Abstract
Self-templating amyloid forms of Sup35 constitute the yeast prion [PSI+]. How the protein-remodelling factor, Hsp104, collaborates with other chaperones to regulate [PSI+] inheritance remains poorly delineated. Here, we report how the Ssa and Ssb components of the Hsp70 chaperone system directly affect Sup35 prionogenesis and cooperate with Hsp104. We identify the ribosome-associated Ssb1:Zuo1:Ssz1 complex as a potent antagonist of Sup35 prionogenesis. The Hsp40 chaperones, Sis1 and Ydj1, preferentially interact with Sup35 oligomers and fibres compared with monomers, and facilitate Ssa1 and Ssb1 binding. Various Hsp70:Hsp40 pairs block prion nucleation by disassembling molten oligomers and binding mature oligomers. By binding fibres, Hsp70:Hsp40 pairs occlude prion recognition elements and inhibit seeded assembly. These inhibitory activities are partially relieved by the nucleotide exchange factor, Fes1. Low levels of Hsp104 stimulate prionogenesis and alleviate inhibition by some Hsp70:Hsp40 pairs. At high concentrations, Hsp104 eliminates Sup35 prions. This activity is reduced when Ssa1, or enhanced when Ssb1, is incorporated into nascent prions. These findings illuminate several facets of the chaperone interplay that underpins [PSI+] inheritance.
Keywords: chaperone, Hsp70, Hsp104, prion, Sup35
Introduction
The yeast prion [PSI+] of Saccharomyces cerevisiae is composed of self-templating amyloid forms of Sup35, a translation termination factor. Sup35 prions function poorly in translation termination, leading to nonsense suppression and exposure of hidden genetic variation, which can confer phenotypic diversity and selective advantages (Shorter and Lindquist, 2005). Sup35 is a modular protein comprised of three domains: N (residues 1–123), which is enriched in particular uncharged polar amino acids (Gln, Asn, Tyr) and confers prionogenicity (Ter-Avanesyan et al, 1994; Glover et al, 1997; King and Diaz-Avalos, 2004); M (residues 124–253), which is highly charged and stabilizes the soluble non-prion state (Liu et al, 2002); and C (residues 254–685), which is a GTPase essential for translation termination (Stansfield et al, 1995). Synthetic Sup35 prions can be generated in vitro, by assembling Sup35 or N-terminal fragments of Sup35, including NM (N and M domains of Sup35), into amyloid fibres. Introducing these fibres into [psi−] cells transforms them to [PSI+] (King and Diaz-Avalos, 2004; Tanaka et al, 2004; Shorter and Lindquist, 2006). The facile generation of synthetic prions de novo and the ability to assess their in vivo activity makes [PSI+] a powerful system to investigate how protein-remodelling factors and molecular chaperones directly regulate prion formation and elimination.
We have begun to address this with regard to the protein-remodelling factor and hexameric AAA+ ATPase, Hsp104 (Wendler et al, 2007; Shorter, 2008). Seminal genetic studies revealed that deletion or overexpression of Hsp104 eliminates [PSI+], implying that intermediate levels of Hsp104 are optimal for [PSI+] inheritance (Chernoff et al, 1995). Subsequently, we defined the direct effects of Hsp104 on fibrous and soluble Sup35 using pure proteins (Shorter and Lindquist, 2004, 2006). At low concentrations, Hsp104 promotes Sup35 fibrillization by accelerating the formation of amyloidogenic oligomers that nucleate Sup35 fibrillization. This eliminates or reduces the lag phase observed when NM or Sup35 fibrillize on their own (Shorter and Lindquist, 2004, 2006; Doyle et al, 2007). Hsp104 also severs nascent Sup35 fibres (Shorter and Lindquist, 2004, 2006). Because Sup35 fibres grow from both ends (Scheibel et al, 2001), this generates extra surfaces for conformational replication and accelerates fibrillization (Shorter and Lindquist, 2004, 2006; Doyle et al, 2007). By contrast, at high concentrations, Hsp104 couples ATP hydrolysis to the elimination of amyloidogenic oligomers and self-templating Sup35 fibres (Shorter and Lindquist, 2004, 2006; Doyle et al, 2007; Wendler et al, 2007). Hsp104 converts Sup35 prions to an ensemble of two distinct forms. One form is SDS-soluble, whereas the other is SDS-resistant amyloid-like material with diminished self-templating activity (Shorter and Lindquist, 2006). Importantly, these diverse Hsp104 activities were verified in vivo, by transforming reaction products into [psi−] cells (Shorter and Lindquist, 2006). Together, these activities provide an explanation for the unusual dosage relationship between Hsp104 and [PSI+] inheritance (Shorter, 2008).
The prion-remodelling activities of Hsp104 are unusual because they do not require Hsp70 and Hsp40 in vitro (Shorter and Lindquist, 2004, 2006). However, Hsp104 is unlikely to operate in isolation in vivo. Indeed, Hsp104 collaborates with Hsp70 and Hsp40 to dissolve and renature the aggregated proteome after environmental stress (Parsell et al, 1994; Glover and Lindquist, 1998). Hsp70 and Hsp40 present denatured aggregated substrates to Hsp104 and refold newly disaggregated polypeptides (Glover and Lindquist, 1998; Haslberger et al, 2007). Yet, their requirement can be bypassed altogether, at least for some aggregates, by asymmetric deceleration of Hsp104 ATPase activity (Doyle et al, 2007) or by the presence of the small heat shock protein, Hsp26, in the aggregate (Haslbeck et al, 2005). Nonetheless, shifting the cellular balance of various Hsp70s and Hsp40s influences [PSI+] induction and propagation (Chernoff et al, 1999; Newnam et al, 1999; Jung et al, 2000; Kushnirov et al, 2000; Kryndushkin et al, 2002; Jones et al, 2004; Allen et al, 2005), although none as stringently as Hsp104. Yet, how they might directly affect the conformational states of the prion protein is uncertain and remains virtually unaddressed.
In yeast, two cytosolic Hsp70 subfamilies affect [PSI+] inheritance: Ssa and Ssb. Ssa can promote or antagonize [PSI+], whereas Ssb antagonizes [PSI+] inheritance (Chernoff et al, 1999; Jung et al, 2000; Kushnirov et al, 2000; Kryndushkin et al, 2002; Jones et al, 2004; Allen et al, 2005). The Ssa subfamily has four members, at least one of which must be expressed for viability (Werner-Washburne et al, 1987). They have crucial functions in protein folding, translocation and prevention of aggregation (Deshaies et al, 1988; Albanese et al, 2006). The Ssb subfamily has two members that interact with nascent polypeptides emerging from ribosomes, although a fraction is found throughout the cytoplasm (Nelson et al, 1992; Albanese et al, 2006). The functions of these proteins in [PSI+] inheritance have been inferred almost entirely by genetic analyses. High levels of Ssa1 or Ssb1 cure weak [PSI+] variants, as do high levels of the Hsp40s, Sis1 and Ydj1. However, strong [PSI+] variants are largely unaffected by these treatments (Kushnirov et al, 2000; Kryndushkin et al, 2002). Ssa1 mutants with enhanced substrate binding destabilize [PSI+], as does deletion of the Ssa1 nucleotide exchange factors (NEFs), Fes1 or Sse1 (Jung et al, 2000; Jones et al, 2004; Sadlish et al, 2008). Ssa1 or Sse1 overexpression promotes de novo [PSI+] induction, even in the absence of [PIN+] (Allen et al, 2005; Sadlish et al, 2008). Spontaneous [PSI+] appearance increases 10-fold when Ssb1 and Ssb2 are deleted (Chernoff et al, 1999). Intriguingly, Ssa1 antagonizes [PSI+] curing by Hsp104 overexpression, whereas Ssb1 enhances it (Chernoff et al, 1999; Newnam et al, 1999). These opposing in vivo effects are mediated by the Ssa and Ssb substrate-binding domains (Allen et al, 2005).
It remains unclear whether the foregoing genetic data reflect direct effects on Sup35 folding or are more indirect, complex pleiotropic effects, such as alterations in the levels of other chaperones or subtle alterations in chaperone networks or other factors that affect [PSI+]. A physical interaction between Sup35 and Ssa1 or Ssb1 has been observed in crude lysates (Allen et al, 2005) and Ssa1 and Ssa2 are major components of ex vivo Sup35 prions (Bagriantsev et al, 2008). An in vitro study revealed that His-tagged Ssa1:Sis1 or Ssa:Ydj1 antagonizes His-tagged Sup35 fibre assembly and could interact with His-tagged Sup35 oligomers (Krzewska and Melki, 2006). However, several major limitations preclude definitive conclusions being drawn from this study. First, Sup35 was purified in the absence of GTP (Krzewska and Melki, 2006), making it prone to forming off-pathway, non-prion aggregates (Shorter and Lindquist, 2006). Second, because no protein transformation assays were performed, it is unclear whether the events truly reflected prionogenesis (Krzewska and Melki, 2006). Third, the full repertoire of Hsp104 prion-remodelling activities were not reconstituted, because under the conditions employed, the interaction between Sup35 and Hsp104 was drastically altered (Krzewska and Melki, 2006; Shorter and Lindquist, 2006). It thus remains unclear how Hsp104 interfaces with the Hsp70 chaperone system in prion inheritance. Furthermore, these studies (Krzewska and Melki, 2006) did not consider the function of the Ssb Hsp70 family or how NEFs, an integral part of the Hsp70 ATPase cycle, affect how Hsp70 might regulate Sup35 fibrillization. Here, we address these issues and define the direct effects of the Ssa and Ssb (including Ssz) components of the Hsp70 chaperone system, various Hsp40 s (Sis1, Ydj1, Zuo1) and NEFs (Fes1) on N, NM and Sup35 conformational conversion. We then describe how the Hsp70 chaperone system combines with Hsp104 to regulate Sup35 prions. Throughout, we verify our findings in vivo, by transforming reaction products into prion-minus yeast cells.
