates a binding site for the HP1 family of transcriptional repressor proteins (Lachner et al, 2001; Smallwood et al, 2007), we also wished to determine whether recruitment of HP1α is disrupted in the absence of G9a.
ChIP experiments using chromatin isolated from TT2 and G9a−/− cells and antisera specific for H3K9me2, H3K9me3a, HP1α and unmodified H3 revealed an ∼2-fold reduction in H3K9me2 in the G9a−/− line in the LTR regions of IAP and MusD elements, in the absence of a significant change in H3 occupancy (Figure 6A and B). Consistent with a previous report indicating that H3K9me2 is decreased in the pericentromeric compartment in G9a−/− ES cells (Rice et al, 2003), an ∼2-fold decrease in H3K9me2 was also observed at major satellite repeats, indicating that G9a activity is not confined to the euchromatic compartment and perhaps explaining why major satellite repeats show a DNA methylation defect in G9a−/− cells. As we are simultaneously surveying the modification status of histones associated with multiple copies of each repeat, we cannot discriminate between complete loss of H3K9me2 at approximately half of the repeats, or a uniform ∼2-fold decrease in this mark across all repeats. Nevertheless, these observations indicate that a significant number of IAP and MusD retrotransposons are direct targets of G9a.
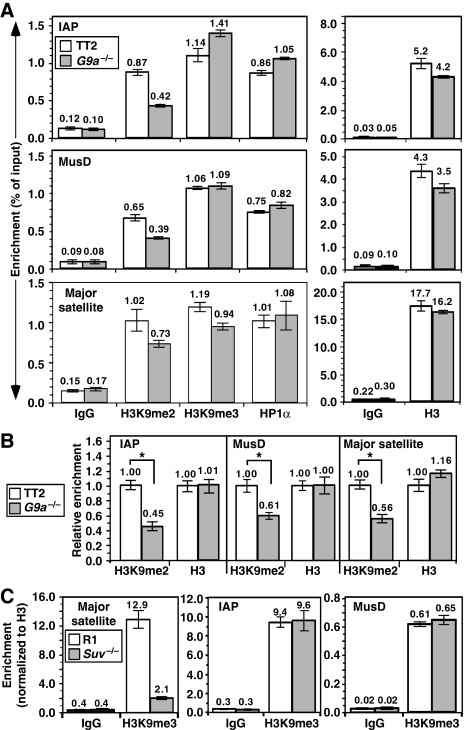
Figure 6.
ERVs show a reduction in H3K9 dimethylation in G9a−/− cells, but no reduction in H3K9 trimethylation or HP1α binding. TT2 wt and G9a−/− ES cells were analysed using ChIP with specific for H3K9me2, H3K9me3, HP1α, unmodified H3 and nonspecific IgG (IgG) as a control. Quantitative real-time PCR was conducted using primers specific for IAP or MusD retrotransposons or major satellite repeats. (A) Mean enrichment values are presented as percentage of input precipitated (±s.d.), relative to the input in the representative experiment shown. (B) Plotting the mean relative enrichment (±s.d.) of H3K9me2 and H3 from three independent experiments reveals an ∼2-fold decrease in H3K9me2 in the G9a−/− line relative to the parent line (*P<0.05, by Student's t-test) but no difference in H3 occupancy at these elements. (C) Relative to the R1 wt parent line, Suv39h1/2−/− (Suv−/−) ES cells show a dramatic decrease in H3K9me3 only at major satellite repeats.
