Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is caused by an unusual deletion with neomorphic activity. This deletion derepresses genes in cis; however which candidate gene causes the FSHD phenotype, and through what mechanism, is unknown. We describe a novel genetic tool, inducible cassette exchange, enabling rapid generation of isogenetically modified cells with conditional and variable transgene expression. We compare the effects of expressing variable levels of each FSHD candidate gene on myoblasts. This screen identified only one gene with overt toxicity: DUX4 (double homeobox, chromosome 4), a protein with two homeodomains, each similar in sequence to Pax3 and Pax7. DUX4 expression recapitulates key features of the FSHD molecular phenotype, including repression of MyoD and its target genes, diminished myogenic differentiation, repression of glutathione redox pathway components, and sensitivity to oxidative stress. We further demonstrate competition between DUX4 and Pax3/Pax7: when either Pax3 or Pax7 is expressed at high levels, DUX4 is no longer toxic. We propose a hypothesis for FSHD in which DUX4 expression interferes with Pax7 in satellite cells, and inappropriately regulates Pax targets, including myogenic regulatory factors, during regeneration.
Keywords: cassette exchange, DUX4, facioscapulohumeral muscular dystrophy, myoblast
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is the third most common inherited myopathy, affecting approximately 1/20 000 individuals. It is caused by a deletion within the large tandem D4Z4 repeat sequence near the telomere of 4q. The normal chromosome 4 carries approximately 150 tandem copies of the 3.3 kb D4Z4 repeat, whereas FSHD-associated chromosomes carry 10 or fewer (Wijmenga et al, 1992). Although caused by a deletion, the disease is dominantly inherited. This is not due to haploinsufficiency of 4qter as a large deletion removing all D4Z4 repeats and extending proximally into chromosome 4 does not cause FSHD, implying that at least one copy of D4Z4 or nearby sequence is necessary for disease-related pathology (Gabellini et al, 2002). The D4Z4 repeat binds a YY-1-containing transcriptional repressor complex and it has been proposed that a repeat array of greater than 10 tandem units can silence nearby genes, whereas a deleted array allows inappropriate activation of nearby genes (Gabellini et al, 2002). Consistent with this, 4q35 sequences are hypo-methylated on FSHD-associated chromosome 4 variants compared with controls (van Overveld et al, 2003).
The region immediately proximal to the D4Z4 repeats harbours a number of candidate genes, including FRG1 (FSHD-related gene) (van Deutekom et al, 1996), which encodes a nucleolar protein involved in RNA biogenesis (van Koningsbruggen et al, 2004), TUBB4Q, a β-tubulin family member, and FRG2, a predicted transcript of four exons comprising an open reading frame (ORF) with no significant homology to any known protein. The sequence proximal to these three genes is gene poor, with no predicted gene for the next 250 kb (van Geel et al, 1999). Past this proximal region, the gene ANT1 (adenine nucleotide transporter) has attracted attention due to its function in apoptosis (Doerner et al, 1997; Gabellini et al, 2002). The D4Z4 repeat itself was originally cloned in a low-stringency screen for novel homeobox genes (Wijmenga et al, 1992). Remarkably, each D4Z4 repeat contains two homeoboxes within a single predicted ORF (Gabriels et al, 1999). This predicted gene is referred to as DUX4 (double homeobox, chromosome 4). In addition to copies within each unit of the tandem array, there is one copy of DUX4 just proximal of FRG2, embedded within a truncated, inverted D4Z4 repeat (Dellavalle et al, 2007). This gene, referred to as DUX4c, is identical to DUX4 from the N terminus through the homeodomains, but the last 82 amino acids have been substituted for an unrelated 32 amino-acid sequence. Thus, DUX4 and DUX4c are also FSHD candidate genes.
As FSHD is a dominant disease resulting from a gain-of-function mutation, modelling disease-related pathology in animals or cells requires testing candidate genes in gain-of-function genetic models. Three candidate genes, FRG1, FRG2, and ANT1, have been tested by overexpression in transgenic mice using the muscle-specific human skeletal actin promoter. Of these three, only FRG1 had a deleterious effect (Gabellini et al, 2002); however, the relevance of this model to FSHD is unclear as the pathology was only seen in mice expressing exceptionally high, non-physiological levels of FRG1. Furthermore, conventional transgenic studies suffer from high variability due to integration site-specific background effects; therefore, it is difficult to conclude that FRG1 is truly more myotoxic when compared with FRG2 or ANT1: expression patterns and absolute levels of expression in each transgenic strain differ. For such comparative studies, the ideal expression system would express each candidate gene from precisely the same genetic locus, and would be conditional and variable, so that low as well as high levels of expression could be tested. With this application in mind, we have therefore developed a novel, portable, cre-mediated gene targeting system, referred to as inducible cassette exchange (ICE). We have used this system to derive a murine myoblast ICE acceptor cell line, targeted this cell line with each FSHD candidate gene, and directly compared the effect of each on proliferating and differentiated myoblasts. We identify disease-related pathological changes from only one candidate, DUX4, and perform a detailed molecular analysis of the downstream effects of its expression during myoblast proliferation and differentiation.
Results
Generation of ICE myoblasts
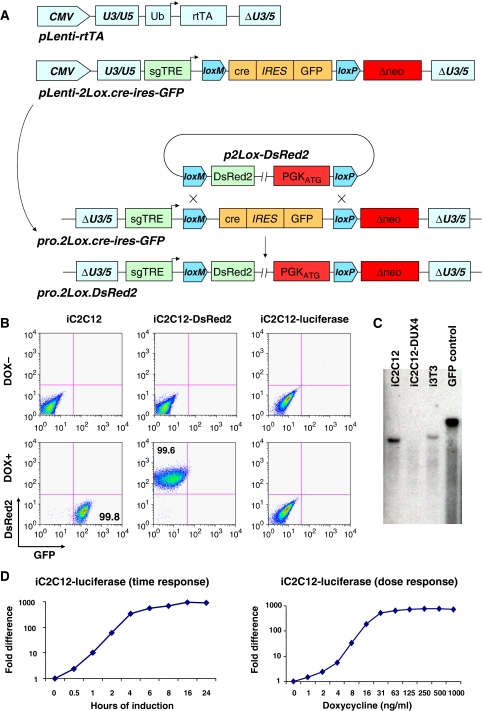
We have described earlier an inducible gene expression system for ES cells (Kyba et al, 2002) in which a circular plasmid carrying a gene of interest is targeted to a doxycycline (dox)-regulated locus. Cre-mediated recombination places a promoter and initiation codon in frame upstream of a G418 resistance gene, ensuring correct targeting (Fukushige and Sauer, 1992) while simultaneously placing the gene of interest under the control of the dox-regulated promoter. To enable site-directed integration in any cell type, we have modified this system to improve efficiency and adapted it for lentiviral transduction. First, to improve recombination efficiency, we have inserted a mutant version of loxP (lox2272 which self-recombines but does not recombine with loxP (Canales et al, 2006), referred to as loxM in Figure 1A) upstream of loxP, which now allows cassette exchange recombination. As the DNA in between loxM and loxP is replaced by the integrating plasmid, we have placed cre recombinase followed by ires–GFP into this space (Figure 1A). Finally, we have inserted the second-generation tetracycline response element (sgTRE, mutated to eliminate basal leakiness; Agha-Mohammadi et al, 2004) upstream of the floxed cre and placed the entire construct onto a self-inactivating lentiviral vector. A companion lentiviral vector carrying the ubiquitin C promoter (Lois et al, 2002) was used to express the reverse tetracycline transactivator (rtTA2(s)-m2; Urlinger et al, 2000). In cells transduced with both constructs, cre is induced with dox, and catalyses its own removal through cassette exchange recombination, placing the gene of interest under the control of dox (Figure 1A). Derivative cell lines can be made in parallel, each with a different gene inserted into the same locus. We refer to this system as ICE.