Results
Sis1 and, to a lesser extent, Ydj1 inhibit spontaneous NM fibrillization
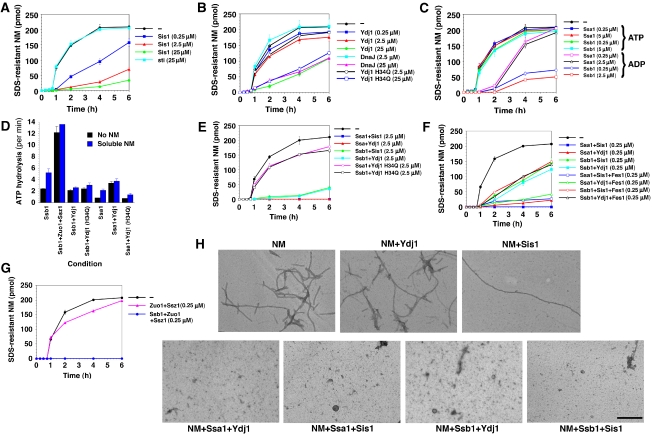
First, we tested how two of the major cytosolic Hsp40s, Ydj1 and Sis1, affected spontaneous NM fibrillization. In the absence of other proteins, NM fibres formed after a lag phase of ∼45 min and assembly was complete after 4 h as determined by acquisition of SDS resistance (Figure 1A) and Thioflavin-T (ThT) fluorescence (data not shown). These two measures were used throughout and gave very similar results in all cases. A 10-fold molar excess of a non-specific protein, soybean trypsin inhibitor (sti), had no effect (Figure 1A). By contrast, Sis1 inhibited NM fibrillization even when NM was in 10-fold excess (Figure 1A). Equimolar or excess Sis1 allowed little NM fibrillization (Figure 1A). Ydj1 also inhibited spontaneous NM fibrillization, but not as potently as Sis1. A 10-fold excess of Ydj1 was required for appreciable inhibition, but this was a relatively non-specific effect because an Escherichia coli Hsp40, DnaJ, also inhibited at these concentrations (Figure 1B). Ydj1 H34Q, a J-domain mutant that binds unfolded polypeptide but cannot stimulate the ATPase activity of Ssa1 (Lu and Cyr, 1998b), also inhibited NM assembly (Figure 1B). Electron microscopy (EM) confirmed that NM formed fibres in the presence of equimolar Ydj1, whereas fibres were few and long in the presence of Sis1 (Figure 1H). Thus, Sis1 reduces the number of amyloidogenic nuclei that form.
Figure 1.
Effects of Hsp70 and Hsp40 on spontaneous NM fibrillization. (A–C) Kinetics of unseeded, rotated (80 r.p.m.) NM (2.5 μM) fibrillization in the presence or absence of either soybean trypsin inhibitor (sti), Sis1 (A), Ydj1, DnaJ (0.25–25 μM) (B), Ssa1 or Ssb1 (0.25–5 μM) (C). All reactions contained ATP (5 mM), except for some reactions in (C) that contained ADP (5 mM). Fibrillization was monitored by the amount of SDS-resistant NM. Values represent means±s.d. (n=3). (D) Ssb1 or Ssa1 (1 μM) were incubated for 30 min at 25°C with or without Ydj1 or Ydj1 H34Q (1 μM) in the presence or absence of soluble NM (2.5 μM). The amount of ATP hydrolysis was then determined. Values represent means±s.d. (n=3). (E–G) NM (2.5 μM) fibrillization as in (A) in the presence or absence of either Ssa1:Ydj1 H34Q, Ssb1:Ydj1 H34Q (2.5 μM) (E), Ssa1:Sis1, Ssa1:Ydj1, Ssb1:Sis1, Ssb1:Ydj1, (0.25-2.5 μM) (E, F), Ssa1:Sis1:Fes1, Ssa1:Ydj1:Fes1, Ssb1:Sis1:Fes1, Ssb1:Ydj1:Fes1 (0.25 μM) (F), Zuo1:Ssz1 or Ssb1:Zuo1:Ssz1 (0.25 μM) (G). Values represent means±s.d. (n=3). (H) EM of unseeded, rotated (80 r.p.m.) NM (2.5 μM) fibrillization after 6 h in the absence or presence of either Ydj1, Sis1, Ssa1:Sis1, Ssa1:Ydj1, Ssb1:Sis1 or Ssb1:Ydj1 (2.5 μM). Bar, 0.5 μm.
Ssa1 and Ssb1 inhibit spontaneous NM fibrillization in the presence of ADP
Next, we determined how the two major classes of cytosolic Hsp70 chaperones affected spontaneous NM fibrillization in the presence of ATP. Neither Ssa1 nor Ssb1 inhibited spontaneous NM fibrillization, even when twice as abundant as NM (Figure 1C). However, if ATP was replaced with ADP, then Ssa1 extended the lag phase of NM fibrillization, but did not affect the amount of assembly after 6 h (Figure 1C). Similarly, Ssb1+ADP extended the lag phase of NM assembly, but also greatly reduced assembly after 6 h even at concentrations 10-fold lower than NM (Figure 1C). Thus, both Ssa1 and Ssb1 have an intrinsic ability to antagonize NM fibrillization, but this is regulated by their nucleotide state.
Hsp70s initially engage substrates in an ATP-bound ‘open' state and typically recognize short peptide segments (∼7 residues) that are exposed in unfolded proteins. With conformational change induced by ATP hydrolysis, substrate peptides are clamped in an extended form through a groove in the Hsp70 substrate-binding domain (Mayer et al, 2001). We investigated how soluble NM affected Ssa1 and Ssb1 ATPase activity. In the absence of NM, Ssb1 hydrolysed ∼3-fold more ATP than Ssa1 (Figure 1D). Soluble NM stimulated Ssa1 ATPase activity ∼4-fold and Ssb1 ATPase activity ∼2-fold. However, this stimulated ATPase activity was not coupled to productive interactions able to inhibit NM fibrillization (Figure 1C).
Effects of Hsp70:Hsp40 pairs on spontaneous NM fibrillization
Through their N-terminal J-domain, specific Hsp40s stimulate the ATPase activity of specific Hsp70s (Mayer et al, 2001). Therefore, we considered the effects of Hsp70:Hsp40 pairs on NM assembly. Both Sis1 and Ydj1 stimulate Ssa1 ATPase and chaperone activity (Lu and Cyr, 1998a). Indeed, Ydj1 stimulated Ssa1 ATPase ∼4.5-fold in the absence of NM and ∼2-fold in the presence of soluble NM (Figure 1D). Both Ssa1:Ydj1 and Ssa1:Sis1 blocked NM assembly, even when NM was in 10-fold excess (Figure 1E and F). These synergistic effects were eliminated when ATP was replaced with ADP or the non-hydrolysable ATP analogue, AMP-PNP (data not shown). Furthermore, the Ydj1 J-domain mutant, Ydj1 H34Q, defective in stimulating Ssa1 ATPase activity (Figure 1D) (Lu and Cyr, 1998b), failed to synergize with Ssa1 (Figure 1E). Thus, synergy between Ydj1 or Sis1 and Ssa1 in preventing NM fibrillization requires Ssa1 ATPase activity.
The ATPase activity and chaperone activity of Ssb1 towards some model substrates is not activated by Sis1 or Ydj1 (Lopez-Buesa et al, 1998; Lu and Cyr, 1998a). Inhibition of NM fibrillization by Ssb1:Sis1 could be accounted for almost entirely by inhibition by Sis1 alone (Figure 1A and F), whereas Ssb1:Ydj1 inhibited NM fibrillization in a manner that could not be explained entirely by additive effects (Figure 1B and F). Ssb1 ATPase activity was required for Ssb1:Ydj1 to inhibit NM assembly, as inhibition was relieved by replacing ATP with AMP-PNP (data not shown). Moreover, the Ydj1 J-domain mutant, Ydj1 H34Q, reduced the inhibition by Ssb1 (Figure 1E). Ydj1 did not stimulate Ssb1 ATPase activity in the presence of soluble NM and neither did Ydj1 H34Q (Figure 1D). Thus, even though Ydj1 does not stimulate global Ssb1 ATPase activity, it productively couples Ssb1 ATPase activity to an inhibition of NM fibrillization and this requires a functional J-domain. One possibility is that Ydj1 stimulates Ssb1 ATPase only in the presence of specific NM conformers. We will return to this point.