Strikingly, no decrease in H3K9me3 or HP1α binding was detected at IAP or MusD retrotransposons in the G9a−/− line (Figure 6A), indicating that an alternative H3K9 HMTase is responsible for the deposition of the H3K9me3 mark at these elements. Although the HMTases Suv39h1 and Suv39h2 are required for H3K9me3 of major satellite repeats (Peters et al, 2003), they do not have a major function in the deposition of the H3K9me3 mark at IAP or MusD retrotransposons, as H3K9me3 enrichment is not reduced at these elements in Suv39h1/2−/− ES cells either (Figure 6C). In contrast, the promoter region of the G9a-regulated Mage-a2 gene, which is significantly enriched for H3K9me2 in wt but not G9a−/− cells (Tachibana et al, 2002; Supplementary Figure S6), showed only very low levels of H3K9me3 enrichment in either line. Taken together, these results indicate that an HMTase with specificity for H3K9 other than G9a, Suv39h1 or Suv39h2 is responsible for H3K9 trimethylation of ERVs and that the deposition of this mark occurs independent of the DNA methylation state of these elements.
In contrast to G9a−/− cells, Dnmt1−/− cells show no decrease in H3K9me2 across the LTR regions of IAP and MusD elements when normalized to unmodified histone H3 (Figure 7), indicating that Dnmt1 is not required for the deposition of this mark by G9a. Surprisingly, HP1α binding and H3K9me3 across the LTR region of IAP and MusD elements also show only a modest or no reduction, respectively, despite the fact that these elements are hypomethylated and aberrantly expressed in Dnmt1−/− cells. The simplest explanation for this observation is that only a small number of proviruses of each class are actually transcribed in Dnmt1−/− cells and in turn depleted of H3K9me3 and HP1α. Alternatively, severely hypomethylated IAP and MusD elements may be transcriptionally active despite the presence of these repressive marks. In either case, these observations clearly demonstrate that H3K9me3 and HP1α binding at these elements are not dependent on the presence of dense DNA methylation.
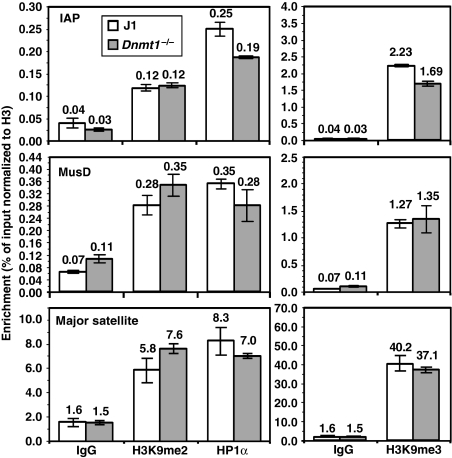
Figure 7.
H3K9 methylation and HP1α binding at MusD, IAP and major satellite repeats in Dnmt1−/− cells. ChIP was conducted on J1 wt and Dnmt1−/− ES cells using antibodies specific for H3K9me2, H3K9me3, HP1α and unmodified H3. Nonspecific IgG was used as a control. Real-time PCR of reverse-crosslinked material using primers specific for IAP, MusD or major satellite repeats was conducted in triplicate and enrichment (±s.d.) is presented as the mean percentage of input material immunoprecipitated, normalized to unmodified H3. IAP elements show a modest reduction in H3K9me3 and HP1α binding in the Dnmt1−/− line, but no change in H3K9me2 enrichment. No significant difference in any of these features was detected at MusD or major satellite repeats.
Introduction of a catalytically inactive G9a transgene partially rescues the DNA methylation defect observed in G9a−/− cells
The observation of a decrease in DNA methylation of ERVs in G9a−/− cells, despite the presence of a high level of residual H3K9 methylation, is surprising, given the known function of K9 methylation in controlling DNA methylation in plants and filamentous fungi (Jackson et al, 2002; Freitag and Selker, 2005). To directly address whether DNA methylation of repetitive elements in mammalian cells is dependent on the catalytic activity of G9a, we took advantage of the previously described observation that the DNA methylation defect observed in G9a−/− cells is rescued by the introduction of a wt G9a transgene (see Figure 1).