Figure 1.
Generation of ICE-recipient cell lines. (A) Schematic representation of the lentiviral constructs carrying the components of the ICE system, and the proviral ICE locus before and after recombination. (B) Flow cytometry of iC2C12 cells, which carry an inducible cre–ires–GFP proviral locus before recombination, and derivative cell lines carrying DsRed2 or luciferase, after recombination. (C) Southern blot analyses with GFP-specific probe DNA to detect copy number and recombination status of the ICE locus in iC2C12, iC2C12-DUX4 and i3T3 cell lines. Note that a single band hybridized with the GFP probe in iC2C12 and i3T3 cells but was missing following recombination (in iC2C12-DUX4 cells, in which GFP was replaced with DUX4). DNA from a spermatogonial cell line carrying GFP was used as a positive control. (D) Dose–response and time course (500 ng/ml) of luciferase gene expression in response to doxycycline.
As the principle virtue of an ICE cell line is that multiple genes of interest can be compared directly at various levels of expression, it is essential that such a cell line carry only a single copy of the ICE locus. We transduced murine C2C12 myoblasts sequentially, first with rtTA at high titre, then with the 2Lox.cre–ires–GFP construct over a titration series. After these two transductions, a transient dose of dox allowed cotransduced cells to be identified, enumerated, and sorted through GFP fluorescence. To derive a single-copy integration clone, we single cell sorted from a transduction in which <1% of cells were GFP+, expanded multiple clones, and retested each for efficiency of GFP expression. We derived several clones in which GFP was detectable in 100% of cells after 24 h of dox, and undetectable by flow cytometry in the absence of dox. Each clone was tested for ICE by dox treatment to induce cre, followed by transfection of an exchange plasmid carrying DsRed2 and selection in G418. Single-copy integrants show dox-inducible green fluorescence converting to dox-inducible red fluorescence (Figure 1B), whereas multi-copy integrants acquire inducible red fluorescence but do not lose green fluorescence (not shown). We selected one clone, referred to as iC2C12, and confirmed that it has a single ICE site by Southern blot analysis. Using a GFP-specific probe, we detected a single band in iC2C12 cells but not in iC2C12-DUX4 where cre–ires–GFP was substituted with DUX4 (Figure 1C). To evaluate dox sensitivity and gene expression kinetics, we targeted iC2C12 with luciferase. Luciferase expression could be titrated across a three-log range by varying the dose of dox, and abundant gene expression was observed within 2 h of dox addition (Figure 1D). These kinetics and dynamic range are sufficient in this context for both a comprehensive comparative analysis of FSHD candidate genes at different levels of expression, and for the molecular evaluation of cell physiological responses to the expression of particular genes of interest.
Head-to-head comparison of FSHD candidate genes
We targeted iC2C12 cells with each FSHD candidate gene: FRG1, FRG2, TUBB4q, ANT1, DUX4, and DUX4c. The derivative cell lines were then exposed to various doses of dox and effects on morphology and viability were evaluated. After 24 h of expression, viability, as measured by ATP content, was reduced by over 80% at 100 ng/ml and over 90% at 500 ng/ml dox in the iC2C12-DUX4 cells, whereas no changes in viability were observed for any other candidate gene (Figure 2A). At this time point, iC2C12-DUX4 cells appeared dead and began lifting from the plate (Figure 2B), whereas cells expressing other candidate genes remained unchanged (not shown). As antibodies are not available to the majority of FSHD candidate genes, to show that these could also be regulated by dox, we generated FLAG tag fusions of several other candidates, and determined by western blotting that they were expressed strictly in response to dox (Figure 2C). As the amount of accumulated protein depends on factors in addition to transcription (RNA and protein stability), absolute protein levels are likely to vary between cell lines; however, at the maximum level possible for each gene, overt toxicity was seen only with DUX4.
Figure 2.
Identification of DUX4-specific cell pathological phenotypes. (A) Cell viability (ATP content), after 24 h in 100 or 500 ng/ml doxycycline-treated cells. DUX4 has a unique dose-dependent effect on viability. (B) Morphology of iC2C12 myoblasts 24 h after DUX4 induction with 500 ng/ml dox. (C) Western blotting analysis of FLAG tag fusion protein expression in FLAG–DUX4, FLAG–ANT1, FLAG–TUBB4q and FRG1-3xFLAG cell lines. Cells were induced with 125 and 500 ng/ml doxycycline for 14 h. (D) Immunofluorescence showing DUX4 (red) accumulation in the nucleus and cell morphological changes (lamin, green) in iC2C12-DUX4 cells over a 16 h time course (500 ng/ml). (E) Dose–response at 20 h (upper panels) and time course (in 500 ng/ml doxycycline, lower panels) of DUX4 expression in iC2C12-DUX4 cells. (F) Western blot analyses of DUX4 expression in differentiated and proliferating cells. iC2C12-DUX4 cells were differentiated into myotubes for 4 days and DUX4 was induced for 14 h for comparison to proliferating cells. (G) Western blots for p21 and cyclin E during a time course of DUX4 induction at 500 ng/ml dox. (H) FACS analyses of DUX4-induced apoptosis and cell death using annexin V (x axis) and 7AAD (y axis) staining. Early apoptotic cells are annexin V+, dead cells are annexin V+/7AAD+. (I) Western blots for activated caspases 6, 8 and 9 in DUX4-expressing myoblasts over a time course. (J) Morphology of cells expressing DUX4 at low levels while proliferating, and viability (K) of these cells by ATP assay. (L) Morphology of DUX4-expressing myotubes and their viability (M).
DUX4 toxicity
The inducible system provides rapid and synchronous gene expression (Figure 2E), which enabled us to visualize the temporal phenotypic progression from normal to non-viable. After varying periods of induction, cells were stained with a monoclonal antibody to DUX4 and counterstained with laminin to visualize morphology. Intranuclear localization of DUX4 protein was apparent in virtually all cells within 2 h after high-level (500 ng/ml) induction (Figure 2D). By 4 h (500 ng/ml), morphological changes were detectable, as cells began to stretch and nuclei acquired an ovoid shape, a change that became more extreme with time. A dose-dependent decrease of Ki67 was also observed (Supplementary Figure S1A). Expression of p21 was markedly elevated as soon as 2 h after induction, and cyclin E levels were reduced from 6 h (Figure 2G). Markers of apoptosis, including activated caspases 6, 8, and 9 were apparent by 6 h, and cleaved caspase 3 and annexin V staining cells by 12 h, and increased thereafter (Figure 2H and I; Supplementary Figure S1C). At lower levels of expression, DUX4 reduced proliferation and induced morphological changes, but did cause obvious cell death (Figure 2J and K). At 30 ng/ml dox, the elongated shape and ovoid nuclei were apparent, but cells were viable for as long as 8 days (not shown). A 1-h pulse of DUX4 expression at 500 ng/ml followed by a 19 h chase resulted in a morphology similar to that of cells induced with a low dose of dox for 24 h (Supplementary Figure S2A). To analyse the effect of DUX4 on myotubes, we differentiated myoblasts into myotubes for 4–6 days using conventional differentiation medium (switch to 2% horse serum). When induced at high levels for 24 h in terminally differentiated myotubes, loss of myotubes was evident; however, large numbers of differentiated cells were still viable (Figure 2L), and by ATP assay we did not detect severe cell death even at relatively high levels of expression (Figure 2M). We confirmed that DUX4 protein is expressed at relatively similar levels in differentiated myotubes and proliferating myoblasts by western blotting (Figure 2F).