Ssb1 ATPase activity is stimulated ∼5-fold by a heterodimer of Zuo1, an Hsp40, and Ssz1, a ribosome-associated Hsp70 (Figure 1D) (Huang et al, 2005). Accordingly, Ssb1:Zuo1:Ssz1 eliminated NM fibrillization, whereas Zuo1:Ssz1 had no effect (Figure 1G). Thus, Ssb1 can interface with Ydj1 or more potently with Zuo1:Ssz1 to antagonize spontaneous NM fibrillization.
When considering Hsp70 action, it is important to account for the entire Hsp70 ATPase cycle. Hsp70 releases substrate upon exchange of ADP for ATP, which is catalysed by NEFs. Hence, we tested the effect of Fes1, a NEF for Ssa1 and Ssb1 (Kabani et al, 2002; Dragovic et al, 2006), on these inhibitory activities of Ssa1 and Ssb1. Fes1 slightly reduced the inhibition of NM assembly by Ssa1:Ydj1, Ssa1:Sis1 and Ssb1:Ydj1 (but not Ssb1:Sis1, as this effect was mediated by Sis1 alone) (Figure 1F). Thus, it appears that stable interaction of Hsp70 with NM is required for maximal inhibition of fibrillization.
Effects of Hsp70 and Hsp40 on N and Sup35 prion assembly
Hsp70 and Hsp40 prevent aggregation by binding short hydrophobic patches on polypeptide surfaces (Mayer et al, 2001). However, NM contains a paucity of hydrophobic peptides predicted to interact with Hsp70. One algorithm (Rüdiger et al, 1997) predicts that DnaK, an E. coli Hsp70, might bind a highly conserved region of M spanning residues 138–152 (PKPKKTLKLVSSSGI). Although Ssa1 and Ssb1 are likely to have different substrate specificities than DnaK, we wondered whether binding to this region is important for the inhibition of NM fibrillization. However, NM fibrillization is driven by sequences in the N domain (Glover et al, 1997; Krishnan and Lindquist, 2005), and so it is not obvious how chaperone binding to this stretch might compromise assembly. Thus, we tested whether Hsp70 and/or Hsp40 could inhibit the rapid amyloidogenesis of N.
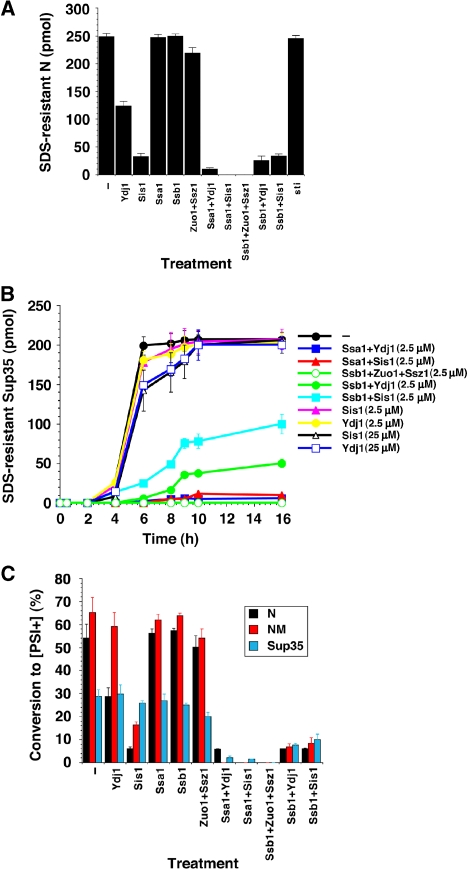
N assembles into amyloid fibres within a few minutes (Glover et al, 1997) and was unaffected by sti, Ssa1+ATP or Ssb1+ATP (Figure 2A). Ydj1 was much more effective in antagonizing N fibrillization than NM fibrillization (Figures 1B and 2A). As with NM, Sis1 potently inhibited N assembly, as did combinations of Ssa1:Ydj1, Ssa1:Sis1 Ssb1:Zuo1:Ssz1, Ssb1:Ydj1 and Ssb1:Sis1 (Figure 2A). Therefore, Hsp70 and Hsp40 recognize determinants within N to prevent fibrillization.
Figure 2.
Effects of Hsp70 and Hsp40 on spontaneous N, NM and Sup35 prionogenesis. (A) Unseeded, rotated (80 rpm) His–N (2.5 μM) fibrillization with ATP (5 mM) after 20 min in the presence or absence of sti, Ydj1, Sis1, Ssa1, Ssb1, Ssa1:Sis1, Ssa1:Ydj1, Ssb1:Sis1 or Ssb1:Ydj1 (2.5 μM). Fibrillization was monitored by the amount of SDS-resistant N. Values represent means±s.d. (n=3). (B) Kinetics of unseeded, rotated (80 r.p.m.) Sup35 (2.5 μM) fibrillization with ATP (5 mM) in the presence or absence of Ydj1, Sis1 (2.5–25 μM), Ssa1:Sis1, Ssa1:Ydj1, Ssb1:Zuo1:Ssz1, Ssb1:Sis1 or Ssb1:Ydj1 (2.5 μM). Fibrillization was monitored by the amount of SDS-resistant Sup35. Values represent means±s.d. (n=3). (C) His–N (2.5 μM) was assembled for 20 min, or NM (2.5 μM) or Sup35 (2.5 μM) were assembled for 6 h in the presence or absence of Ydj1, Sis1, Ssa1:Sis1, Ssa1:Ydj1, Ssb1:Zuo1:Ssz1, Ssb1:Sis1 or Ssb1:Ydj1 (2.5 μM). Reaction products were concentrated and transformed into [psi−] cells. The proportion of [PSI+] colonies was then determined. Values represent means±s.d. (n=3).
Next, we tested how Hsp70 and/or Hsp40 affected assembly of full-length Sup35, purified under native conditions (Shorter and Lindquist, 2006). Here, Sis1 or Ydj1 alone had no effect, even at a 10-fold molar excess over Sup35 (Figure 2B). Ssa1+ATP and Ssb1+ATP alone were also ineffective (data not shown). Ssa1:Ydj1 and Ssa1:Sis1 potently inhibited Sup35 assembly (Krzewska and Melki, 2006), as did Ssb1:Zuo1:Ssz1 (Figure 2B). Surprisingly, so did Ssb1:Sis1 and Ssb1:Ydj1, although not as strongly as the Ssa1 pairs (Figure 2B). Thus, Ydj1 or Sis1 can synergize with Ssa1 or Ssb1 to antagonize Sup35 fibrillization.
Although N, NM and Sup35 fibrillization is clearly inhibited in vitro, have we actually inhibited prion formation? To test this, we determined whether reaction products could transform [psi−] cells to [PSI+] (Shorter and Lindquist, 2006). The ability of an Hsp70, Hsp40 or Hsp70:Hsp40 pair to inhibit N, NM or Sup35 fibrillization translated directly into an ability to inhibit the formation of infectious conformers (Figure 2C). Thus, in this minimal system, Hsp70 and Hsp40 directly regulate prion formation.
Kinetic sensitivity of NM and Sup35 fibrillization to Hsp70 and Hsp40
Next, we tested which stage of NM fibrillization was sensitive to Hsp70 and Hsp40. Various combinations of Hsp70 and/or Hsp40 were added to fibrillization reactions at designated times, and these reactions were then allowed to proceed for a total time of 6 h. At the same designated times, other NM fibrillization reactions proceeding in the absence of chaperones were stopped or treated with reaction buffer. This allowed us to determine the latest stage of NM fibrillization that is sensitive to disruption by any given chaperone.
NM assembled into fibres after a lag phase of ∼45 min and an assembly phase of ∼195 min (Figure 1A). In control reactions, fibrillization was insensitive to buffer (Figure 3A). Hsp104 reversed NM fibrillization regardless of how far the reaction had progressed (Figure 3A), consistent with the ability of Hsp104 to disassemble NM fibres and amyloidogenic oligomers at this concentration (Shorter and Lindquist, 2004). Equimolar Ydj1 failed to significantly inhibit NM assembly at any stage (Figure 1B; data not shown). Sis1 only antagonized NM fibrillization if added during the incipient 30 min of assembly (Figure 3A). Thus, Sis1 specifically antagonizes reaction intermediates that accumulate during lag phase. Once fibres begin to form, Sis1 is unable to antagonize conformational conversion. Addition of any combination of Hsp70 and Hsp40 at any stage of NM fibrillization arrested further assembly, suggesting an inhibition of seeded assembly (Figure 3A). In contrast to Hsp104, Hsp70 and Hsp40 did not disassemble NM fibres after they had formed (Figure 3A, 4 h time point). Very similar results were obtained using full-length Sup35 except that Sis1 alone did not antagonize Sup35 fibrillization (Figure 3B).
Figure 3.
Kinetic sensitivity of NM and Sup35 fibrillization to Hsp70 and Hsp40. (A) Unseeded, rotated (80 r.p.m.) NM (2.5 μM) fibrillization reactions were performed with ATP (5 mM), and at the indicated times, reactions were either terminated or treated with buffer, Hsp104, Sis1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (2.5 μM) and incubated for a total time of 6 h. Fibrillization was assessed by sedimentation analysis. Values represent means±s.d. (n=3). (B) Unseeded, rotated (80 r.p.m.) Sup35 (2.5 μM) fibrillization reactions were performed with ATP (5 mM) and at the indicated times reactions were either terminated or treated with buffer, Hsp104, Sis1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (2.5 μM), and incubated for a total time of 6 h. Fibrillization was assessed by ThT fluorescence. Values represent means±s.d. (n=3).
