Abstract
Small RNAs exert an effect through diverse RNA interference pathways to transcriptionally or post-transcriptionally silence their targets. The Piwi-interacting RNAs (piRNAs) represent a germline-specific small RNA pathway where Piwi proteins themselves are thought to mediate piRNA biosynthesis. Here, we provide strong evidence for a piRNA amplification loop in zebrafish, in which Ziwi and Zili bind piRNAs of opposite polarity. Furthermore, we describe a function for Zili in transposon defense and germ cell differentiation, as well as a crucial function in meiosis, significantly extending the function of Piwi proteins beyond the control of transposable elements in vertebrates.
Keywords: germ cell differentiation, meiosis, piRNAs, Piwi, zebrafish
Introduction
Non-coding RNAs are increasingly recognized as major players in the regulation of gene activity. Arguably one of the best-studied classes of such regulatory RNA molecules are the small RNAs that are central in processes like RNA interference and microRNA-mediated gene silencing. Argonaute proteins are well characterized for their function in these RNA-mediated gene-silencing processes. They directly bind the small RNA guides to target complementary sequences in DNA or RNA with diverse results, such as the induction of histone and DNA methylation, mRNA breakdown and inhibition of translation (Zamore and Haley, 2005).
In animals, the Argonaute protein family consists of two distinct subclasses: Ago and Piwi (Carmell et al, 2002). The small RNA guides of the Ago proteins can be microRNAs (miRNAs), or small interfering RNAs (siRNAs), which are about 21 nucleotides long, and are derived from double-stranded RNA, through processing by the RNaseIII enzyme Dicer (Bernstein et al, 2001). The Piwi proteins are named after the founding member of this class: Drosophila Piwi (Cox et al, 1998) and are mostly expressed specifically in the germ line. The small RNA-binding partners of Piwi proteins are distinct from miRNAs and typical siRNAs. These Piwi-interacting RNAs (piRNAs) are longer than miRNAs, they are germ line specific and are not processed by Dicer (Aravin et al, 2006; Girard et al, 2006; Grivna et al, 2006a; Lau et al, 2006; Vagin et al, 2006; Houwing et al, 2007). Piwi proteins and piRNAs have been shown to have an important function in transposon control. In Drosophila, loss of Piwi results in transposon activation, which leads to both germline and embryonic defects (Aravin et al, 2001; Sarot et al, 2004; Vagin et al, 2004, 2006; Kalmykova et al, 2005; Saito et al, 2006; Savitsky et al, 2006). In mammals, loss of MILI and MIWI2 results in transposon activation (Aravin et al, 2007; Carmell et al, 2007), which may cause meiotic arrest before pachytene, resulting in germ cell loss. Both transcriptional as well as post-transcriptional silencing mechanisms have been proposed for this transposon regulatory function, as Piwi proteins are active small RNA-guided nucleases (Gunawardane et al, 2007) and loss of Piwi proteins has been correlated to defects in heterochromatin formation (Pal-Bhadra et al, 2004) and de novo DNA methylation in germ cells (Aravin and Bourc'his, 2008; Kuramochi-Miyagawa et al, 2008). Piwi proteins have also been found to be associated with ribosomes, suggesting that they may have an important function in translational regulation (Grivna et al, 2006b).
Furthermore, Piwi proteins have been implicated in germline/stem cell maintenance (Cox et al, 2000; Deng and Lin, 2002; Kennerdell et al, 2002; Pal-Bhadra et al, 2002; Kuramochi-Miyagawa et al, 2004; Aravin et al, 2007; Carmell et al, 2007). In Drosophila, Piwi is necessary for self-renewing divisions of germ-line stem cells in both males and females (Lin and Spradling, 1997; Cox et al, 1998). In mice, MIWI and MILI are involved in the regulation of spermatogenesis but have not been shown to affect the maintenance of mitotic germ cells (Deng and Lin, 2002; Kuramochi-Miyagawa et al, 2004), whereas MIWI2 seems to be essential for maintaining the germline stem cells (Carmell et al, 2007), resembling Drosophila Piwi function. In zebrafish, Ziwi is necessary for the maintenance of the germ line, and loss of Ziwi causes pre-meiotic germ cells to die due to apoptosis (Houwing et al, 2007). Furthermore Piwi proteins regulate neoblasts, which are stem cells that allow extraordinary regenerative capacity in the planaria Schmidtea mediterranea (Reddien et al, 2005).