To determine whether DUX4 is specifically toxic for myoblasts, we generated DUX4-inducible fibroblasts i3T3-DUX4 from ICE-modified 3T3 cells (manuscript in preparation) and DUX4-inducible embryonic stem cells iES-DUX4 (manuscript in preparation). In 3T3 fibroblasts, low-dose induction of DUX4 resulted in similar cell morphological changes as seen in C2C12 myoblasts, and at high levels DUX4 induced rapid cell death (Supplementary Figure S3A). DUX4 impaired cell viability in a dose-dependent manner, as confirmed by the ATP assay (Supplementary Figure S3B and C). Similar toxic effects were observed when DUX4 was induced in mouse ES cells (Supplementary Figure S4B); however, cell death was not as rapid as observed in C2C12 and 3T3 cells. Interestingly, ES cells progressively lost their characteristic morphology and colony structure and acquired a fibroblastic cell shape on induction with DUX4 (Supplementary Figure S4B). To determine whether DUX4 is toxic for all cell types, we induced it in embryoid bodies (EBs, which contain cell types representing all three germ layers) derived from these ES cells. The toxic effect of DUX4 during EB differentiation was distinguished by a decrease in EB size, increased cell death and apoptosis seen by annexin V staining, and decreased outgrowth of the cells when plated in monolayer (Supplementary Figure S4D–F). However, all cell types were not equally sensitive to the DUX4 expression, as some cells remained viable even after 5 days of induction (500 ng/ml) (Supplementary Figure S4D–F). In support of the in vitro assay, we measured teratoma formation in immunodeficient mice and found that mice injected with iES-DUX4 cells and treated with dox from the first day of transplantation did not develop tumors; however, when dox was initiated at day 14 post-injection, tumors regressed but did not disappear (Supplementary Figure S4G). We conclude that different cell types have different sensitivities to DUX4.
Gene expression changes provoked by DUX4
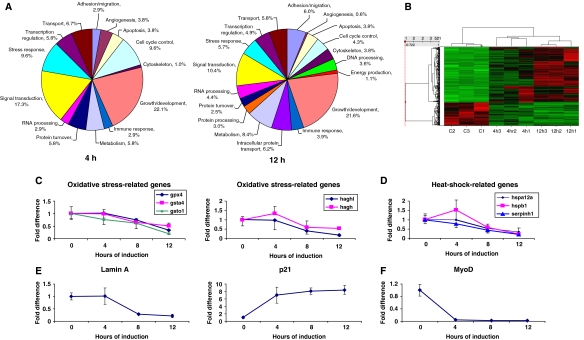
Because DUX4 is a homeodomain protein and rapidly transits to the nucleus after expression, we reasoned that its toxic effects were mediated through changes in gene expression. We therefore performed microarray transcriptional profiling experiments comparing uninduced cells to those expressing DUX4 for 4 or 12 h. We identified 156 genes differentially expressed at 4 h (107 upregulated and 49 downregulated) and 1011 genes at 12 h (576 upregulated and 435 downregulated). The majority of the genes (more than two-third) that were altered at 4 h were also changed in the 12 h sample. Representatives of a variety of functional gene ontology classes were identified (Figure 3A). Moreover, these alterations were consistent and reproducible across replicates, as demonstrated by hierarchical clustering (Figure 3B). For both time points, approximately one-third of the altered genes were uncharacterized cDNAs or genes with unknown functions (Supplementary Tables S1 and S2). Of the genes with known or suspected functions, the greatest numbers of genes at both 4 and 12 h were involved in cell cycle control or regulation of growth/development. Signal transduction and stress response were the next largest functional categories. Genes identified as differentially expressed at 12 h exhibited a similar ontological profile, with the addition of DNA packaging/processing, intracellular protein transport, and energy production categories represented. The gene lists in their entirety are shown in Supplementary Tables S1 and S2.
Figure 3.
DUX4 target genes. (A) DUX4 target genes (left chart, 4 h post-induction, right chart 12 h) represented by gene ontology. (B) Hierarchical clustering. Signal values for genes differentially expressed at 4 or 12 h were normalized by Z score and clustered using Spotfire DecisionSite software. Bright green represents low signal values, bright red indicates very high signal values, and black represents median signal values. Experiments are clustered by columns, and individual genes by rows. C1 through C3 represent 0 h controls, and 4 and 12 h represent samples treated with doxycycline for the indicated times. As shown, all controls clustered together as did the 4 and 12 h replicates. The majority of changes in gene expression increased over time (i.e. became brighter red by 12 h, the latest time point test. (C–F) Real-time PCR confirmation of expression changes for key FSHD-related genes: (C) glutathione redox pathway components, (D) heat-shock proteins, (E) Lamin A and p21, and (F) MyoD, which made the cutoff only in one replicate due to sensitivity of the microarray.
Previous microarray studies on FSHD patient biopsies have identified two classes of genes uniquely altered in FSHD when compared with other muscular dystrophies: MyoD target genes and oxidative stress response genes (Winokur et al, 2003b; Celegato et al, 2006). A proteomics study on patient biopsies has confirmed that oxidative stress response proteins, several proteins regulated by MyoD, and MyoD itself, are present at much reduced levels in FSHD biopsies when compared with unaffected controls (Celegato et al, 2006). We therefore evaluated the expression of these genes in our data and observed numerous examples of oxidative stress response gene downregulation. These changes were significant only in the 12-h sample, suggesting that they may be secondary targets of DUX4. The genes we identified, similar to the studies on FSHD patient biopsies, are primarily enzymes involved in glutathione redox metabolism. We confirmed these results independently by real-time PCR (Figure 3C). In addition to oxidative stress/glutathione redox genes, a number of heat-shock genes were downregulated, including Hspb1, Hsp12a, Serpinin, confirmed by real-time PCR (Figure 3D). We also noted repression of LaminA, the protein product of which is altered in several other muscular dystrophies (Figure 3E). Many upregulated genes were also observed, for example p21 (Figure 3E). Although MyoD was downregulated in one microarray, it was excluded from the final data set as its signal was undetectable from the other two replicates. Transcription factors are often expressed at mRNA levels that are not detectable with microarrays (Canales et al, 2006), so we performed real-time PCR to evaluate MyoD expression. This clearly demonstrated that MyoD was rapidly downregulated by DUX4 (Figure 3F). The fact that the two unique classes of genes altered in FSHD are downstream targets of DUX4 suggests a potential role for this candidate gene in FSHD pathology.