Hsp70 and Hsp40 target NM and Sup35 oligomers
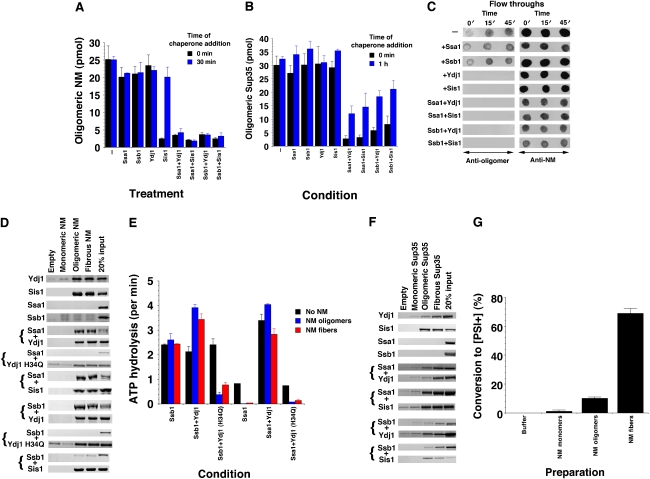
How do Hsp70 and Hsp40 inhibit fibrillization during lag phase? During this phase, structurally malleable NM oligomers rapidly assemble and gradually metamorphose into structured, amyloidogenic oligomers that elicit rapid fibrillization and concomitant conformational replication (Serio et al, 2000; Shorter and Lindquist, 2004). Adding these malleable oligomeric intermediates at the beginning of NM fibrillization shortens the lag phase and accelerates assembly in a dose-dependent way. These malleable NM oligomers possess a hydrodynamic radius of 50–130 nm and can be recovered by ultracentrifugation (Scheibel and Lindquist, 2001; Scheibel et al, 2004). Their formation was unaffected by equimolar concentrations of Ssa1, Ssb1 or Ydj1 (Figure 4A, black bars). Importantly, Sis1, Ssa1:Sis1, Ssa1:Ydj1, Ssb1:Ydj1 and Ssb1:Sis1 reduced oligomer formation ∼10-fold (Figure 4A, black bars). Very similar results were obtained with full-length Sup35, except that Sis1 failed to inhibit Sup35 oligomer formation (Figure 4B, black bars). This inhibition of molten oligomer formation explains how these chaperone combinations inhibit spontaneous NM and Sup35 fibrillization.
Figure 4.
Hsp70 and Hsp40 target NM oligomers and fibres to inhibit fibrillization. (A) NM (2.5 μM) with ATP (5 mM) was rotated for 5 min (80 r.p.m.) in the presence or absence of Ssa1, Ssb1, Ydj1, Sis1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (2.5 μM) (black bars). Alternatively, NM (2.5 μM) with ATP (5 mM) alone was rotated (80 r.p.m.) for 30 min and then Ssa1, Ssb1, Ydj1, Sis1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (2.5 μM) were added. This reaction was continued for 10 min (blue bars). Oligomeric NM was recovered by centrifugation at 436 000 g for 30 min and the amount in the pellet fraction determined. Values represent means±s.d. (n=3). (B) Sup35 (2.5 μM) with ATP (5 mM) was rotated for 1 h (80 r.p.m.) in the presence or absence of either Ssa1, Ssb1, Ydj1, Sis1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (2.5 μM) (black bars). Alternatively, Sup35 (2.5 μM) with ATP (5 mM) alone was rotated (80 r.p.m.) for 1 h and then Ssa1, Ssb1, Ydj1, Sis1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (2.5 μM) were added. This reaction was continued for 10 min (blue bars). Oligomeric Sup35 was recovered by centrifugation at 436 000 g for 30 min and the amount in the pellet fraction determined. Values represent means±s.d. (n=3). (C) NM (2.5 μM) with ATP (5 mM) was rotated (80 r.p.m.) for 0–50 min. At 0, 15 and 45 min, His–tagged Ssa1, Ssb1, Ydj1, Sis1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 were added. At 50 min, reactions were depleted of chaperones using Ni-NTA agarose. Depleted reactions were spotted onto nitrocellulose and probed with anti-oligomer or anti-NM antibody. (D) Ssa1, Ssb1, Ydj1, Sis1, Ssa1:Ydj1, Ssa1:Ydj1 H34Q, Ssa1:Sis1, Ssb1:Ydj1, Ssb1:Ydj1 H34Q or Ssb1:Sis1 (2.5 μM) were incubated in the presence of ATP (5 mM) with Ni-NTA beads (empty) or beads bound to NM–His monomers, oligomers or fibres. Washed beads were eluted and eluates were fractionated by SDS–PAGE and Coomassie-stained or processed for immunoblot (to detect Sis1). (E) Ssb1 or Ssa1 (1 μM) were incubated for 30 min at 25°C with or without Ydj1 or Ydj1 H34Q (1 μM) in the presence or absence of NM oligomers or fibres (2.5 μM). The amount of ATP hydrolysis was then determined. Values represent means±s.d. (n=3). (F) Ssa1, Ssb1, Ydj1, Sis1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (2.5 μM) were incubated in the presence of ATP (5 mM) with Ni-NTA beads (empty) or beads bound to His–Sup35 monomers, oligomers or fibres. Washed beads were eluted, and eluates were fractionated by SDS–PAGE and Coomassie-stained. (G) NM–His monomers, oligomers or fibres were transformed into [psi−] cells. The proportion of [PSI+] colonies was then determined. Values represent means±s.d. (n=3).
Next, we determined how Hsp70 and/or Hsp40 affect preformed molten NM oligomers. Thus, we allowed molten oligomers to form for 30 min and then added the indicated Hsp70 and/or Hsp40 and incubated for a further 10 min. Ssa1+ATP, Ssb1+ATP and Ydj1 had no effect on preformed NM oligomers (Figure 4A, blue bars). Neither did Sis1 (Figure 4A), which suggests that the ability of Sis1 to inhibit NM assembly when added at 30 min may be due to simple binding to NM conformers. By contrast, combinations of Hsp70 and Hsp40 disassembled preformed oligomers and reduced their abundance ∼10-fold (Figure 4A, blue bars). Similar results were obtained with full-length Sup35, although disassembly was not as effective as with NM (Figure 4B, blue bars). Hence, Hsp70 and Hsp40 potently inhibit NM and Sup35 assembly even when added late in lag phase because they can disassemble molten oligomers.
These data suggest that Sis1, Ssa1:Sis1, Ssa1:Ydj1, Ssb1:Ydj1 and Ssb1:Sis1 would prevent the appearance of amyloidogenic NM oligomers that nucleate NM assembly (Serio et al, 2000). These obligate intermediates can be tracked using a conformation-specific antibody that recognizes mature NM oligomers, but not NM fibres, monomers or immature oligomers (Shorter and Lindquist, 2004). Unfortunately, this approach was not viable because Hsp70 and Hsp40 cross-reacted with the anti-oligomer antibody but not with an anti-NM antibody (data not shown). Thus, to determine if Hsp70 and Hsp40 could interact with these intermediates, we added His-tagged chaperones at various times during lag phase. We then depleted reactions of chaperones using Ni-NTA agarose and probed the flow-through (unbound) fractions for amyloidogenic NM oligomers. In the absence of chaperones, flow-through fractions contained amyloidogenic NM oligomers that were more abundant at the end of lag phase (Figure 4C). Neither Ssa1+ATP nor Ssb1+ATP depleted reactions of these forms (Figure 4C). By contrast, Ydj1 or Sis1 alone completely depleted amyloidogenic NM oligomers, as did any combination of Hsp70 and Hsp40 (Figure 4C). Surprisingly, NM was still abundant in the flow-through, which is consistent with a selective depletion of oligomers that constitute only ∼10% of the total NM (Scheibel and Lindquist, 2001). Hence, Ydj1 and Sis1 interact with amyloidogenic oligomers, but only Sis1 inhibits assembly directly. Ydj1 requires the additional presence of Ssa1 or Ssb1.
These data suggest that Hsp70 and Hsp40 interact selectively with particular NM conformers. We tested this directly by attaching NM–His monomers, oligomers or fibres to Ni-NTA (see Materials and methods) and determining whether they bound Hsp70 and/or Hsp40. Intriguingly, no Hsp70 and/or Hsp40 bound monomeric NM stably enough to be recovered in this manner, with the exception of Ydj1 alone, which bound in small amounts (Figure 4D). This suggests that NM monomers may not display motifs that are strongly recognized by Hsp70 and Hsp40. By contrast, NM oligomers and fibres bound Ydj1 or Sis1 alone, but not Ssa1+ATP or Ssb1+ATP alone (Figure 4D). However, Ydj1 enabled retrieval of Ssa1 and Ssb1 with NM oligomers or fibres, as did Sis1 (Figure 4D). The Ydj1 H34Q J-domain mutant failed to recruit Ssa1 or Ssb1 to NM oligomers or fibres, even though Ydj1 H34Q bound these conformers itself (Figure 4D). NM oligomers and fibres strongly inhibited Ssa1 ATPase activity in the absence of Ydj1 (Figure 4E). Ydj1, however, greatly stimulated Ssa1 ATPase activity under these conditions, whereas Ydj1 H34Q did not (Figure 4E). Ssb1 ATPase activity was unaffected by NM fibres and oligomers in the absence of Ydj1 (Figure 4E), but Ydj1 stimulated Ssb1 ATPase ∼1.6-fold in the presence of oligomers or fibres (Figure 4E). By contrast, Ssb1 ATPase activity was inhibited in the presence of Ydj1 H34Q and NM oligomers or fibres (Figure 4E). Thus, Ydj1 with a functional J-domain couples Ssb1 ATPase activity to NM oligomer or fiber binding. This ability to selectively target Ssb1 to NM oligomers explains why Ydj1 synergizes with Ssb1 in inhibiting spontaneous NM fibrillization (Figure 1F).