Here, we show that Zili, one of the two Piwi proteins in zebrafish, has a very dynamic nucleocytoplasmic distribution. Furthermore, we provide evidence for a piRNA amplification loop in zebrafish involving both Ziwi and Zili. Such an amplification loop has been proposed earlier on the basis of piRNA analysis in Drosophila where different Piwi proteins bind to piRNAs that are derived from transcripts from opposite strands of the same loci (Brennecke et al, 2007; Gunawardane et al, 2007). So far it has remained unclear whether such a clear functional separation between different Piwi paralogues exists in other species besides Drosophila. Our data show that, at least in zebrafish, this is a conserved feature. Additionally, we show that loss of Zili results in an increased level of transposon transcripts and the inability of germ cells to differentiate, ultimately leading to the loss of these germ cells around 7 weeks of age. Finally, we show that Zili has a function in meiosis, independent from its function in transposon silencing, extending the function of Piwi proteins beyond transposon defense.
Results and discussion
Zili protein has a dynamic nucleocytoplamic distribution
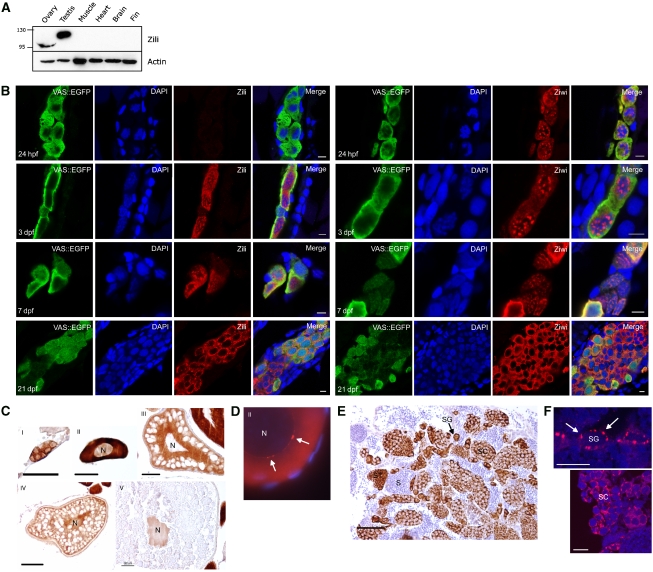
The Piwi protein family has two members in Danio rerio (zebrafish): Ziwi and Zili (ENSDARG00000041699 and ENSDARG00000062601, respectively). Ziwi has been the focus of previous work (Houwing et al, 2007) and was found to be specifically expressed in the germ line either in a granular fashion or diffuse in the cytoplasm, depending on the developmental stage of the animal. Here, we analyse Zili in more detail. Full-length zili cDNA was cloned using 5′ RACE and sequenced (Supplementary Figure 1; GenBank accession number FJ168029). With an antibody raised against the N terminus of Zili, Zili protein can be detected in primordial germ cells (PGCs) from 3 days post-fertilization (dpf) onwards (Figure 1). Thus, unlike Ziwi, maternal contribution of Zili is undetectable. In PGCs, in the first week of development, Zili protein localizes to the nucleus and is diffusely present in the cytoplasm. At 3 dpf, nuclear Zili seems not to colocalize with DAPI bright spots but rather seems to be excluded from them. From 7 dpf, Zili protein starts localizing more to the cytoplasm, whereas at this time point Ziwi protein leaves the granules around the nucleus and is more diffusely distributed in the cytoplasm. At 3 weeks post-fertilization (wpf), Zili is present in a granular distribution in the cytoplasm, whereas Ziwi remains diffuse. In mice, genomes of PGCs undergo extensive epigenetic reprogramming through genome-wide de- and remethylation (Sasaki and Matsui, 2008), with transposon sequences being de novo-methylated during the fetal prospermatogonium stage (Kato et al, 2007). MILI and MIWI2 have been suggested to have an important function in DNA remethylation (Aravin et al, 2007; Carmell et al, 2007; Kuramochi-Miyagawa et al, 2008). Although it is not known how and when epigenetic changes are set up in the newly formed germ line of the zebrafish, it is possible that Zili mediates similar processes in the developing zebrafish germ line during the first week of development.
Figure 1.
Zili is a nucleocytoplasmic protein. (A) Western blot showing specific expression of Zili in testis and ovary. The migration path of the Zili protein is distorted by highly abundant yolk proteins, resulting in a lower band for Zili in ovary than testis. (B) At 24 h post-fertilization (hpf), Zili protein cannot be detected in PGCs, whereas Ziwi protein is maternally provided and localizes to perinuclear granules. At 3 days post-fertilization (dpf), Zili protein is present in the nucleus of PGCs, as well as in the cytoplasm. From 7 dpf onwards, Zili protein starts localizing to the cytoplasm, whereas at this point, Ziwi protein exits the perinuclear granules and becomes diffuse in the cytoplasm. At 3 weeks post fertilization (wpf), Zili displays a granular distribution in the cytoplasm, whereas Ziwi is diffuse. Scale bar is 5 μm. (C) Zili (brown) in ovary is present in all stages of oogenesis in cytoplasm and/or nucleus (N). Stages of oogenesis are oogonia (I), stage I oocytes (7–140 μm; II), stage II oocytes (140–340 μm; III), stage III oocytes (340–690 μm; IV) and stage IV oocytes (0.69–0.73 mm; V) (Selman et al, 1993). Scale bar ia 50 μm. (D) Zili (red) in granules (white arrows) around the nucleus (N) of stage I oocyte. (E) Zili (brown) in testis is present predominantly in the cytoplasm of all stages of spermatogenesis, except the fully differentiated sperm. Stages of spermatogenesis are spermatogonia (SG), spermatocytes (SC), spermatids (ST) and sperm (S). Scale bar is 50 μm. (F) Zili (red) in granules around the nucleus of spermatogonia (upper panel) and spermatocytes (lower panel). Scale bar is 10 μm.