DUX4 and oxidative stress
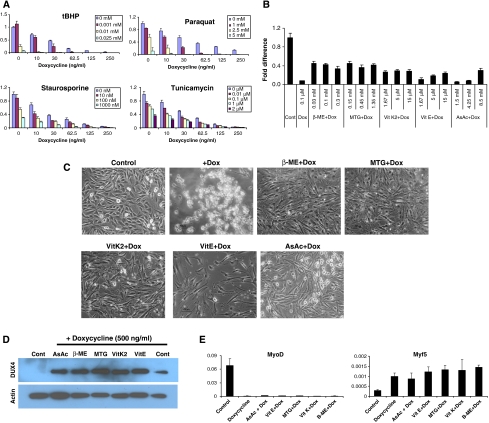
As genes involved in the glutathione redox cycle were repressed, we assumed that DUX4-expressing cells would have lower capacity to buffer oxidative stress. Additionally, a previous study has found that myoblasts from FSHD patients are more sensitive to oxidative stress than are control myoblasts from unaffected individuals (Winokur et al, 2003a). We therefore tested the sensitivity of DUX4-expressing cells to a variety of stress-inducing reagents at different concentrations. We found in all cases that DUX4 expression, even at levels only weakly detectable by western blot (10 ng/ml dox), enhanced the sensitivity of stress-inducing compounds (Figure 4A). In the case of tBHP and Paraquat, compounds that specifically induce oxidative stress, this enhancement was highly synergistic. Concentrations of these compounds, which have no effect or a weak effect on viability in the absence of DUX4, have a significant effect on viability when DUX4 is expressed (tBHP at 1 μM or Paraquat at 1 mM, for example). Concentrations that reduce but do not eliminate viability, combined with DUX4 expression, completely abolish viability with low levels of DUX4 expression. On the other hand, the dose–response curves for staurosporine and tunicamycin, non-oxidative stressors, although shifted, do not show threshold changes when DUX4 is expressed.
Figure 4.
Effect of antioxidants on DUX4-expressing myoblasts. (A) ATP assay for cell survival of DUX4-expressing myoblasts exposed to different stress-inducing agents. Oxidative stressors (tBHP and Paraquat) show a synergistic interaction with DUX4 at low levels. (B) Cell rescue with various concentrations of antioxidants demonstrated by ATP assay after 24 h of DUX4 induction (500 ng/ml dox). (C) Cell morphology of iC1C12-DUX4 myoblasts induced with 500 ng/ml dox for 24 h and treated with various antioxidants. (D) Western blot demonstrating that DUX4 was still expressed in the rescued cells (24 h post-induction). (E) qRT–PCR analyses of MyoD and Myf5 in antioxidant-rescued, DUX4-expressing cells. Data represent the fold difference compared with the level of GAPDH, error bars are STDEV (n=3). Antioxidants have no effect on the expression of these downstream genes.
As cells experience significant oxidative stress simply by being cultured in vitro, the enhanced oxidative sensitivity of DUX4-expressing myoblasts points to a possible reason for cell death. We therefore tested whether antioxidants would have any effect on viability of DUX4-expressing myoblasts. Each compound tested (β-mercaptoethanol, monothioglycerol, ascorbic acid, vitamin K2 and vitamin E) enabled myoblasts to grow in the presence of toxic levels of DUX4 (Figure 4B–D). Antioxidants also inhibited DUX4 toxicity in i3T3-DUX4 fibroblasts (Supplementary Figure S3D). Quantificationrevealed that this rescue reached a plateau at approximately 40%, meaning that other consequences of DUX4 expression were preventing cells from growing at the wild-type rate (Figure 4B). Indeed, antioxidants did not prevent the characteristic morphological change caused by DUX4 (Figure 4C) and had no effect on MyoD or Myf5 expression (Figure 4E); therefore, they rescue by buffering DUX4 toxicity, rather than by inactivating DUX4 protein.
DUX4 and myogenesis
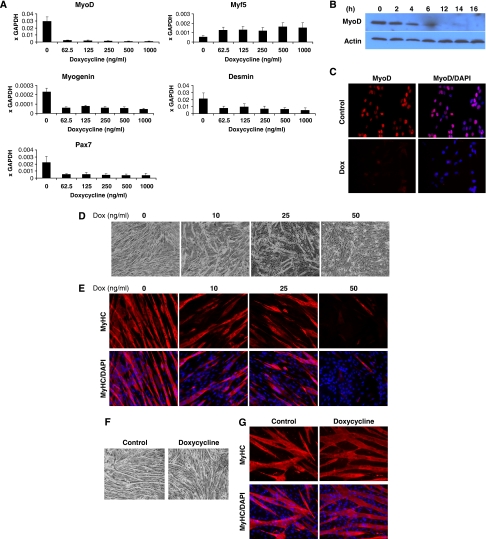
Because of the effect of DUX4 on MyoD expression, we tested the expression of other myogenic regulatory factors (MRFs) and myogenic genes. We observed reduced levels of Pax7, myogenin, and desmin mRNA, together with moderately increased levels of Myf5 (Figure 5A). This result was seen at sublethal induction levels for all MRFs (62.5 ng/ml dox; Figure 5A). These real-time PCR results were confirmed by immunohistochemistry (Figure 5B and C).
Figure 5.
DUX4 interferes with myogenic regulators and diminishes myotube formation. (A) qRT–PCR analyses of myogenic-specific genes in iC1C12-DUX4. DUX4 was induced with various concentrations of dox for 12 h. (B) Western blot analyses for the expression of MyoD and Myf5 in iC2C12-DUX4 over 16 h. (C) Immunofluorescence for MyoD (red) in 16 h DUX4-induced cells. Nuclei were contrastained with DAPI (blue). (D) Morphology of iC2C12-DUX4 after 4 days of differentiation in 2% horse serum. Note that DUX4-expressing cells, depending on the dox concentration, showed impaired to diminished differentiation. (E) Immunofluorescence for MyHC (red) on day 4 of differentiation of iC2C12-DUX4 cells. (F) Morphology of control (iC2C12 target) cells after 4 days of differentiation induced with 50 ng/ml doxycycline and evaluated by immunofluorescence for MyHC (G). Note that doxycycline alone does not have any significant effect on iC2C12 differentiation.
To determine whether interference with MyoD and other MRFs led to a myogenic differentiation defect, we evaluated myogenic differentiation at low levels of DUX4 expression in which there was no significant cell death. iC2C12-DUX4 myoblasts grown in the absence of dox were switched to differentiation conditions containing 10, 25 or 50 ng/ml dox. In contrast to the ubiquitous presence of multinucleated myotubes when DUX4 was not induced, only sporadic differentiation was present in the DUX4-induced samples, with less seen at 25 than at 10 ng/ml dox (Figure 5D). This diminished morphological differentiation was supported by decreased levels of MyHC by immunohistochemistry (Figure 5E). The effect of DUX4 on differentiation to myotubes was more severe if DUX4 induction was started before the switch to differentiation conditions, while cells were still proliferating (not shown). To confirm that alteration of myogenic gene expression and inhibition of differentiation in these cells were due to DUX4 expression and not due to dox treatment alone, we performed similar experiments on iC2C12 cells (the parent cell line before targeting with DUX4) and observed no changes (Figure 5F and G). As effects on differentiation and MRF expression are seen at low doses of DUX4 expression and as early as 4 h after induction, they are unlikely to be due to nonspecific apoptotic effects.