We were concerned that the observed interactions (Figure 4D) may be due to tethering NM through its C-terminal tag; however, N-terminally tagged His–NM gave identical results (data not shown). Moreover, similar results were obtained using full-length Sup35, except that some faint interactions were now detected with Sup35 monomers (Figure 4F). These were considerably less robust than the interactions with oligomers and fibres (Figure 4F) and might be due to a slight instability of Sup35's C-terminal domain. Apparently, NM and Sup35 oligomers and fibres expose conformational epitopes not found in monomeric forms, which are recognized by Hsp40. This facilitates the recruitment of Hsp70. Our NM oligomer preparations converted [psi−] cells to [PSI+]. However, they did not do so as effectively as NM fibres (Figure 4G) and it might be that a fraction of the oligomers converted to fibres during the incubations required for transformation. Taken together, these data suggest that Hsp70 and Hsp40 exert their inhibitory effects, at least in part, by targeting infectious conformers.
Hsp70:Hsp40 pairs inhibit seeded assembly by occluding prion recognition elements
The binding of Hsp70 and Hsp40 to NM and Sup35 fibres (Figure 4D and F) and the temporal sensitivity of NM and Sup35 assembly to Hsp70:Hsp40 pairs (Figure 3) suggested an ability to inhibit assembly seeded by preformed fibres. Hsp70 or Hsp40 alone were unable to inhibit seeded assembly (Figure 5A). By contrast, Hsp70:Hsp40 pairs inhibited seeded assembly, with Ssa1 pairs inhibiting more than Ssb1 pairs (Figure 5A). Fes1, a NEF for Ssa1 and Ssb1 (Kabani et al, 2002; Dragovic et al, 2006), reduced this inhibition (Figure 5A). Very similar results were observed with full-length Sup35 (Figure 5B). These data suggest that Hsp70:Hsp40 pairs might interact with fibres in a manner that blocks their growing ends.
Figure 5.
Hsp70 and Hsp40 inhibit seeded NM fibrillization and occlude prion recognition elements. (A) Seeded (2% wt/wt) NM (2.5 μM) fibrillization with ATP (5 mM) after 16 h in the presence or absence of sti, Ydj1, Sis1, Ssa1, Ssb1, Fes1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1, Ssb1:Sis1, Ssa1:Ydj1:Fes1, Ssa1:Sis1:Fes1, Ssb1:Ydj1:Fes1 or Ssb1:Sis1:Fes1 (2.5 μM). Fibrillization was monitored by the amount of SDS-resistant NM. Values represent means±s.d. (n=3). (B) Seeded (2% wt/wt) Sup35 (2.5 μM) fibrillization with ATP (5 mM) after 16 h in the presence or absence of sti, Ydj1, Sis1, Ssa1, Ssb1, Fes1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1, Ssb1:Sis1, Ssa1:Ydj1:Fes1, Ssa1:Sis1:Fes1, Ssb1:Ydj1:Fes1 or Ssb1:Sis1:Fes1 (2.5 μM). Fibrillization was monitored by ThT fluorescence. Values represent means±s.d. (n=3). (C) Excimer fluorescence of NM G38C-pyrene (black) or G96C-pyrene (blue) (2.5 μM) after 16 h seeded (2% wt/wt) assembly with ATP (5 mM) in the absence or presence of sti, Ydj1, Sis1, Ssa1, Ssb1, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (2.5 μM). The ratio of excimer fluorescence to non-excimer fluorescence (I465 nm/I375 nm) is plotted. Values represent means±s.d. (n=3).
Prion recognition elements termed the ‘Head' (residues ∼25–38) and ‘Tail' (residues ∼91–106 in NM fibres formed at 25°C) make homotypic intermolecular contacts such that NM fibres are held together by an alternating sequence of ‘Head-to-Head' and ‘Tail-to-Tail' contacts (Krishnan and Lindquist, 2005; Tessier and Lindquist, 2007). Hsp70:Hsp40 could inhibit either ‘Head-to-Head' or ‘Tail-to-Tail' contact formation or both to arrest seeded fibrillization. To test these possibilities, we employed two different NM single cysteine mutants labelled with pyrene in either the head (G31C) or the tail region (G96C) (Krishnan and Lindquist, 2005). Upon intermolecular contact formation and fibrillization, pyrene molecules form excimers (excited-state dimers) that produce a strong red shift in fluorescence. Hsp70 or Hsp40 alone were unable to inhibit seeded ‘Head-to-Head' or ‘Tail-to-Tail' contact formation. By contrast, Hsp70:Hsp40 pairs inhibited both intermolecular contacts, and Ssa1 pairs inhibited more strongly than Ssb1 pairs (Figure 5C). Thus, Hsp70:Hsp40 pairs bind to NM fibres and occlude prion recognition elements to inhibit seeded assembly.
Combined effects of Hsp104, Hsp70 and Hsp40 on Sup35 assembly
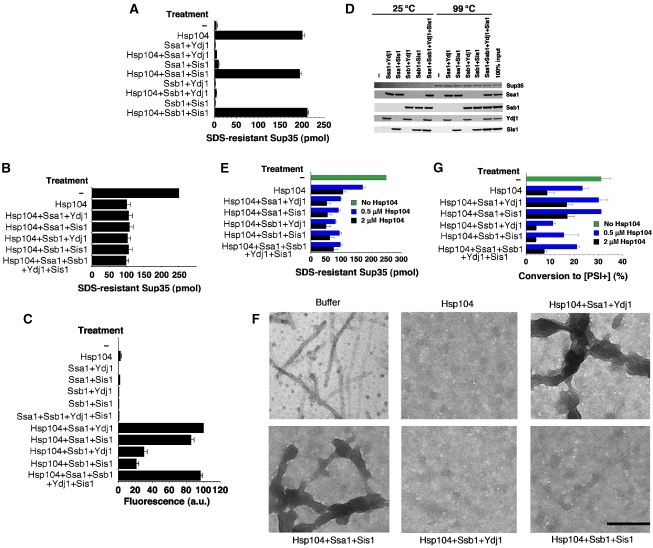
Clearly, Hsp70 and Hsp40 strongly inhibit both de novo and templated assembly of Sup35 prions. How do these activities interface with the prion-remodelling activities of Hsp104? At low concentrations, Hsp104 stimulates the de novo assembly of Sup35 prions (Shorter and Lindquist, 2004, 2006) (Figure 6A). This stimulation of Sup35 prion assembly was inhibited by Ssa1:Ydj1 and Ssb1:Ydj1 pairs (Figure 6A), but permitted by Ssa1:Sis1 and Ssb1:Sis1 pairs, which would ordinarily inhibit Sup35 assembly in the absence of Hsp104. Thus, the stimulatory activity of Hsp104 is prevented by some Hsp70:Hsp40 combinations but permitted by others.
Figure 6.
Hsp70 and Hsp40 modulate the prion-remodelling activities of Hsp104. (A) Unseeded, rotated (80 r.p.m.) Sup35 (2.5 μM) fibrillization with ATP (5 mM) after 2 h in the presence or absence of Hsp104 (0.03 μM) plus or minus Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 or Ssb1:Sis1 (0.25 μM). Fibrillization was assessed by the amount of SDS-resistant Sup35. Values represent means±s.d. (n=3). (B) Sup35 fibres (2.5 μM monomer) were incubated with or without the indicated combination of Hsp104, Ssa1, Ssb1, Ydj1 and Sis1 (2 μM) plus ATP (5 mM). Disassembly was assessed by the amount of SDS-resistant Sup35. Values represent means±s.d. (n=3). (C) Disaggregation of heat-aggregated GFP (0.45 μM) in the presence of the indicated combination of Hsp104, Ssa1, Ssb1, Ydj1 and Sis1 (2 μM) plus ATP (5 mM). Disaggregation was assessed by the amount of GFP fluorescence. Values represent means±s.d. (n=3). (D) Sup35 (2.5 μM) fibres were assembled with rotation (80 r.p.m.) for 8 h with or without various combinations of Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1, Ssb1:Sis1 or Ssa1:Ssb1:Ydj1:Sis1 (0.15 μM) plus ATP (5 mM). Fibres were then collected by sedimentation at 436 000 g for 10 min and treated with SDS–PAGE sample buffer at either 25 or 99°C and processed for immunoblot. (E–G) Sup35 (2.5 μM) fibres were assembled with rotation (80 r.p.m.) for 8 h with or without various combinations of Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1, Ssb1:Sis1 or Ssa1:Ssb1:Ydj1:Sis1 (0.15 μM) plus ATP (5 mM). Hsp104 (0.5 μM or 2 μM) was then added and reactions incubated for a further 30 min. Remodelling was assessed by the amount of SDS-resistant Sup35 (E). Values represent means±s.d. (n=3). (F) Alternatively, products from 2 μM Hsp104 reactions were viewed by EM. Bar, 150 nm. (G) Other reactions were concentrated and transformed into [psi−] cells. The proportion of [PSI+] colonies was then determined. Values represent means±s.d. (n=3).