In the adult, Zili is found in both the female and male gonad (Figure 1). In the ovary, the protein is present in all stages of germ cell differentiation, with a distinct subcellular localization. In mitotic and early meiotic cells, Zili protein is predominantly cytoplasmic. In primary oocytes, the protein is found in a granular distribution around the nucleus. Later, in maturing oocytes, the protein localizes to the nucleus. In testis, Zili protein is present in the cytoplasm of mitotic and meiotic germ cells, where a distinct granular distribution around the nucleus is observed. No Zili protein has been detected in the fully differentiated sperm cell.
Ziwi- and Zili-bound piRNAs
Zebrafish piRNA populations in ovary and testis tissue have been characterized (Houwing et al, 2007), although it remained unclear whether Ziwi and Zili each bind to specific piRNA populations. To elucidate the respective functions of each of the two Piwi proteins, Ziwi and Zili proteins from ovary and testis lysates were immunoprecipitated and associated RNAs were purified. 5′-32P labelling of the isolated RNA showed a distinct small RNA species approximately 27 nucleotides long from the Ziwi and Zili immunoprecipitations (Supplementary Figure 2A). Subsequently, these small RNAs from both Ziwi and Zili complexes from ovary and testis were cloned and sequenced. Size distribution of the cloned sequences showed a distinct peak at 27 and 26 nucleotides from the Ziwi and Zili immunoprecipitations, respectively, both in ovary and testis (Supplementary Figure 2B). Over 140 000 sequences were identified in these four libraries, of which 42% map uniquely.
We have previously shown that a large portion of zebrafish piRNAs map to transposon sequences (Houwing et al, 2007). In general, Ziwi and Zili immunoprecipitate-derived libraries yield similar outcomes. Compared with total piRNAs from ovary and testis in zebrafish, we found that Ziwi- and Zili-bound piRNAs cover the same loci. Genomic mapping of cloned sequences shows that most sequences can be regarded as piRNAs, with a small percentage of RNAs with other annotations, such as miRNAs, tRNA, snRNAs and snoRNAs (Supplementary Figure 2C). Of the total number of sequences, 25–55% of piRNAs are comprised of repeat-associated regions, which are often annotated as transposons (Supplementary Figure 2C). piRNAs that are categorized as ‘not annotated' may well hold many transposon-derived sequences, as these are poorly annotated. In fact, the mapping frequency of ‘not annotated' piRNAs more closely resembles that of transposons, than that of coding regions (Supplementary Figure 1D).
Evidence for a piRNA amplification cycle
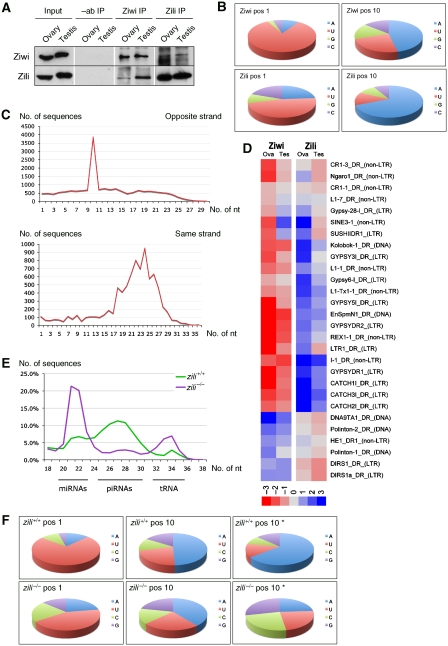
Further analysis showed that Ziwi, in particular, has a strong preference for piRNAs with a uridine nucleotide at their 5′-end (86%) (Figure 2B). Zili piRNAs show strong enrichment for an adenosine nucleotide (62%) at position 10. At most loci, Ziwi and Zili bind piRNAs of opposite polarity (Supplementary Figure 3), and the 5′-ends of complementary Ziwi and Zili piRNAs are separated by 10 nucleotides (Figure 2C), suggesting that the 5′-ends of zebrafish piRNAs are generated through Piwi protein-mediated nucleolytic cleavages. Ziwi and Zili piRNAs with the same polarity show an average overlap of 25 nucleotides, indicating that certain regions within a locus are preferentially producing piRNAs. This could be at the level of primary piRNA production (see below), although no sequence motifs can be detected close to these sequences. It could also mean that certain piRNAs are better in driving target cleavage than others.