Pax3 and Pax7 compete with DUX4 and rescue toxicity
Homeodomain transcription factors can be grouped into subclasses determined by the amino-acid sequence of their respective homeodomains. A sequence dendrogram analysis including the DUX4 homeodomains and representative members of the major homeodomain subtypes is shown in Figure 6A. The DUX4 homeodomains are most similar to each other, and fall out of the tree nearest the Pax family, and far from the canonical clustered Hox genes (HoxAn–HoxDn). Within the Pax family, the DUX4 homeodomains are most similar to those of Pax3 and Pax7. This is provocative, given the important function that these factors have in myogeneic development and regeneration. If the homeodomains were similar enough to compete with Pax3 or Pax7 for binding to common targets, then a simple hypothesis for the pathogenic role of DUX4 in FSHD would be that it interferes with Pax3 or Pax7 function in satellite cells, or inappropriately alters the expression of Pax3 or Pax7 target genes such as MyoD during regeneration. If DUX4 competes with Pax3 or Pax7 for antipodal regulation of the same target genes, then a corollary prediction is that overexpression of Pax3 or Pax7 should compete with DUX4 and reduce or eliminate DUX4 phenotypes in our system. To test this, we transduced iC2C12-DUX4 cells with retroviral constructs expressing Pax3–ires–GFP, Pax7–ires–GFP or ires–GFP alone, and sorted pure populations of GFP+ cells. These derivative cell lines constitutively overexpress Pax genes, and express DUX4 conditionally. Pax3 and Pax7 were able to rescue viability and proliferation fully when DUX4 expression was induced with 100 ng/ml of dox, whereas GFP alone had no effect (Figure 6B). This rescue was indeed competitive as evidenced by the phenotype at higher levels of DUX4 expression. Although at 500 or 1000 ng/ml of dox, Pax3 and Pax7 maintained significant numbers of viable cells, these were fewer in number than at 100 ng/ml dox, and by comparing the 24 h to the 48 h ATP assay it is evident that at high levels of DUX4 expression, the viable, Pax3/Pax7-rescued cells were not proliferating. In addition, at high doses of dox, surviving cells show signs of morphological alteration characterized by a stretched and elongated shape. (Figure 6C, lower panels). This result was not due to reduced levels of DUX4 with Pax3/Pax7 expression, indeed the reverse was observed: high levels of DUX4 inhibited the accumulation of Pax3/Pax7 (Figure 6D). Although some reduction in Pax3/Pax7 transcription was evident, a large part of this reduction is post-transcriptional (Supplementary Figure S6) and likely due to decreased protein stability (Abu Hatoum et al, 1998; Boutet et al, 2007). To demonstrate that rescue from DUX4 toxicity was specific to Pax3/Pax7, we conducted the same experiment using an unrelated homeodomain-containing protein (HoxB4). We did not observe rescue with HoxB4 (Supplementary Figure S7).
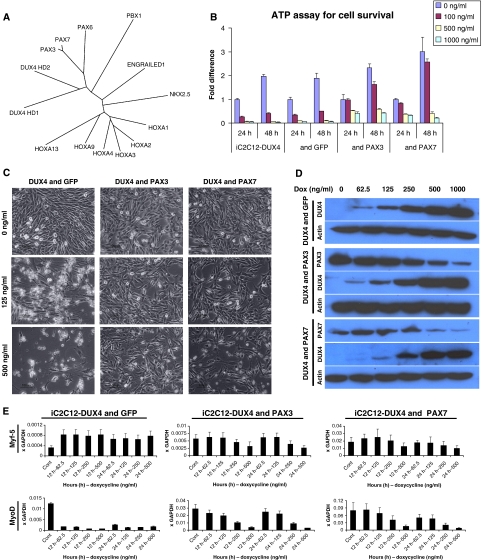
Figure 6.
Pax3 and Pax7 compete with DUX4 and rescue toxicity. (A) Unrooted tree representing the sequence relationship of different homeodomains. The length of arms indicates the distance between a given sequence and the nearest hypothetical sequence in common with the rest of the tree. (B) Cell viability demonstrated by ATP assay at 24 and 48 h post-DUX4 induction iC2C12-DUX4, iC2C12-DUX4 & GFP, iC2C12-DUX4 & Pax3 and iC2C12-DUX4 & Pax7. Note complete cell rescue in Pax3- and Pax7-expressing cells at 100 ng/ml dox. (C) Cell morphology of iC2C12-DUX4, iC2C12-DUX4 & Pax3 and iC2C12-DUX4 & Pax7 24 h after DUX4 induction with 125 and 500 ng/ml dox. Note the elongated cells at 125 ng/ml dox in the control (left) panel versus stellate cells in the Pax3- or Pax7-overexpressing groups (middle and right panels). (D) Western blot analyses for DUX4, Pax3 and Pax7 at 24 h of induction with different concentrations of dox, demonstrating that DUX4 is still expressed in these myoblasts. (E) qRT–PCR for MyoD and Myf5 in rescued cells at 12 and 24 h post-induction with varying levels of dox. Data represent the fold difference compared with the level of GAPDH, error bars are STDEV (n=3).
We then evaluated the expression of MyoD and Myf5 in the Pax3-, Pax7-, and control GFP-transduced iC2C12-DUX4 myoblasts. Without intervention, DUX4 rapidly reduces the levels of MyoD message and protein (Figures 3F, 5A–C and 6E). With Pax3 or Pax7 expression, this effect was significantly and competitively inhibited (Figure 6E). Only at very high levels of DUX4 expression (500 ng/ml dox) was MyoD effectively repressed. In a similar manner, Pax3 and Pax7 eliminated the ability of DUX4 to increase Myf5 expression. Therefore, DUX4 competes with Pax3 and Pax7 to regulate both viability-associated genes and the MRFs, MyoD and Myf5.
Discussion
Many biological problems are amenable to comparative gain-of-function experimental approaches. However, conventional methods of performing such genetic manipulations are confounded by expression variation. Gene targeting by homologous recombination, although efficient in ES cells, is not feasible in most cell lines. By combining conditional Tet-on regulation with selectable cre/Lox cassette exchange in a single-copy integration vector, we show that it is possible to generate cell lines with highly efficient ICE target loci, ideal for comparative gain-of-function studies, such as comparing phenotypic effects of various genes or evaluating mutants of a given gene. The conditional nature of the expression system facilitates work with genes with toxic phenotypes that would otherwise preclude the establishment of stable cell lines. The main technical challenge is identifying the clonal integrant in which the inducible locus is expressed robustly, not silenced, and not leaky. As lentiviral vectors integrate randomly, such integrants can always be recovered, provided a sufficient number of independent clones can be screened. As the targeting construct is generic, the utility of the system increases exponentially with the number of compatible ICE target cell lines available.
Because of the unusual genetics of FSHD, in which one or more of a large number of potentially upregulated candidate genes may cause the phenotype, dissecting the molecular pathology of this disease is ideally suited to this experimental approach. Expression profiling and proteomics have identified oxidative stress and myogenesis as the biological processes principally affected in FSHD muscle (Winokur et al, 2003b; Celegato et al, 2006; Macaione et al, 2007). However, there is no agreement on the underlying molecular mechanism, nor which upregulated gene(s) is (are) responsible for FSHD. By using the ICE system to compare each FSHD candidate gene directly over a range of expression levels, we have identified DUX4 as the only candidate to have general toxic effects. Furthermore, we show that DUX4 also has specific effects on the biological processes affected in FSHD. Given that D4Z4 repeats from FSHD-associated deletion chromosome 4 variants are hypomethylated (van Overveld et al, 2003), and that FSHD-associated transcription of the terminal repeat has been demonstrated (Dixit et al, 2007), and indeed DUX4 protein has been detected in FSHD myoblasts (Dixit et al, 2007), it is reasonable to assume that DUX4 may be specifically expressed in FSHD-affected muscle, and that some of the consequences of its expression, described in the current study, may be responsible for some of the pathology seen in FSHD. However, caution must be exercised, as these studies represent gain-of-function analyses, which address sufficiency but not necessity, and potentially relevant effects of other candidate genes are not ruled out by this study.