Constructs encoding two G9a mutants (G9a−/−Tg(C1168A) and G9a−/−Tg(Y1120V;Y1207F)), each of which harbour amino-acid substitutions in the SET domain that reduce catalytic activity to <1% of that of wt G9a, but do not affect the ability of the encoded protein to form a complex with GLP (see Tachibana et al, this issue), were stably introduced into the G9a−/− line. Western blot analyses of cell lines stably expressing each of these transgenes revealed that the exogenous wt and mutant proteins are produced at the expected molecular weight (Figure 8A). Furthermore, quantitative western blot analysis reveals that GLP is expressed at similar levels in the wt, G9a−/− and G9a−/− Tg lines, confirming that the expression of G9a does not significantly influence the stability of its binding partner GLP (Tachibana et al, 2005) (Figure 8B). As expected, Southern blot analysis with an IAP-specific probe reveals that DNA methylation is reduced in the G9a−/− parent line and restored to wt levels in the G9a−/−Tg(wt) line (Figure 8C). Strikingly, DNA methylation of IAP elements is also increased in cells expressing either of the catalytic mutants, albeit not to the same level as observed for the wt transgene (Figure 8C–E). DNA methylation levels at MusD (Supplementary Figure S7) and L1 repeats (data not shown) were also significantly increased in the G9a−/−Tg(C1168A) line relative to the parent G9a−/− line.
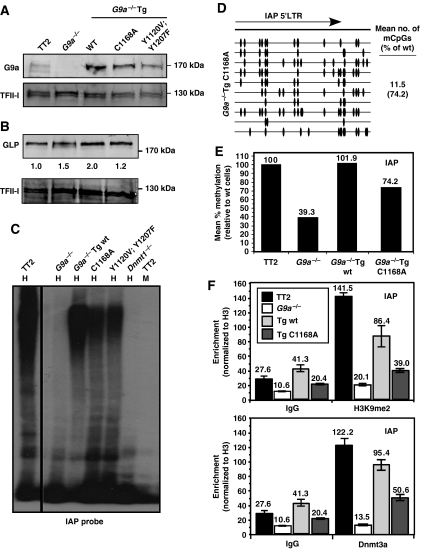
Figure 8.
Stable expression of catalytically inactive G9a transgenes is sufficient to rescue the DNA methylation defect observed in G9a−/− ES cells. The G9a−/− line 2-3 was stably transfected with constructs encoding a wt G9a transgene G9a−/−Tg(wt), or the mutant G9a transgenes G9a−/−Tg(C1168A) and G9a−/−Tg(Y1120V;Y1207F). Each of the latter transgenes harbour amino-acid substitutions in the SET domain that reduce the catalytic activity of the encoded protein to <1% of that of the wt protein. (A) Western blot analysis of cell lines expressing each of these transgenes reveals that the mutant proteins are expressed at the expected molecular weight. An antibody specific for TFII-I was used as a loading control. (B) Quantitative western blot analysis of GLP expression in these lines was determined by normalizing to the signal obtained for endogenous TFII-I on the same blot. (C) Genomic DNA isolated from wt (TT2), G9a−/− (2-3), G9a−/−Tg(wt), G9a−/−Tg(C1168A), G9a−/−Tg(Y1120V;Y1207F) and Dnmt1−/− ES cells was digested with HpaII (H) and subject to Southern blotting using an IAP-specific probe. MspI (M) was used as a control. (D) Bisulphite analysis using primers specific for the IAP 5′LTR confirms that expression of catalytically inactive G9a partially rescues the DNA methylation defect. (E) A bar graph showing the mean no. of mCpGs per molecule sequenced is shown for the bisulphite data presented in (D) and Figure 1D. (F) ChIP was conducted on the wt (TT2), G9a−/− (2-3), G9a−/−Tg(wt) and G9a−/−Tg(C1168A) lines using antisera raised against H3K9me2, Dnmt3a or unmodified H3. Nonspecific IgG was included as a control. Real-time PCR of reverse-crosslinked material using IAP-specific primers was carried out in triplicate and enrichment (±s.d.) is presented as the mean percentage of input material immunoprecipitated, normalized to unmodified H3.