Figure 2.
Amplification cycle for piRNAs. (A) Western blots for Ziwi and Zili protein, showing that Ziwi and Zili co-immunoprecipitate. (B) Ziwi piRNAs show a preference for uracil at their 5′-end, whereas Zili piRNAs have a preference for adenosine at position 10. (C) Sequence overlap of 10 nucleotides for Ziwi and Zili piRNAs of the opposite strand (upper panel), whereas piRNAs of the same strand show a 19–28 nucleotide overlap (lower panel). (D) Heat map showing a strand bias in Ziwi and Zili piRNAs mapping to several transposon species. Red depicts minus reads, whereas blue depicts plus reads. Calculated for loci that have 400+ total reads. Plotted ratio of plus/minus reads in log2 scale. (E) Size distribution of sequences cloned from zili+/+ and zili−/− mutant gonads at 5 weeks post fertilization (wpf), showing a clear reduction in the number of piRNAs in the zili−/− mutant. (F) Nucleotide frequencies at positions 1 and 10. Position 10* indicates all piRNAs without uracil at position 1. When comparing piRNAs that do not have a 5′ uracil preference (*) to all piRNAs from zili+/+, an increased preference for adenosine at position 10 can be detected. In zili−/− piRNAs, the preference for adenosine at position 10 has disappeared in piRNAs without a 5′-uracil preference (*).
The above-described signatures are present in piRNAs derived from all transposon classes (Supplementary Figure 5). They are also present, although weaker, in piRNAs within one library (Supplementary Figure 4), suggesting partial redundancy between the Piwi proteins. Alternatively, we cannot exclude that a minor portion of cloned piRNAs may derive from Zili-bound RNAs in the Ziwi immunoprecipitation and vice versa, as these proteins co-immunoprecipitate (see below). If a more general view of transposon classes is taken, a clear trend can be observed (Figure 2D), where Ziwi, in general, binds antisense piRNAs, whereas Zili binds sense piRNAs. In some cases, Ziwi and Zili seem to switch piRNA polarity. We do not understand the reason behind this, but these cases may be informative in understanding the polarity preference of Piwi proteins.
These data strongly support a conserved amplification loop for piRNAs in zebrafish, analogous to the so-called ‘ping-pong' model, which was described in Drosophila (Brennecke et al, 2007; Gunawardane et al, 2007). In zebrafish, this system is bipartite, involving Ziwi and Zili proteins. This amplification loop starts with a primary set of piRNAs, which is generated through an unknown mechanism. These primary piRNAs are bound to Ziwi, and when confronted with homologous mRNAs, the Zili protein is loaded with a piRNA derived from the targeted mRNA, having a 5′-end being defined by Ziwi-mediated cleavage. Zili may then again target antisense transcripts of the same transposon, resulting in the loading of more Ziwi protein with the corresponding piRNA. Effectively, this triggers an amplification of piRNA levels. This model of piRNA amplification suggests that Ziwi and Zili come in close physical proximity to one another. Accordingly, one may expect that both proteins co-immunoprecipitate, which is indeed what we observe (Figure 2A). Some loci clearly show antisense Ziwi piRNAs and sense Zili piRNAs; as an example we display a piRNA cluster on chromosome 14 (Supplementary Figure 3). Within this cluster, strand bias switches back and forth. These blocks correspond to repeats that switch orientation. Interestingly, whereas Ziwi piRNAs from this locus are present in both ovary and testis, Zili piRNAs seem to be missing in testis. This could reflect the absence of transcriptional activity at this locus in testis, which would take away the substrate for Zili piRNA production. As a result, the amplification cycle would not function, also leading to lower Ziwi piRNA levels in testis, which can indeed be observed (Supplementary Figure 3). Finally, when comparing Ziwi and Zili subpopulations of piRNAs to total piRNAs, we find that Ziwi piRNAs are much more abundant than Zili piRNAs. The simplest explanation for this would be that there is less Zili protein compared with Ziwi protein. Alternatively, it could point to a big reservoir of primary piRNAs searching for targets, or perhaps faster kinetics of the Zili enzyme.