Although DUX4 is expressed at low levels in FSHD-affected muscle (Dixit et al, 2007), induction of high levels of DUX4 expression was useful in identifying its target genes and pathways. Among the notable rapidly induced changes were MyoD downregulation and p21 upregulation, which were seen within 2 h of DUX4 expression. At the protein level, reduced levels of MyoD have been reported in FSHD muscle biopsies (Celegato et al, 2006) and elevated levels of p21 have been reported in FSHD myoblast cultures (Winokur et al, 2003a). At the gene expression level, misregulation of MyoD target genes has also been reported (Winokur et al, 2003b). We observed MyoD repression even with low-level, non-toxic induction of DUX4, along with consequent perturbation of MyoD downstream targets and impaired differentiation. Interestingly, it has recently been reported that myoblasts and mesoangioblasts from FSHD patients exhibit morphological differentiation defects (Dellavalle et al, 2007; Barro et al, 2008). Among the late response changes (12 h, probably secondary responses), we observed downregulation of a number of genes involved in buffering oxidative damage. This has been reported in FSHD biopsy samples at the RNA level (Winokur et al, 2003b; Celegato et al, 2006) and at the protein level (Celegato et al, 2006; Macaione et al, 2007). Consistent with this, FSHD myoblasts have been shown to be more sensitive to compounds that induce oxidative damage than control myoblasts (Winokur et al, 2003a). In our hands, DUX4 expression rendered iC2C12-DUX4 myoblasts hypersensitive to oxidative stress, even at barely detectable levels of DUX4 induction that otherwise have no effect on viability. We further found that addition of antioxidants to DUX4-expressing cells provided significant rescue from cell death, even at the highest levels of DUX4 expression, demonstrating that inability to buffer oxidative damage is a key part of the toxic phenotype of DUX4 expression. By inducing sensitivity to oxidative damage, repressing MyoD, and upregulating p21, DUX4 expression in the murine myoblast cell culture system recapitulates many of the hallmark RNA and protein changes observed in FSHD muscle. This is consistent with a role for DUX4 in the pathology of FSHD.
High-level expression of DUX4 led to apoptosis, indicated by the presence of activated forms of various caspases and annexin V staining by fluorescence-activated cell sorter (FACS). Consistent with this, it has recently been reported that transient transfection of D4Z4 repeat units into C2C12 cells induces apoptosis (Kowaljow et al, 2007). Activation of caspase 3 has been observed in affected but not unaffected FSHD muscle (Laoudj-Chenivesse et al, 2005), as well as upregulation of caspase 1, 2, 3, and 6 transcripts (Sandri et al, 2001; Winokur et al, 2003b); however, the importance of apoptosis in the FSHD phenotype is controversial (Winokur et al, 2003a). At low levels, DUX4 does not lead directly to apoptosis; however, it may sensitize cells to other apoptotic signals.
We observed that DUX4 was much more toxic to proliferating myoblasts than to their differentiated counterparts. Both the sensitivity of DUX4-expressing myoblasts to oxidative stress and the reduction in their ability to generate terminally differentiated myotubes, if manifest in FSHD muscle, would place greater demand on the resident muscle stem and progenitor cell pool. More myoblasts would be required to perform a given amount of repair if a fraction was lost to oxidative stress and the surviving fraction was less efficient at differentiating into functional muscle. Depending on the severity, this would lead to the eventual exhaustion of muscle regenerative potential. However, in addition to the greater demand placed on the stem cell pool, our results suggest that DUX4 expression may deplete or incapacitate the stem cell pool directly. We have shown that DUX4 competes with both Pax3 and Pax7 for the regulation of myogenic target genes. This genetic interaction is intriguing, given that the nearest phylogenetic neighbours of the DUX4 homeodomains are those of Pax3 and Pax7. These two paired-box transcription factors have important, and partially redundant, functions in the development of skeletal muscle and dorsal neural tube (for review, see Buckingham and Relaix, 2007). In adult muscle, Pax7 is expressed in quiescent and activated satellite cells as well as in proliferating myogenic progenitors, and is downregulated prior to myoblast differentiation and fusion. Expression of Pax3 in adult muscle is localized to a subset of cells located in the interstitial space of the skeletal muscle (Kuang et al, 2006), in a small percentage of satellite cells in particular muscles, for example, diaphragm (Montarras et al, 2005; Cerletti et al, 2008) and transiently in activated myoblasts (Conboy and Rando, 2002; Cerletti et al, 2008). MyoD is a well-known target of Pax3 activation during embryogenesis (Tajbakhsh et al, 1997) and it is transcriptionally dependent on the expression of Pax7 in adult myogenesis (Olguin and Olwin, 2004; Relaix et al, 2006). Divergent induction/repression kinetics of MyoD is probably dependent on the level of Pax7 overexpression and on the cell type (Olguin and Olwin, 2004; Relaix et al, 2006). In our system, MyoD is rapidly downregulated by DUX4 and this downregulation is reversed by Pax3 or Pax7 overexpression. In the adult, Pax7 is required for satellite cell self-renewal and is thus essential for ongoing muscle regeneration—in Pax7 knockout mice, muscle develops, but satellite cell number drops precipitously after birth and mice live only a few weeks (Mansouri et al, 1996; Seale et al, 2000; Oustanina et al, 2004). Therefore, if DUX4 showed the same competitive interaction with Pax7 in satellite cells, it would be expected to phenocopy the Pax7 mutant, resulting in reduced self-renewal of satellite cells. This hypothesis is consistent with the relatively late onset of FSHD and with both the latency in outgrowth of FSHD myoblast cultures from affected muscles and the reduced proliferative potential of FSHD myoblast cultures once established (Vilquin et al, 2005).
Although FSHD may be caused by the combinatorial action of multiple overexpressed 4q35.2 candidate genes, we have shown that DUX4 expression alone is sufficient to recapitulate key aspects of the FSHD molecular phenotype. Critically, this phenotype is obtained even with very low levels of DUX4 expression, such as have been reported in FSHD myoblasts (Dixit et al, 2007). The competitive interaction between DUX4 and Pax7 predicts satellite cell and regeneration defects, thus we hypothesize that FSHD may be the first example of a muscular dystrophy with a stem cell aetiology. To substantiate this hypothesis, it will be critical to evaluate the expression of DUX4 in satellite cells of FSHD-affected muscle and to evaluate the effect of knocking down DUX4 expression in primary cultures derived from these satellite cells.
Materials and methods
Cloning of retro and lenti-virus plasmids
pLenti-rtTA was made as follows: The lenti vector FUGW (Lois et al, 2002) was cut with BamHI and EcoRI; rtTA2SM2 from pUHDrtTA2SM2 (Urlinger et al, 2000) (gift of H Bujard) was removed as an EcoRI/HindIII fragment, which included an SV40 polyA; the EcoRI site was destroyed and replaced with BglII, and a new EcoRI site was added immediately downstream of the HindIII site, and the resulting BglII/EcoRI fragment was inserted into the BamHI/EcoRI digested FUGW, replacing GFP and destroying the upstream BamHI site. The SV40 polyA was then removed by EcoRI/BamHI digestion, blunting, and relegation. Pax3 cDNA from pSPORT-Pax3 (BC048699; Open Biosystems) and Pax7 cDNA from pBRIT-Pax7d (Seale et al, 2004) were subcloned as EcoRI and XhoI fragments into pMSCV-ires-GFP to generate pMSCV-Pax3-ires-GFP and pMSCV-Pax7-ires-GFP, respectively. pMSCV-HOXB4-ires-GFP was described earlier (Kyba et al, 2002).