Hsp70 and Hsp40 assist Hsp104 in the disaggregation and reactivation of denatured protein aggregates (Glover and Lindquist, 1998), but how would they affect the disassembly of Sup35 prions that occurs at high Hsp104 concentrations? Sup35 prions were assembled and then incubated with various combinations of Hsp104, Hsp70 and Hsp40. Hsp70 and Hsp40 alone were unable to disassemble Sup35 fibres (Figure 3; data not shown). Hsp104 alone promoted the disassembly of Sup35 prions, but surprisingly no combination of Hsp70 and Hsp40 increased or decreased this activity (Figure 6B). The ability of these Hsp70 and Hsp40 proteins to assist Hsp104 in the solubilization of thermally denatured GFP aggregates was confirmed in separate reactions (Figure 6C). Here, Hsp104 alone or Hsp70 plus Hsp40 alone were inactive (Figure 6C).
Hsp70 and Hsp40 may exert an effect upstream of Hsp104 in the disaggregation of some substrates (Haslberger et al, 2007). Therefore, we tested whether Ssa1 or Ssb1 might promote disassembly by Hsp104 if they were incorporated into nascent Sup35 prions. We assembled Sup35 fibres in the presence of non-inhibitory levels of Hsp70 and Hsp40. In the absence of Sup35, Hsp70 and Hsp40 remained soluble and were not found in the pellet fraction (data not shown). By contrast, Hsp70 and Hsp40 coaggregated with the Sup35 fibres. SDS (2%) readily disrupted this interaction, whereas Sup35 fibres remained SDS insoluble (Figure 6D). Sup35 fibres with Ssa1 and Ssb1 bound could still seed the assembly of soluble NM (data not shown) and were better substrates for disassembly by Hsp104 (Figure 6E). This was particularly evident at lower Hsp104 concentrations (Figure 6E). At higher concentrations, Hsp104 converted up to ∼80% of Sup35 to an SDS-soluble form, indicating that Hsp70 and Hsp40 render fibres more susceptible to remodelling by Hsp104 (Figure 6E). EM revealed that Hsp104 converted Sup35 fibres assembled alone or in the presence of Ssb1 to amorphous species (Figure 6F). By contrast, numerous Sup35 fibres assembled in the presence of Ssa1 persisted after Hsp104 treatment, but the fibres now had an altered morphology and wider diameter (Figure 6F). Remarkably, these fibres are very similar in appearance to ex vivo Sup35 prions (Bagriantsev et al, 2008).
The SDS-resistance assay does not distinguish between non-infectious amyloid-like material and prions, and thus cannot be used to infer effects solely about Sup35 prions. Therefore, we determined whether the disassembly products formed in the presence of Hsp104 or Hsp104, Hsp70 and Hsp40 were able to induce [PSI+]. Surprisingly, Sup35 fibres disassembled in the presence of Ssa1 retained more [PSI+]-inducing activity, whereas Ssb1 caused further reductions in [PSI+]-inducing activity (Figure 6G). Indeed, at lower Hsp104 concentrations, Ssa1 prevented elimination of Sup35 prions (Figure 6G). This is despite the increased amount of disassembly catalysed by Hsp104, suggesting that Ssa1 helps to maintain infectious prions after Hsp104 remodelling, whereas Ssb1 promotes the formation of non-infectious, SDS-resistant amyloid-like material.
Discussion
Here, we delineate for the first time how both Ssa and Ssb components of the Hsp70 chaperone system combine with Hsp104 to influence the conformational transitions of Sup35. Various Hsp70:Hsp40 pairs restrict the de novo formation and seeded assembly of Sup35 prions. However, other factors including the NEF, Fes1 and Hsp104 alleviate aspects of this inhibition. Importantly, Hsp70 and Hsp40 profoundly alter the consequences of prion remodelling by Hsp104. They can either help to preserve (Ssa) or eliminate (Ssb) the self-replicating state.
Hsp40s alone (Ydj1 or Sis1) directly antagonize N and NM fibrillization. Surprisingly, Sis1 was more effective than Ydj1. This was unexpected, as Ydj1 is more effective than Sis1 in counteracting the aggregation of Ure2 and heat-denatured luciferase (Lu and Cyr, 1998a; Lian et al, 2007). Hence, the effectiveness of Sis1 and Ydj1 in preventing aggregation may be substrate specific. Sis1 was effective only in preventing NM fibrillization during lag phase and did not affect seeded assembly, indicating that it specifically antagonizes prefibrillar intermediates en route to fibre assembly. Accordingly, Sis1 bound NM oligomers directly. However, so did Ydj1, suggesting that the consequences of binding were different for each Hsp40. Curiously, neither Ydj1 nor Sis1 could antagonize fibrillization of full-length Sup35. A possible explanation is that the large C-terminal GTPase domain of Sup35 titrates Ydj1 or Sis1 away from the prion domain. Another study found that His-tagged Ydj1 could inhibit N-terminally His-tagged Sup35 aggregation at 10°C (Krzewska and Melki, 2006). However, it is unclear if this was due to the tags or because His–Sup35 was purified without GTP and prone to forming off-pathway, non-prion aggregates that (like heat-denatured luciferase) were sensitive to Ydj1.
Ssa1+ATP or Ssb1+ATP had no effect on NM or Sup35 fibrillization. However, Ssa1+ADP or Ssb1+ADP both inhibited NM assembly, especially Ssb1, indicating an intrinsic ability to antagonize NM fibrillization. Both Ssa1 and Ssb1 in combination with Ydj1 or Sis1 strongly antagonized spontaneous and seeded NM fibrillization. Ydj1 and Sis1 stimulate Ssa1 ATPase activity and synergize with Ssa1 to prevent aggregation of heat-denatured luciferase (Lu and Cyr, 1998a). Indeed, inhibition of NM and Sup35 assembly required Ssa1 ATPase activity, as it did not occur in the presence of AMP-PNP. Although a genetic interaction between Ssb1 and Sis1 has been uncovered (Ohba, 1997), neither Ydj1 nor Sis1 were known to enhance Ssb1 chaperone activity anymore than additively (Lu and Cyr, 1998a). Nonetheless, Ydj1 or Sis1 and Ssb1 synergized to antagonize Sup35 fibrillization. This required Ssb1 ATPase activity and was compromised by AMP-PNP. Ydj1 (but not Ydj1 H34Q) and Sis1 enabled Ssb1 retrieval with NM oligomers and fibres in the presence of ATP. Moreover, Ydj1 (but not Ydj1 H34Q) stimulated Ssb1 ATPase activity in the presence of NM oligomers or fibres. Thus, Ydj1 with a functional J-domain productively couples Ssb1 ATPase activity to the inhibition of NM fibrillization. Intriguingly, Ssb1 and Sis1 or Ydj1 assisted Hsp104 in the disaggregation and reactivation of heat-aggregated GFP. In vivo, Ssb1 is often tethered to ribosomes, where it cooperates with the Hsp40, Zuo1 and atypical Hsp70, Ssz1, to prevent misfolding of nascent polypeptides (Nelson et al, 1992; Huang et al, 2005). Zuo1:Ssz1 heterodimers stimulate Ssb1 ATPase activity and Ssb1:Zuo1:Ssz1 potently inhibited NM and Sup35 fibrillization. Thus, upon emergence from the ribosome, nascent Sup35 is likely shielded from prion conformers. However, the ability of Ssb1 to cooperate with Ydj1 and Sis1, as well as Hsp104, suggests underappreciated ribosome-independent functions for Ssb1.
Various Hsp70:Hsp40 pairs antagonized nucleation and polymerization of NM fibres (Figure 7A), and this explains their ability to cure particular [PSI+] variants when overexpressed (Kushnirov et al, 2000). These effects were exerted through specific interactions with either NM oligomers or fibres. Various Hsp70:Hsp40 pairs prevented the formation and even disassembled molten NM oligomers, thereby blocking spontaneous fibrillization. They also bound to NM fibres in a manner that prevented ‘Head-to-Head' and ‘Tail-to-Tail' contact formation, which drives NM polymerization (Krishnan and Lindquist, 2005). With the exception of Ydj1, we could not detect robust interactions between Hsp70 and Hsp40 and NM or Sup35 monomers. This is surprising given that NM is predominantly natively unfolded (Glover et al, 1997; Scheibel and Lindquist, 2001). Hsp70 and Hsp40 might interact transiently with NM monomers to regulate their folding. Yet, it seems likely that the Hsp70 chaperone system recognizes conformational epitopes displayed by NM oligomers and fibres. Indeed, Ssa1 and Sis1 can be retrieved with Sup35 from [PSI+] but not [psi−] cells, and Ssb1 and Ydj1 are found associated with ex vivo Sup35 prions (Bagriantsev et al, 2008). Ssb1 also coimmunoprecipitates with Sup35 from [psi−] cells, but it is unclear whether this reflects a direct interaction (Bagriantsev et al, 2008).