Genic piRNAs
A striking 18% of piRNAs, on average, map to exons of about 5000 genes in both sense and antisense orientation (Supplementary Table 1). Intronic piRNAs were excluded from this analyses, as introns often contain repetitive sequences. However, sense piRNAs were observed overlapping intron–exon boundaries, suggesting that these genic piRNAs are pre-splicing products. The mapping frequency of these exonic piRNAs is usually one (Supplementary Figure 1D), indicating that repeat-associated piRNAs are excluded from this set. To further reduce the chance that transposon-like sequences are leading to a misinterpretation of our results, we further narrowed the exonic piRNA set by selecting for unique mapping positions and an absence of alternative splicing, as this may lead to the annotation of intronic sequences as exonic. In this way, 1911 genes with sense and 366 genes with antisense reads were identified. The intersection of both sets defines 187 genes, to which in total 1923 piRNAs were mapped: 1206 sense and 717 antisense. Even within this limited set, we can clearly detect the ping-pong signature, where sense and antisense piRNAs tend to overlap 10 bases at their 5′-ends (Supplementary Figure 6). This may indicate a function of piRNAs in the regulation of gene transcripts, although no specific gene class stands out.
Zili is essential for germ cell maintenance and differentation
To study the function of Zili protein in the zebrafish germ line, we screened for mutant alleles in an ENU-mutagenized library (Wienholds et al, 2003) and identified a nonsense allele in exon 1 of zili (G51STOP). Using immunostainings, Zili protein could not be detected in these zili−/− gonads at 6 weeks of age, whereas in wild-type siblings, the protein was abundant (Supplementary Figure 7A). As there is no evidence for N-terminal splice variants, we assume this ziliG51STOP allele (hu3173) to be a null allele.
First, we characterized piRNAs from zili−/− gonads. Small RNAs were cloned and sequenced from mutant gonads as well as wild-type gonads at 5 weeks of age. In total, 73 000 reads for the wild-type library and 36 000 reads for the mutant library were obtained, showing a clear size distribution of miRNA and piRNA peaks as well as a peak corresponding mostly to fragmented tRNAs (Figure 2E). When germ cell numbers were normalized using vasa, a germ cell-specific transcript (Yoon et al, 1997), a 70-fold drop in vasa levels compared with ef1α transcripts was observed (data not shown), indicating that germ cell numbers are greatly reduced in mutant gonads. Consistent with the loss of Zili, we see no evidence for Zili-bound piRNAs in the zili−/− gonads. To address this, we analysed the population of piRNAs containing nucleotides other than uridine at the 5′-end to determine whether there was a preference for adenosine at position 10. In the wild-type sample, a clear bias was observed but the signature disappeared completely in the mutant. A preference for adenosine at position 10 in piRNAs that start with uridine can still be seen, consistent with a possible redundancy between Ziwi and Zili (Figure 2F). This evidence suggests that, although there is a slight ping-pong signature present that is expressed by the adenosine preference at position 10 in all piRNAs, this does not originate from Zili piRNAs but rather from Ziwi piRNAs. This may indicate that Ziwi alone is sufficient for low levels of amplification. Alternatively, these piRNAs could represent the maternal pool of piRNAs, which also shows a preference for adenosine at position 10 in all piRNAs as well as piRNAs without a 5′-uridine (data not shown).
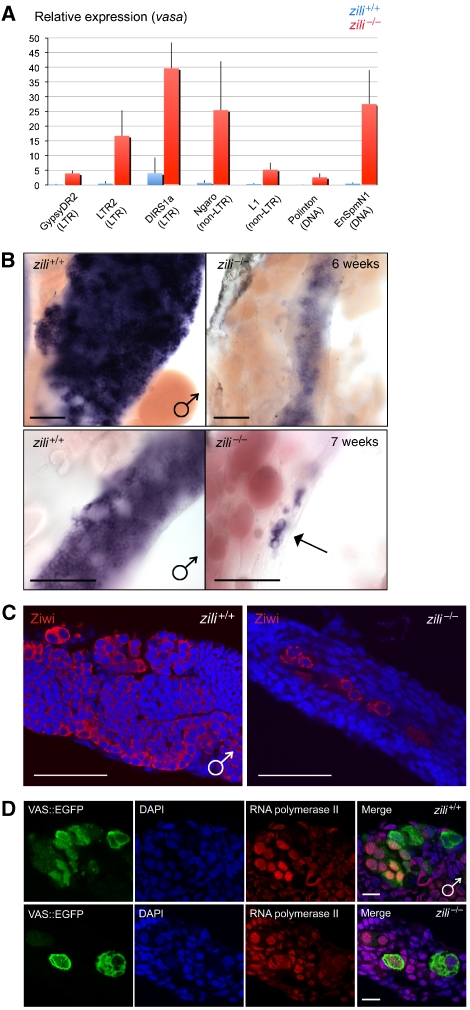
We next looked at transposon transcript levels. Transposon transcripts were reported to be upregulated in Drosophila and mice following perturbation of the piRNA pathway (Aravin et al, 2007; Carmell et al, 2007). In accordance with this observation, we observe increased levels of several LTR, non-LTR and DNA elements in zili−/− germ cells at 5 weeks of age, compared with their siblings, as determined by quantitative PCR (Figure 3A). Transposon transcripts were found to be present in the germ cells of fish gonads by in situ hybridization, and not in the somatic cells (Supplementary Figure 7B). Vasa transcript levels were therefore used to normalize expression levels (Yoon et al, 1997). Results of quantitative PCR were supported by in situ hybridizations (Supplementary Figure 8). Chromogenic detection shows a low level or absence of transposon transcripts in wild-type gonads at 5 weeks of age, whereas in zili−/− germ cells, an elevated level of transcripts can be detected. It is important to note, however, that zili−/− germ cells are phenotypically quite distinct from their wild-type counterparts at 5 weeks of age (see below). Consequently, we cannot exclude that the observed upregulation is secondary to this developmental defect.