Generating p2Lox plasmid with DsRed2 and luciferase reporter genes
P2Lox plasmid was generated by inserting the Lox2272 sequence immediately downstream of LoxP in the previously reported pLox plasmid by Kyba et al (2002) and replacing the sequence from SalI to NotI with a SalI/NotI fragment from pBS-GFP, in which GFP is inserted at the EcoRV site. DsRed2 was subcloned from pIRES2-DsRED2 (Clontech, Palo Alto, CA, USA) as a NcoI (blunt) and NotI fragment and cloned into SmaI and NotI digested p2Lox. Luciferase was subcloned directionaly as an XhoI/BamHI fragment from pGL3-Basic Vector (Promega) was cloned directionally into P2Lox.
Cloning of FSHD candidate genes into p2Lox
DNA sequence from the terminal D4Z4 repeat (2.7 kb) containing DUX4 was obtained from pClneo-DUX4 (Gabriels et al, 1999) and subcloned into XhoI/NotI cloning sites of p2Lox, generating p2lox-DUX4. DUX4c was subcloned from pCLneo-DUX4c as XbaI/EcoRI fragment into pBS to acquire XhoI and NotI cloning sites, which were then used for subcloning into p2lox to generate p2lox-DUX4c. cDNAs for FRG1 and ANT1 were obtained from Open Biosystems. Genes were excised by EcoRI/NotI digestion and cloned directionally into p2lox to generate p2lox-FRG1 and p2lox-ANT1. To clone FRG2 cDNA, total RNA was harvested from differentiated human FSHD myoblasts (Rijkers et al, 2004) using Trizol (Invitrogen) and cDNA was transcribed using the ThermoScript RT–PCR System (Invitrogen) and random hexamer primers. FRG2 was amplified by forward (atgggaaagggaaatgaagactcccga) and reverse (tcattcccagagctgcatctctgct)-specific primers using FastStart High Fidelity PCR (Roche). Whole DNA sequence of TUBB4q was generated by PCR using BAC as a template and forward (atgagggagcttgtgctcacgc) and reverse (tcagactcctcctccctcggc) primers. Both PCR products were subcloned into p2lox using SalI and NotI as these sites were incorporated into the primers. DYKDDDDK peptide sequence was fused at N-terminal to generate FLAG tag version of the FLAG–DUX4, FLAG–ANT1 and FLAG–TUBB4q. 3 × FLAG tag at C terminus of FRG1 was generated by cloning FRG1 in frame with p2Lox-3xFLAG. The integrity of all of the genes cloned into p2Lox was confirmed by sequencing.
Generating the iC2C12 targeting cell line
C2C12 myoblasts were transduced with Lenti-rtTA virus at high titre to generate C2C12-rtTA cells. Expanded C2C12-rtTA cells were transduced with the serial dilutions of Lenti-i2lox-cre to obtain single-copy integration. To test the titration and to purify the target population, 2 days after the second infection, cre–ires–GFP was induced with 500 ng/ml dox (Sigma) for 24 h and cells were analysed for GFP on a FACS Aria (BD). GFP+ cells from the lowest dilution (less than 1%) were sorted and expanded in the absence of dox. After several passages, doubly transfected cells were induced once again for cre–ires–GFP and single GFP+ cells were single cell-sorted into 96-well plates. At 1 week after sorting, the colonies obtained from single cells were split in two 96-well replica plates and expanded for 2 days. One of the 96-well replica plates was induced with dox once and GFP expression was evaluated by FACS. Clones showing 100% GFP+ expression were expanded and further tested. Each clone was tested for a non-silencing, single-copy integration site by targeting the inducible locus with DsRed2 (p2lox-DsRed2). At 24 h before targeting, the endogenous cre was induced with 500 ng/ml dox, and 2 h before the transfection the medium containing dox was replaced with fresh medium without dox. p2lox-DsRed2 plasmid was transfected using FUGENE 6 and selection with 800 μg/ml G418 (Gibco) was started the following day. G418-resistant cultures were established within 10 days, at which point DsRed2 was induced by dox and cells were analysed by FACS 24 h later. One non-silencing clone that was capable of efficient replacement of GFP with DsRed2 was selected for these studies and designated iC2C12.
Illumina microarray analysis
Microarray gene expression analyses were performed on iC2C12 cells induced with 500 ng/ml dox. Each culture was divided equally into three arms, and each arm was harvested at the same time 24 h later. One arm was not treated, the second was treated 4 h before harvest, and the third was treated 12 h before harvest. Total RNA (100 ng) isolated by Trizol (Invitrogen) was amplified using the Illumina RNA Amplification kit and labelled by incorporation of biotin-16-UTP. Samples were hybridized to llumina BeadChips, which were scanning with an Illumina BeadArray Reader. Array data processing and normalization were performed using Illumina Bead-Studio software. Three arrays (0, 4 and 12 h) were repeated on three different BeadChips in three independent experiments. Signal values were normalized by global mean and log transformed using GeneSifter software (VizX Labs, Seattle, WA, USA). Pairwise comparisons and Student's t-test were subsequently performed, and a difference of at least two-fold with a P-value of less than 0.05 was considered as a statistically significant change in gene expression. Spotfire DecisionSite 9.0 (Spotfire Inc., Somerville, MA) and significance analysis of microarrays were also used to perform pairwise comparisons, two-class paired comparisons, and one-way analysis of variance to confirm statistical significance of expression differences. Finally, individual comparisons among the experimental groups (0 versus 4 and 0 versus 12) on each chip were done independently, and only those differences that were consistent (i.e. occurred for all three experiments) were retained.
Quantitative real-time RT–PCR (qRT–PCR)
Total RNA was extracted with Trizol (Invitrogen) and cDNA was generated using 1 μg DNase-treated RNA with oligo-dT primer and ThermoScript (Invitrogen). PCR was performed by using TaqMan or SYBR green real-time PCR premixture on 7500 real-time PCR System (Applied Biosystems). For muscle-related genes (Pax3, Pax7, MyoD, Myf5, myogenin, desmin and MCK), pre-made probes were purchased from Applied Biosystems. For validation of microarray data, the list of the primers used in the qPCR is shown in Supplementary Methods. Actin or glyceraldehyde phosphate dehydrogenase were used as internal standards. All reactions were performed at least in triplicate and the data were normalized and analysed by 7500 System Software (Applied Biosystems).
Statistical analyses
All experiments were done at least three times. Data shown for real-time PCR are the mean±s.d. Difference between means was compared by the two-tailed Student's t-test (GraphPad Prism 5) and was considered significantly different at P<0.05.
Supplementary Material
Supplementary Figures S1–S7
Supplementary Table S1
Supplementary Table S2
Supplementary Figure Legends
Supplementary Materials and Methods
Acknowledgments
This study was generously supported by the Dr Bob and Jean Smith Foundation. DB was supported by a Muscular Dystrophy Association Development Grant (MDA 4361) and by a fellowship supplement from Facioscapulohumeral Muscular Dystrophy (FSHD) Society. We thank Dr Kent Hamra and Karen Chapman for the help with the Southern blot. We thank Jennifer Cheeseman for help in preparing the paper.