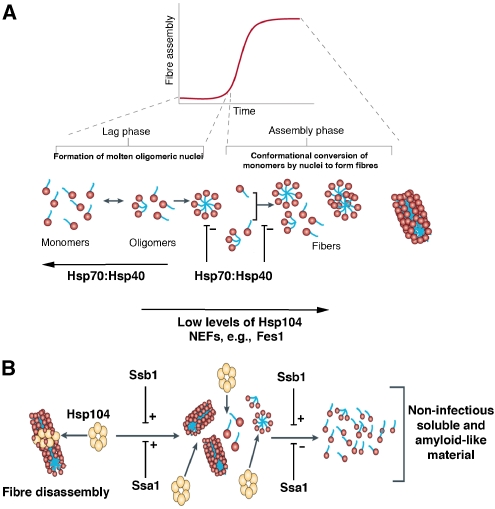
Figure 7.
Model of Hsp104, Hsp70 and Hsp40 interplay during Sup35 prionogenesis. (A) Sup35 prions assemble after a lag phase during which a dynamic ensemble of monomeric and molten oligomeric species form. The intermolecular contacts that nucleate prion assembly are likely established within molten oligomers. Once formed, fibres stimulate their own assembly by recruiting and converting monomers at their ends. The Hsp70:Hsp40 pairs, Ssa1:Ydj1, Ssa1:Sis1, Ssb1:Ydj1 and Ssb1:Sis1 inhibit this process by disassembling molten oligomers and by binding mature nuclei to occlude prion recognition elements and prevent seeded assembly. This inhibition is partially relieved by the NEF, Fes1. At low concentrations, Hsp104 stimulates prionogenesis and can relieve inhibition by Ssa1:Sis1 and Ssb1:Sis1, but not Ssa1:Ydj1 and Ssb1:Ydj1. (B) At high concentrations, Hsp104 disassembles Sup35 fibres by a fragmentation process that is stimulated by the incorporation of Ssa1 and Ssb1 (plus Hsp40) into Sup35 fibres. In the absence of Hsp70 and Hsp40, excessive disassembly by Hsp104 ultimately converts Sup35 fibres to a collection of soluble and non-infectious amyloid-like conformers. This step is promoted by Ssb1 and inhibited by Ssa1.
Importantly, the Ssa1 and Ssb1 NEF, Fes1, partially relieved the inhibitory effects of Hsp70 and Hsp40 (Figure 7A). This is consistent with genetic data that Fes1 deletion destabilizes [PSI+], whereas Fes1 overexpression promotes [PSI+] (Jones et al, 2004). Fes1 NEF activity promotes substrate release by Ssa1 and Ssb1, which may remove Hsp70 from NM oligomers and fibres and allow conformational conversion to proceed. Thus, yeast possesses factors that promote or relieve the inhibition of prion assembly by Hsp70.
Another such factor is Hsp104. At low concentrations, Hsp104 accelerates Sup35 prion assembly (Shorter and Lindquist, 2006). Here, we find that this stimulatory Hsp104 activity can overcome the inhibition of Sup35 fibrillization by Ssa1:Sis1 and Ssb1:Sis1 (Figure 7A). Thus, under some circumstances, Hsp104 may override the inhibitory effects of Hsp70. However, Ssa1:Ydj1 and Ssb1:Ydj1 prevented prion assembly even in the presence of stimulatory Hsp104 concentrations. Hence, yeast cells harbour conflicting Hsp40-dependent chaperone machineries that balance the promotion or inhibition of Sup35 prionogenesis.
Surprisingly, Hsp70 and Hsp40 could not stimulate Sup35 prion disassembly by Hsp104, if added after prion assembly. This is in contrast to the disaggregation of denatured protein aggregates by Hsp104, Hsp70 and Hsp40 (Glover and Lindquist, 1998). Sup35 fibres that had been co-assembled with non-inhibitory levels of Hsp70 and Hsp40 were more effectively disassembled by Hsp104 (Figure 7B). When these proteins are incorporated into nascent Sup35 prions, they may help present specific polypeptide loops of Sup35 fibres to Hsp104, thereby allowing it to achieve better access to the fibre core (Haslberger et al, 2007; Wendler et al, 2007). Alternatively, Hsp70 and Hsp40 may subtly alter fibre architecture to a form more amenable to disassembly by Hsp104. This is consistent with genetic data that Hsp70 assists prion fragmentation by Hsp104 (Jung et al, 2000; Jones et al, 2004). Importantly, the consequences of Hsp104 remodelling were different depending on which Hsp70 was incorporated into Sup35 prions. Ssb1 promoted the elimination of Sup35 prions, whereas Ssa1 inhibited it (Figure 7B). Thus, Ssb1 likely increases the Hsp104 activity that converts Sup35 prions to non-replicating amyloids, whereas Ssa1 prevents it, perhaps by maintaining active fibre ends. These data may explain why overexpression of Ssa1 inhibits curing by overexpression of Hsp104, whereas overexpression of Ssb1 promotes it (Chernoff et al, 1999; Newnam et al, 1999). The protection of self-replicating activity by Ssa1 despite remodelling by Hsp104 helps to explain genetic correlates that suggest that Ssa1 promotes [PSI+] (Allen et al, 2005; Sadlish et al, 2008).
Overall, our findings illuminate a sophisticated regulatory network of molecular chaperones that restrict or promote access to and from the Sup35 prion state. This network likely responds to environmental cues, such as stress, to either promote or eliminate [PSI+] and associated phenotypes. Indeed, many different types of stress promote [PSI+] induction (J Tyedmers, L Madriaga, and S Lindquist, submitted). When Sup35 is in the prion state, ribosomes read-through stop codons and this produces a wide array of new phenotypic states (True and Lindquist, 2000). Under stress conditions, it is likely advantageous for a subset of individuals to access hidden genetic variation through [PSI+] and sample new heritable phenotypes that might promote survival (Shorter and Lindquist, 2005; King and Masel, 2007). How the levels of various chaperones change in response to various stresses is complex, but Ssb1, a consistent [PSI+] antagonist, is downregulated by many stresses (Albanese et al, 2006), which may facilitate [PSI+] induction. Further, stress may allow Sup35 prionogenesis to proceed unfettered as chaperones are diverted to buffer various misfolded proteins. The multifarious activities of various chaperones on Sup35 prion formation and propagation may help to ensure that not every cell gains or loses [PSI+] with environmental stress, thereby ensuring a diversity of phenotypes in the culture. However, deeper systems analyses are needed to understand how chaperone networks respond to the environment to promote or antagonize [PSI+].
Materials and methods
Proteins
Untagged NM, Sup35, Hsp104, Ydj1, Sis1, Ssa1 and Ssb1 were purified as described by Shorter and Lindquist (2004, 2006). GTP (1 mM) was present throughout the entire Sup35 purification. After purification, Sup35 was filtered through a 0.2 μm filter and used immediately for fibrillization reactions. Freezing and thawing were completely avoided, as this promotes the formation of non-prion aggregates, which complicates analysis. NM G31C and NM G96C were purified and labelled with pyrene-maleimide (Molecular Probes) as described by Krishnan and Lindquist (2005). For some experiments, N-terminally His-tagged N, NM, Ydj1, Sis1, Ssa1, Ssb1 and C-terminally His-tagged NM were prepared as described (Glover et al, 1997; Shorter and Lindquist, 2004; Tessier and Lindquist, 2007). The Ydj1 H34Q mutant was generated by Quikchange mutagenesis (Stratagene) and purified as performed for Ydj1. Fes1 was purified as a GST-tagged protein as described by Kabani et al (2002). His–Zuo1 and Ssz1 complexes were purified from yeast as described by Huang et al (2005). DnaJ was from Assay Designs and soybean trypsin inhibitor was from Sigma.
Fibrillization and disassembly
His–N and NM (2.5 μM) fibrillization were conducted in assembly buffer (AB) (40 mM HEPES-KOH, pH 7.4, 150 mM KCl, 20 mM MgCl2, 1 mM DTT). For Sup35 (2.5 μM) fibrillization, AB was supplemented with 1 mM GTP and 10% glycerol (Shorter and Lindquist, 2006). Unseeded reactions were rotated (80 r.p.m. on a Mini-Rotator, Glas-Col) for 0–16 h at 25°C. Seeded reactions were unrotated and performed at 25°C. In reactions containing ATP (5 mM), a regeneration system was included comprising creatine phosphate (40 mM) and creatine kinase (0.5 μM). For reactions containing AMP-PNP (5 mM) or ADP (5 mM), the ATP regeneration system was omitted. Fibre assembly and disassembly were conducted in the presence or absence of the indicated proteins.
The extent of fibre assembly or disassembly was determined by EM, SDS resistance, sedimentation analysis or ThT fluorescence as described by Shorter and Lindquist (2004). Briefly, to assess SDS resistance, the amount of SDS-soluble (2% SDS, 25°C) NM or Sup35 was determined by quantitative densitometry of immunoblots (for reactions containing NM and Sis1 or DnaJ) or Coomassie-stained gels in comparison with known quantities of SDS-soluble NM or Sup35. From this value, the amount of SDS-resistant NM or Sup35 was calculated. Sedimentation was at 436 000 g for 10 min at 25°C. Supernatant and pellet fractions were resolved by SDS–PAGE and stained with Coomassie or processed for immunoblot (for reactions containing Sis1 and NM). The quantity of NM or Sup35 in the pellet was determined by densitometry and comparison with known quantities of NM or Sup35. For ThT, reactions were diluted 10-fold in AB containing ThT, to yield final concentrations of ThT (20 μM) and NM or Sup35 (0.5 μM). After a 10 min incubation, reactions were then excited at 450 nm (bandwidth 5 nm) and emission at 482 nm (bandwidth 10 nm) was recorded. Pyrene excimer formation was monitored as described by Krishnan and Lindquist (2005). Renaturation of heat-aggregated GFP was performed as described by Doyle et al (2007).