Figure 3.
Zili is required for germ cell differentiation. (A) Relative expression of transposon transcripts compared with vasa transcripts in zili+/+ and zili−/− gonads at 5 wpf. Several LTR (GypsyDR2, LTR2, DIRS1a), non-LTR (Ngaro, L1) and DNA elements (Polinton, EnSpmN1) show an increased transcript level in zili−/− gonads compared with wild-type siblings. Error bars show average±s.d. for three replicates. (B) Whole mount in situ hybridization of wild type and zili−/− (G51STOP) gonads at 6 wpf (upper panel) and 7 wpf (lower panel), using an antisense probe for vasa. Loss of Zili leads to a strong reduction in the amount of vasa-positive cells (arrow), which at 8 wpf are completely lost. Scale bar is 100 μM. (C) Ziwi (red) protein is still present in germ cells of zili−/− gonads (6 wpf). Ziwi protein displays a granular distribution, reminiscent of earlier-stage germ cells. Scale bar is 100 μM. (D) zili+/+ germ cells (5 wpf) (upper panel), as well as zili−/− germ cell (5 wpf) (lower panel), are positive for active RNA polymerase II (red). Scale bar is 10 μM.
Loss of Ziwi results in apoptotic pre-meiotic germ cells at 3 weeks of age (Houwing et al, 2007). To determine at which developmental stage germ cells are lost in zili mutant fish, we used vasa as a marker for the germ line (Yoon et al, 1997). At 6 weeks of age, we observe a reduction in the number of germ cells in zili mutants compared with wild-type siblings, whereas at 7 weeks of age, almost all germ cells disappeared in zili−/− gonads (Figure 3B). After 7 weeks, no germ cells can be detected. At 6 weeks of age, Ziwi protein is still abundantly present in zili−/− germ cells (Figure 3C), although germ cells are clearly reduced in number. Ziwi protein displays a granular distribution in the zili−/− germ cells, reminiscent of earlier-stage mitotic germ cells. Additionally, whereas in wild-type gonads at 6 weeks of age, several stages of germ cell differentiation can be observed, in mutant gonads, no differentiated germ cells are detected. Nanos in situ hybridization shows an absence of this oocyte marker in zili−/− germ cells, suggesting that germ cells are not undergoing female development (Supplementary Figure 9A).
Consistent with ziwi−/− fish, zili−/− fish are always phenotypically male. As zebrafish do not have sex chromosomes, this is probably secondary to the loss of germ cells, as ablation of PGCs in fish results in male development (Weidinger et al, 2003). All fish first develop oocytes from 10 dpf till 25 dpf (Uchida et al, 2002). By 35 dpf, ovaries can be distinguished from testis. This differentiation occurs much earlier than the loss of germ cells we observe in zili−/− fish. Perhaps the presence of differentiated oocytes around 2–3 weeks of age is required for female development. In zili−/− fish, germ cells are not differentiating and oocytes are not present at this time point, resulting in an obligatory male development. Possibly this, rather than the complete absence of germ cells, may also be the cause of obligatory male development in Ziwi mutants and dead-end morphants (Weidinger et al, 2003; Houwing et al, 2007). Furthermore, zili−/− germ cells are still expressing proliferating cell nuclear antigen (PCNA) (Supplementary Figure 9B) and are positive for active RNA polymerase II (Figure 3D), indicating that these cells have not arrested and that transcription is still taking place. Additionally, zili−/− germ cells are negative for cleaved caspase-3 (Supplementary Figure 9C), suggesting that whereas ziwi−/− germ cells are entering apoptosis (Houwing et al, 2007), zili−/− germ cells are lost by another mechanism, possibly due to their inability to differentiate.