Importantly, the level of H3K9me2 enrichment in the G9a−/−Tg(C1168A) line remains significantly below that observed for the TT2 and G9a−/−Tg(wt) lines (Figure 8F). Furthermore, rescue of the DNA methylation defect in the G9a−/−Tg(C1168A) line is accompanied by recruitment of Dnmt3a to the IAP LTR, although at a level lower than that observed in the wt parent line or the G9a−/− line rescued with the wt G9a transgene (Figure 8F). Taken together, these results reveal that independent of its catalytic activity, G9a promotes de novo DNA methylation through enhancing recruitment of Dnmt3a.
Discussion
We demonstrate that G9a is required for DNA methylation of representative LTR and non-LTR retrotransposons and a number of CpG-rich promoters in murine ES cells. However, unlike the H3K9 HMTases in plants and filamentous fungi (Jackson et al, 2002; Freitag and Selker, 2005), G9a promotes DNA methylation of repetitive elements independent of its catalytic activity. How might G9a influence DNA methylation, given that H3K9 methylation per se does not seem to be the predominant trigger? Although it is possible that deletion of G9a influences DNA methylation through an indirect mechanism, given that H3K9me2 is decreased at each of the genomic regions analysed, we favour the possibility that G9a–GLP exert an effect in cis to promote DNA methylation. Indeed, two groups recently reported that in somatic cells, G9a interacts directly with Dnmt1 in a complex that includes PCNA (Esteve et al, 2006; Sharif et al, 2007), indicating that Dnmt1 and G9a coordinate H3K9 methylation and maintenance DNA methylation at the replication fork.
Our survey of the DNA methylation status of a number of repetitive and single-copy genomic sequences reveals that the extent of the defect in G9a−/− ES cells is generally not as severe as that in Dnmt1−/− ES cells. Although we did detect an interaction between G9a and GLP through co-immunoprecipitation, as described earlier (Tachibana et al, 2005), we were unable to detect an interaction between G9a and any of the DNMTs in ES cells (MCL and SL, unpublished data). On the other hand, we did find that Dnmt3a recruitment to the promoter regions of LTR retrotransposons was reduced in the G9a−/− line relative to the wt control.
Thus, although we cannot confirm whether G9a influences Dnmt1 activity in ES cells, we propose that G9a regulates DNA methylation in these cells at least in part by promoting de novo DNMT activity in cis. In support of this model, we find that introduced MLV-based retroviral vectors, which are unmethylated at the time of integration, are not efficiently de novo methylated in G9a−/− cells and show a defect in silencing similar to that observed in Dnmt3a/b−/− ES cells infected with the same virus (KBD and MCL, in preparation). As maintenance methylation by Dnmt1 is reported to be an inefficient process in ES cells (Liang et al, 2002), continual de novo methylation may be required to preserve DNA methylation homoeostasis. Alternatively, active demethylation by an as yet unidentified DNA demethylase may necessitate ongoing de novo methylation by Dnmt3a and/or Dnmt3b to maintain steady-state levels of this epigenetic mark.
Intriguingly, despite the fact that the potentially active IAP and MusD retrotransposons show a dramatic reduction in DNA methylation in G9a−/− cells, these interspersed repetitive elements remain transcriptionally inactive. As G9a−/− cells show a somewhat higher level of residual DNA methylation than Dnmt1−/− ES cells, in which these elements are aberrantly expressed, it is possible that potentially active ERVs are expressed only when DNA methylation density drops below a critical threshold. Consistent with this model, unlike Dnmt1-null mice, which show high levels of IAP expression (Walsh et al, 1998), compound heterozygous mice carrying a hypomorphic Dnmt1 allele over a null allele show genome-wide hypomethylation but no detectable IAP expression (Howard et al, 2008).