References
- Abu Hatoum O, Gross-Mesilaty S, Breitschopf K, Hoffman A, Gonen H, Ciechanover A, Bengal E (1998) Degradation of myogenic transcription factor MyoD by the ubiquitin pathway in vivo and in vitro: regulation by specific DNA binding. Mol Cell Biol 18: 5670–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agha-Mohammadi S, O'Malley M, Etemad A, Wang Z, Xiao X, Lotze MT (2004) Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J Gene Med 6: 817–828 [DOI] [PubMed] [Google Scholar]
- Barro M, Carnac G, Flavier S, Mercier J, Vassetzky Y, Laoudj-Chenivesse D (2008) Myoblasts from affected and non affected FSHD muscles exhibit morphological differentiation defects. J Cell Mol Med; E-pub ahead of print 24 May 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA (2007) Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell 130: 349–362 [DOI] [PubMed] [Google Scholar]
- Buckingham M, Relaix F (2007) The role of pax genes in the development of tissues and organs: pax3 and pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol 23: 645–673 [DOI] [PubMed] [Google Scholar]
- Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, Ma Y, Maqsodi B, Papallo A, Peters EH, Poulter K, Ruppel PL, Samaha RR, Shi L, Yang W, Zhang L et al. (2006) Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol 24: 1115–1122 [DOI] [PubMed] [Google Scholar]
- Celegato B, Capitanio D, Pescatori M, Romualdi C, Pacchioni B, Cagnin S, Vigano A, Colantoni L, Begum S, Ricci E, Wait R, Lanfranchi G, Gelfi C (2006) Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics 6: 5303–5321 [DOI] [PubMed] [Google Scholar]
- Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, Wagers AJ (2008) Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell 134: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Rando TA (2002) The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell 3: 397–409 [DOI] [PubMed] [Google Scholar]
- Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, Innocenzi A, Galvez BG, Messina G, Morosetti R, Li S, Belicchi M, Peretti G, Chamberlain JS, Wright WE, Torrente Y, Ferrari S, Bianco P, Cossu G (2007) Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol 9: 255–267 [DOI] [PubMed] [Google Scholar]
- Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Matteotti C, van Acker AM, Leo O, Figlewicz D, Barro M, Laoudj-Chenivesse D, Belayew A, Coppee F, Chen YW (2007) DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA 104: 18157–18162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner A, Pauschinger M, Badorff A, Noutsias M, Giessen S, Schulze K, Bilger J, Rauch U, Schultheiss HP (1997) Tissue-specific transcription pattern of the adenine nucleotide translocase isoforms in humans. FEBS Lett 414: 258–262 [DOI] [PubMed] [Google Scholar]
- Fukushige S, Sauer B (1992) Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci USA 89: 7905–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110: 339–348 [DOI] [PubMed] [Google Scholar]
- Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A (1999) Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236: 25–32 [DOI] [PubMed] [Google Scholar]
- Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Matteotti C, Arias C, Corona ED, Nunez NG, Leo O, Wattiez R, Figlewicz D, Laoudj-Chenivesse D, Belayew A, Coppee F, Rosa AL (2007) The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord 17: 611–623 [DOI] [PubMed] [Google Scholar]
- Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA (2006) Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol 172: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, Daley GQ (2002) HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109: 29–37 [DOI] [PubMed] [Google Scholar]
- Laoudj-Chenivesse D, Carnac G, Bisbal C, Hugon G, Bouillot S, Desnuelle C, Vassetzky Y, Fernandez A (2005) Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J Mol Med 83: 216–224 [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D (2002) Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295: 868–872 [DOI] [PubMed] [Google Scholar]
- Macaione V, Aguennouz M, Rodolico C, Mazzeo A, Patti A, Cannistraci E, Colantone L, Di Giorgio RM, De Luca G, Vita G (2007) RAGE-NF-kappaB pathway activation in response to oxidative stress in facioscapulohumeral muscular dystrophy. Acta Neurol Scand 115: 115–121 [DOI] [PubMed] [Google Scholar]
- Mansouri A, Stoykova A, Torres M, Gruss P (1996) Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development 122: 831–838 [DOI] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M (2005) Direct isolation of satellite cells for skeletal muscle regeneration. Science 309: 2064–2067 [DOI] [PubMed] [Google Scholar]
- Olguin HC, Olwin BB (2004) Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol 275: 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T (2004) Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J 23: 3430–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M (2006) Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol 172: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T, Deidda G, van Koningsbruggen S, van Geel M, Lemmers RJ, van Deutekom JC, Figlewicz D, Hewitt JE, Padberg GW, Frants RR, van der Maarel SM (2004) FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD patients. J Med Genet 41: 826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, El Meslemani AH, Sandri C, Schjerling P, Vissing K, Andersen JL, Rossini K, Carraro U, Angelini C (2001) Caspase 3 expression correlates with skeletal muscle apoptosis in Duchenne and facioscapulo human muscular dystrophy. A potential target for pharmacological treatment? J Neuropathol Exp Neurol 60: 302–312 [DOI] [PubMed] [Google Scholar]
- Seale P, Ishibashi J, Scime A, Rudnicki MA (2004) Pax7 is necessary and sufficient for the myogenic specification of CD45+:Sca1+ stem cells from injured muscle. PLoS Biol 2: E130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA (2000) Pax7 is required for the specification of myogenic satellite cells. Cell 102: 777–786 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M (1997) Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89: 127–138 [DOI] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W (2000) Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA 97: 7963–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Lemmers RJ, Grewal PK, van Geel M, Romberg S, Dauwerse HG, Wright TJ, Padberg GW, Hofker MH, Hewitt JE, Frants RR (1996) Identification of the first gene (FRG1) from the FSHD region on human chromosome 4q35. Hum Mol Genet 5: 581–590 [DOI] [PubMed] [Google Scholar]
- van Geel M, Heather LJ, Lyle R, Hewitt JE, Frants RR, de Jong PJ (1999) The FSHD region on human chromosome 4q35 contains potential coding regions among pseudogenes and a high density of repeat elements. Genomics 61: 55–65 [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S, Dirks RW, Mommaas AM, Onderwater JJ, Deidda G, Padberg GW, Frants RR, van der Maarel SM (2004) FRG1P is localised in the nucleolus, Cajal bodies, and speckles. J Med Genet 41: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM (2003) Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 35: 315–317 [DOI] [PubMed] [Google Scholar]
- Vilquin JT, Marolleau JP, Sacconi S, Garcin I, Lacassagne MN, Robert I, Ternaux B, Bouazza B, Larghero J, Desnuelle C (2005) Normal growth and regenerating ability of myoblasts from unaffected muscles of facioscapulohumeral muscular dystrophy patients. Gene Ther 12: 1651–1662 [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, van Ommen G-J B, Padberg GW, Frants RR (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2: 26–30 [DOI] [PubMed] [Google Scholar]
- Winokur ST, Barrett K, Martin JH, Forrester JR, Simon M, Tawil R, Chung SA, Masny PS, Figlewicz DA (2003a) Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul Disord 13: 322–333 [DOI] [PubMed] [Google Scholar]
- Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM (2003b) Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet 12: 2895–2907 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S7
Supplementary Table S1
Supplementary Table S2
Supplementary Figure Legends
Supplementary Materials and Methods