Zili has a crucial function in meiosis
In addition to a nonsense allele, we identified a missense allele, with a leucine to proline substitution after the PAZ domain (L590P). This ziliL590P allele (hu2087) permits germ cell development both in a homozygous situation as well as in combination with the zili null allele. Consequently, these genotypes support both male and female development. Males develop normally and are fertile. General morphology of the ovary and the oocytes appears normal, and an oocyte-specific structure called the Balbiani body (Kloc et al, 2004) also appears to be present in stage I oocytes.
Although Zili and Ziwi proteins are localized normally in the oocytes (data not shown), ziliL590P/L590P and ziliL590P/− females are sterile. The earliest defect we have been able to observe in ziliL590P/− oocytes is a nuclear positioning defect in stage IV oocytes, where the nucleus is peripheral instead of centrally localized (Figure 4D). Ultimately, all oocytes display a defect in meiosis I. Although wild-type females lay eggs that complete meiosis I immediately after laying and subsequently complete meiosis II upon sperm entry, we see neither event in eggs laid by ziliL590P/− females (Figure 4B and C), although these are fertilized. As ziliL590P/− males can fertilize eggs and do not display any meiotic defects, female meiosis seems to be more dependent on Zili function. We find no evidence that this phenotype is linked to increased levels of transposon transcripts (Figure 4A) as determined by quantitative PCR. Results of quantitative PCR were supported by in situ hybridizations (Supplementary Figure 8). Several LTR, non-LTR and DNA elements show no increase in transcript levels in this mutant ovary, indicating that Zili protein has a crucial function in germ cell development independent of its function in transposon defense. These observations suggest a function of piRNAs in chromosome segregation or in early meiotic processes.
Figure 4.
Zili has a function in meiosis. (A) Relative expression of transposon transcripts compared with vasa transcripts in ziliL590P/− ovaries. Several LTR (GypsyDR2, LTR2, DIRS1a), non-LTR (Ngaro, L1) and DNA (Polinton, EnSpmN1) elements were tested and show no difference in ziliL590P/− ovaries compared with wild-type siblings. Error bars show average±s.d. for three replicates. (B) DAPI staining on embryos before fertilization, at 10 min post-fertilization (mpf) and 30 mpf show a meiotic arrest in ziliL590P/−. In wild-type embryos, meiosis I is completed immediately after egg-laying, and meiosis II is completed after sperm entrance (arrow indicates male pronucleus), and both polar bodies are extruded (1 asterisk is first polar body, 2 asterisk is second polar body). The male and female pronuclei fuse and form a two-cell embryo at 30 mpf. When the embryo is unfertilized, meiosis II is completed, although the DNA eventually starts breaking down. In eggs laid by ziliL590P/− females, the egg is fertilized (arrow), but meiosis I is not completed, a first polar body is not formed and the cell fails to divide. Scale bar is 25 μm. (C) After 15 mpf, the male and female pronuclei have fused and the second polar body (2 asterisk) is surrounded by tubulin. In eggs laid by ziliL590P/− females, tubulin surrounds part of the arrested female pronucleus, but no polar body is formed. Scale bar is 25 μm. (D) vasa mRNA in ziliL590P/− localizes normally to Balbiani body (asterisk), whereas vitellogenic oocytes (IV) display a peripheral position of the nucleus (arrow). Diagram shows quantification of nuclear position by ratio of the longest distance and shortest distance from the nuclear membrane to the cell membrane. Error bars are s.e.m. (t-test P<0.01).
Concluding remarks
In summary, we provide evidence for a piRNA amplification loop in zebrafish driven by two different Piwi proteins, Ziwi and Zili (Supplementary Figure 10). So far such a bipartite system was described only in Drosophila (Brennecke et al, 2007; Gunawardane et al, 2007) but is now shown to be a conserved mechanism in zebrafish and possibly other vertebrates. This piRNA amplification loop is compromised when Zili protein is absent. Our data suggest that Ziwi alone may be sufficient for low levels of amplification. In mice, MIWI and MILI mutants display meiotic defects, but the maintenance of mitotic germ cells appears to be unaffected (Deng and Lin, 2002; Kuramochi-Miyagawa et al, 2004). MIWI2 seems to be essential for maintaining the germline stem cells, much similar to zebrafish Piwi function, although in MIWI2 mutants, germ cell differentiation is still observed (Carmell et al, 2007). In planaria, Piwi functions to maintain the regeneration ability of stem cells (Reddien et al, 2005). It will be interesting to explore to what extent the function of Piwi proteins in planaria neoblasts and the function of Zili in germ cell differentiation are mechanistically related. The fact that we find Zili to be nuclear suggests that this mechanism, at least in fish, may exert an effect through chromatin or DNA modifications. Similar effects on the chromosomes in oocytes undergoing meiosis may be the basis for the block in meiosis we see in ziliL590P/− oocytes. In MIWI2 mutant mice, male germ cells arrest in early prophase I and show increased transposon levels. These mice resemble DNMT3L mutant mice, which fail to methylate transposons in the male germ line, indicating that increased transposon activity is most likely the underlying cause of the meiotic arrest (Bourc'his and Bestor, 2004; Carmell et al, 2007), unlike the meiotic arrest we observe in ziliL590P/− oocytes. Interestingly, in rice, loss of the Ago protein MEL1 results in arrested germ cells in early prophase I. Meiotic chromosomes fail to undergo normal condensation and lack hypomethylated histone H3K9. However, the initiation and establishment of germ cells themselves is unaffected (Nonomura et al, 2007). It will be interesting to investigate whether here also there may be a related mechanism at the basis of these observations. Importantly, the function of Zili in meiosis can be separated from its function in transposon silencing, indicating that the Piwi pathway reaches beyond the silencing of repetitive elements in zebrafish.