Alternatively, as H3K9me3 enrichment and HP1α binding at IAP and MusD elements are not dramatically reduced in G9a−/− or Dnmt1−/− cells, it is possible that an alternative repressive pathway maintains the vast majority of ERVs in a silent state independent of DNA methylation. Our observations clearly show that an HMTase with specificity for H3K9 other than G9a, GLP, Suv39h1 or Suv39h2 marks LTR retrotransposons in ES cells, leaving SETDB1, which shows di- and tri-methyl HMTase activity towards H3K9 in vitro and in vivo (Wang et al, 2003), or the closely related SETDB2, as the remaining candidates in the Suv39 family of HMTases for this activity.
Given the well-documented deleterious effects of retrotransposition on genomic integrity, the existence of a DNA methylation-independent silencing pathway may serve to minimize proviral expression during those stages in embryonic development when DNA methylation levels are relatively low, such as following fertilization or in the developing germ line. Intriguingly, deletion of the Piwi protein Mili leads to derepression of L1 and IAP ERVs and the loss of DNA methylation at L1 elements (Aravin et al, 2007), revealing that Piwi-interacting RNAs (piRNAs) generated by transposable elements in the germ line are required to maintain these elements in a silent state. Such piRNAs may have a function in the targeting of H3K9 HMTase activity to homologous repetitive elements prior to de novo DNA methylation of these elements.
We show that G9a is required for DNA methylation in ES cells not only of repetitive elements but also of the CpG-rich promoter regions of a number of genes that are normally densely methylated in ES cells. These results are consistent with those reported by Tachibana and colleagues (see accompanying paper by Tachibana et al), and indicate that in addition to the deposition of the H3K9me2 mark, the G9a–GLP complex may have a genome-wide influence on DNA methylation homoeostasis in ES cells. As GLP expression is downregulated in primordial germ cells, coincident with genome-wide DNA demethylation in these cells (Seki et al, 2005, 2007), it is possible that the G9a–GLP heteromeric complex has a function in the programmed changes in DNA methylation that occur not only following fertilization but also in the developing germ line.
Materials and methods
Cell lines
J1 wt (129S4/SvJae), Dnmt1c/c (Dnmt1−/−) (Lei et al, 1996), Dnmt3a and Dnmt3b double-negative (Dnmt3a/b−/−) (Okano et al, 1999), TT2 wt (c57BL/6xCBA), G9a−/− (clones 2-3 and 22-10), G9a−/−Tg (clone 15-3) (Tachibana et al, 2002), GLP−/−, GLP−/−Tg (Tachibana et al, 2005), G9a−/−Tg(wt), G9a−/−Tg(C1168A) (clone G4), G9a−/−Tg(Y1120V;Y1207F) (clone G7), R1 wt (129X1/SvJ × 129S1) and Suv39h1 and Suv39h2 double-negative (Suv39h1/2−/−) (Peters et al, 2001) ES cells were passaged every 48–72 h in DMEM supplemented with 15% FBS (HyClone), 20 mM HEPES, 0.1 mM non-essential amino acids, 0.1 mM 2-mercaptoethanol, 100 U/ml penicillin, 0.05 mM streptomycin, leukaemia-inhibitory factor and 2 mM glutamine on gelatinized plates.
Bisulphite sequencing and MeDIP analyses
Genomic DNA was subject to sodium bisulphite conversion using the EZ DNA Methylation-Gold kit (Zymo Research) as described earlier (Appanah et al, 2007). MeDIP was conducted as described earlier (Weber et al, 2007). Detailed protocols are provided in the Supplementary data.
Northern and Southern blot analyses
Southern blot analyses, restriction digests, membrane transfers and preparation of the DNA probe were performed by standard methods. A detailed protocol is provided in the Supplementary data.