ATPase assays
Ssa1 or Ssb1 (1 μM) were incubated with Ydj1 or Ydj1 H34Q (1 μM) in the presence or absence of soluble NM, NM oligomers or NM fibres (2.5 μM NM monomer) at 25°C for 30 min in AB plus 1 mM ATP. Background hydrolysis was calculated and subtracted by using reactions performed without Hsp70. The release of inorganic phosphate was determined using a malachite green phosphate detection kit (Innova).
NM and Sup35 oligomer assembly and disassembly
NM (2.5 μM) was incubated in AB plus ATP (5 mM) with rotation (80 r.p.m.) for 5–30 min at 25°C. Sup35 (2.5 μM) was incubated with rotation (80 r.p.m.) for 1 h at 25°C. To assess effects on oligomer assembly, the indicated Hsp70 (2.5 μM) and/or Hsp40 (2.5 μM) were present upon the initiation of oligomer assembly. For oligomer disassembly, NM oligomers were allowed to form for 30 min and Sup35 oligomers for 1 h, and only then was the indicated Hsp70 (2.5 μM) and/or Hsp40 (2.5 μM) added. Disassembly was allowed to proceed for 10 min at 25°C. Oligomeric NM or Sup35 was recovered by centrifugation at 436 000 g for 30 min, resolved by SDS–PAGE and stained with Coomassie or processed for immunoblot (for reactions containing NM and Sis1). The amount of NM or Sup35 in the pellet fraction was determined by densitometry.
Protein transformation
His–N, NM and Sup35 fibre transformation was performed as described (Krishnan and Lindquist, 2005; Shorter and Lindquist, 2006).
Affinity chromatography
NM (2.5 μM) was incubated in AB-βME (AB with 2 mM β-mercaptoethanol instead of 1 mM DTT) plus ATP (5 mM) with rotation (80 r.p.m.) for 50 min at 25°C. At 0, 15 or 45 min, the indicated His–Hsp70 (2.5 μM) and/or His–Hsp40 (2.5 μM) were added. At 50 min, reactions were depleted of His–Hsp70 and/or His–Hsp40 with Ni-NTA agarose. Depleted reactions were spotted onto nitrocellulose and probed with anti-oligomer antibody or anti-NM antibody.
Alternatively, NM–His monomers, oligomers or fibres were attached to Ni-NTA at ∼2 mg/ml. NM–His monomers were generated by dissolution in 8 M urea, 20 mM Tris–HCl, pH 7.4. NM–His oligomers were formed by incubation of NM–His (5 μM) for 30 min at 25°C with rotation (80 r.p.m.) in AB-βME, followed by passage over a Microcon YM-100 (100 kDa molecular weight cutoff) filter (Millipore). The retentate was resuspended in AB-βME and attached to Ni-NTA. This fraction is enriched in amyloidogenic NM oligomers as determined by anti-oligomer immunoreactivity (Shorter and Lindquist, 2004). These oligomers do not bind Congo Red, indicating an absence of NM fibres, which was confirmed by EM. NM–His fibres were generated by incubating NM–His (5 μM) for 16 h at 25°C with rotation (80 r.p.m.). The indicated Hsp70 (2.5 μM) and/or Hsp40 (2.5 μM) in AB-βME plus ATP (5 mM) were incubated for 30 min at 25°C with either empty-, NM–His monomer-, NM–His oligomer-, or NM–His fibre-beads. Recovered beads were washed extensively with AB-βME and eluted with SDS–PAGE sample buffer. Eluates were fractionated by SDS–PAGE and Coomassie-stained or processed for immunoblot (to detect Sis1). The same procedure was followed for full-length, N-terminally His-tagged Sup35 except that His–Sup35 monomers and oligomers were generated by incubating His–Sup35 (5 μM) for 1 h at 25°C with rotation in AB-βME (plus 10% glycerol and 1 mM GTP). Oligomers were then separated from monomers by Superose-6 gel filtration and attached to Ni-NTA. Oligomeric fractions did not react with ThT, indicating an absence of fibres.
Acknowledgments
We thank M Dünnwald, J Tyedmers and A Gitler for comments on the manuscript; MN Knight, E Sweeny, C Glabe, D Cyr, S Wickner, E Craig and J Brodsky for generous provision of reagents. This work was supported by an American Heart Association Scientist Development Grant (JS) and NIH grant (GM25874) (SL).
References
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J (2006) Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124: 75–88 [DOI] [PubMed] [Google Scholar]
- Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO (2005) Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169: 1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW (2008) Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell 19: 2433–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+]. Science 268: 880–884 [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD (1999) Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI+] prion. Mol Cell Biol 19: 8103–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R (1988) A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332: 800–805 [DOI] [PubMed] [Google Scholar]
- Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S (2007) Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol 14: 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic Z, Shomura Y, Tzvetkov N, Hartl FU, Bracher A (2006) Fes1p acts as a nucleotide exchange factor for the ribosome-associated molecular chaperone Ssb1p. Biol Chem 387: 1593–1600 [DOI] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89: 811–819 [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Miess A, Stromer T, Walter S, Buchner J (2005) Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem 280: 23861–23868 [DOI] [PubMed] [Google Scholar]
- Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A (2007) M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 25: 247–260 [DOI] [PubMed] [Google Scholar]
- Huang P, Gautschi M, Walter W, Rospert S, Craig EA (2005) The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol 12: 497–504 [DOI] [PubMed] [Google Scholar]
- Jones G, Song Y, Chung S, Masison DC (2004) Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol 24: 3928–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Jones G, Wegrzyn RD, Masison DC (2000) A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Beckerich JM, Brodsky JL (2002) Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol Cell Biol 22: 4677–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R (2004) Protein-only transmission of three yeast prion strains. Nature 428: 319–323 [DOI] [PubMed] [Google Scholar]
- King OD, Masel J (2007) The evolution of bet-hedging adaptations to rare scenarios. Theor Popul Biol 72: 560–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL (2005) Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV (2002) Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem 277: 23702–23708 [DOI] [PubMed] [Google Scholar]
- Krzewska J, Melki R (2006) Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J 25: 822–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD (2000) Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol 10: 1443–1446 [DOI] [PubMed] [Google Scholar]
- Lian HY, Zhang H, Zhang ZR, Loovers HM, Jones GW, Rowling PJ, Itzhaki LS, Zhou JM, Perrett S (2007) Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J Biol Chem 282: 11931–11940 [DOI] [PubMed] [Google Scholar]
- Liu JJ, Sondheimer N, Lindquist SL (2002) Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc Natl Acad Sci USA 99 (Suppl 4): 16446–16453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Buesa P, Pfund C, Craig EA (1998) The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains. Proc Natl Acad Sci USA 95: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Cyr DM (1998a) Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem 273: 27824–27830 [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM (1998b) The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem 273: 5970–5978 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Brehmer D, Gassler CS, Bukau B (2001) Hsp70 chaperone machines. Adv Protein Chem 59: 1–44 [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA (1992) The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71: 97–105 [DOI] [PubMed] [Google Scholar]
- Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO (1999) Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol 19: 1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba M (1997) Modulation of intracellular protein degradation by SSB1-SIS1 chaperon system in yeast S. cerevisiae. FEBS Lett 409: 307–311 [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S (1994) Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372: 475–478 [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B (1997) Substrate specificity of the DNAK chaperone determined by screening cellulose-bound peptide libraries. EMBO J 16: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H, Rampelt H, Shorter J, Wegrzyn RD, Andreasson C, Lindquist S, Bukau B (2008) Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS ONE 3: e1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T, Bloom J, Lindquist SL (2004) The elongation of yeast prion fibers involves separable steps of association and conversion. Proc Natl Acad Sci USA 101: 2287–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T, Kowal AS, Bloom JD, Lindquist SL (2001) Bidirectional amyloid fiber growth for a yeast prion determinant. Curr Biol 11: 366–369 [DOI] [PubMed] [Google Scholar]
- Scheibel T, Lindquist SL (2001) The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat Struct Biol 8: 958–962 [DOI] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289: 1317–1321 [DOI] [PubMed] [Google Scholar]
- Shorter J (2008) Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals 16: 63–74 [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S (2004) Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304: 1793–1797 [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S (2005) Prions as adaptive conduits of memory and inheritance. Nat Rev Genet 6: 435–450 [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S (2006) Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell 23: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. Embo J 14: 4365–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328 [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN (1994) The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [PSI+] in the yeast Saccharomyces cerevisiae. Genetics 137: 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier PM, Lindquist S (2007) Prion recognition elements govern nucleation, strain specificity and species barriers. Nature 447: 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL (2000) A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483 [DOI] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR (2007) Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell 131: 1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA (1987) Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol 7: 2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]