Quantification of proviral mRNA levels
RNA was isolated using Tri reagent (Sigma) according to the manufacturer's protocol. DnaseI-treated RNA was subject to first-strand cDNA synthesis using RevertAid H Minus kit (Fermentas) in the presence or absence of reverse transcriptase. Quantitative RT–PCR using MLV-, IAP- and MusD-specific primers, or β-actin-specific primers as an internal control (all primer sequences are listed in Supplementary Table 1), was conducted with EvaGreen dye (Biotium) on an Opticon 2 thermal cycler (Bio-Rad). Relative expression levels were determined by normalizing to the β-actin gene.
Antibodies and ChIP experiments
ChIP for histones (Appanah et al, 2007) and non-histone proteins (O'Geen et al, 2007) was conducted as described. Details are provided in the Supplementary data.
Western blot analysis
Nuclear extractions were conducted as described (Tachibana et al, 2002). Western blot analyses were conducted using the Odyssey Infrared Imaging System (LI-COR Biosciences), as described in the Supplementary data.
Supplementary Material
Supplementary data
Acknowledgments
We thank Dr En Li for the Dnmt1−/− and Dnmt3a/b−/− cell lines, Dr Thomas Jenuwein for the Suv39h1/2−/− ES cell line and Dr Stephen Smale for HP1-specific antibodies and the UBC flow cytometry and Nucleic Acid Protein Service facilities. We thank Mark Groudine, James Ellis, Carolyn Brown and Jacob Hodgson for comments on the article. This study was supported by CIHR grant 77805 to MCL, 10825 to DLM and a Grant-in Aid from the Ministry of Education, Science, Technology and Culture of Japan to MT and YS. MCL is a Scholar of the Michael Smith Foundation for Health Research. Research in the laboratory of DS is supported by the Novartis Research Foundation and the European Union (LSHG-CT-2004-503433 and LSHG-CT-2006-037415).
References
- Appanah R, Dickerson DR, Goyal P, Groudine M, Lorincz MC (2007) An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS Genet 3: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E (2003) Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 23: 5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, Li E (2002) A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277: 38746–38754 [DOI] [PubMed] [Google Scholar]
- Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S (2006) Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 20: 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y (2006) G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8: 188–194 [DOI] [PubMed] [Google Scholar]
- Freitag M, Selker EU (2005) Controlling DNA methylation: many roads to one modification. Curr Opin Genet Dev 15: 191–199 [DOI] [PubMed] [Google Scholar]
- Goyon C, Rossignol JL, Faugeron G (1996) Native DNA repeats and methylation in Ascobolus. Nucleic Acids Res 24: 3348–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A (2008) Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27: 404–408 [DOI] [PubMed] [Google Scholar]
- Ikegami K, Iwatani M, Suzuki M, Tachibana M, Shinkai Y, Tanaka S, Greally JM, Yagi S, Hattori N, Shiota K (2007) Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuff EL, Lueders KK (1988) The intracisternal A-particle gene family: structure and functional aspects. Adv Cancer Res 51: 183–276 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA (2002) Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol 22: 480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Mabuchi H, Fujii H, Calin G, Alder H, Negrini M, Rassenti L, Kipps TJ, Bullrich F, Croce CM (2001) Cloning and characterization of CLLD6, CLLD7, and CLLD8, novel candidate genes for leukemogenesis at chromosome 13q14, a region commonly deleted in B-cell chronic lymphocytic leukemia. Cancer Res 61: 2870–2877 [PubMed] [Google Scholar]
- Mager DL, Freeman JD (2000) Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J Virol 74: 7221–7229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL (2006) Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24: 800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, van de Lagemaat LN, Mager DL (2002) Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res 12: 1483–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D (2008) Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- O'Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ (2007) Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet 3: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A (2007) Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell 27: 596–608 [DOI] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y (2005) Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278: 440–458 [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M (2007) Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638 [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi SI, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Smallwood A, Esteve PO, Pradhan S, Carey M (2007) Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev 21: 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancheva I (2005) Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem Cell Biol 83: 385–395 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283 [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y (2003) mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell 12: 475–487 [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Cambareri EB, Kinsey JA (2001) DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Mol Genet Genomics 265: 748–754 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data