Materials and methods
Zebrafish strains and genetics
Zebrafish were kept under standard conditions (Westerfield, 1993). Zebrafish mutants were derived from ENU mutagenized libraries using target-selected mutagenesis as described (Wienholds et al, 2003; Draper et al, 2004). Founders with mutant alleles zili hu3173 and zili hu2087 were out-crossed against wild-type fish (TL) or VAS∷EGFP transgenic fish (Krovel and Olsen, 2002) and subsequently in-crossed against each other to obtain homozygous offspring. DNA was extracted from caudal fin tissue amputated from anesthetized fish. The primers used to amplify and resequence the alleles are as follows: zili_hu3173_F 5′-GGCACCATGGCAGCAGTCG-3′, zili_hu3173_R 5′-CTCGTCCTCGCCCAAAACC-3′, zili_hu2087 F 5′-CTGTAGTGCAGGTTCCATTG-3′ and zili_hu2087 R 5′-TAACCACACCACCAAGTGCAG-3′.
In situ hybridization
In situ hybridization was performed as described before (Houwing et al, 2007). A gypsyDR1 fragment was PCR-amplified using primers 5′-GCCATCACTCTGGTTCACC-3′ and 5′-TTTCCACAATGAAATGTGGG-3′, cloned and used as a template for probe synthesis. An ngaro fragment was PCR-amplified using primers 5′-GGGAGCGATCGAGACCTACC-3′ and 5′-CAATCATATCACGTGCTCCTCTCG-3′, cloned and used as a template for probe synthesis. An LTR2 fragment was PCR-amplified using primers 5′-GAGTGTTTGGTGTCGTTAGAATGCCCTTGAC-3′ and 5′-CAGGTAATACGACTCACTATAGGGGGTTATACCTGTGGGTCAC-3′ and was used as a template for probe synthesis with T7 polymerase. A DIRS1a fragment was PCR-amplified using primers 5′-GCGGGTGCGTCACGCTTGCTTGAC-3′ and 5′-CAGGTAATACGACTCACTATAGGGGTAACCTCGAACGTTCCC-3′ and was used as a template for probe synthesis with T7 polymerase. A nanos probe was obtained from an RZPD clone containing full-length nanos in pme18s-FL. A PCNA probe was obtained from EST-clone fc43g05 containing the last exon of PCNA.
Western blot and immunostaining analysis
Western blot and immunostainings were done as described before (Houwing et al, 2007). Zili antibodies were raised in rabbits with the synthetic peptide MDPKRPTFPSPPGVI+C. Ziwi antibodies were raised in rabbits with the synthetic peptide MTGRARAR+C. Antisera were subsequently purified against the synthetic peptide (Eurogentec). Embryos were fixed in 80% MeOH/20% DMSO at RT for 3 h. Mouse anti-alpha tubulin B512 obtained from Sigma was used. Rabbit anti-Caspase-3 was used as described before (Houwing et al, 2007). Rabbit anti-active RNA polymerase II (ab5131) obtained from Abcam was used.
Immunoprecipitation
Immunoprecipitations were performed with Ziwi and Zili affinity-purified antibodies. Tissues were sonicated for 1 min at 4 °C and centrifuged for 10 min at 12 000 r.p.m. at 4 °C. Supernatant was used for IP. One IP generally contains 30 μl Dynabeads (Invitrogen), half ovary or three testes, and Ziwi antibody 1:200 or Zili antibody 1:250 in a total volume of 500 μl. IP lysis buffer contains 25 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, 1 mM DTT, protease inhibitors. Beads, antibodies and lysate were incubated for 2 h at 4 °C while rotating. Beads were washed three times with wash buffer (25 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1.5 mM MgCl2, 1 mM DTT). Beads were used for Trizol (Invitrogen) RNA isolation or eluted with 0.1 M citrate pH 2.5 for western blot analysis. RNA was 5′-labelled with 32-P and run on 15% denaturing polyacrylamide gels. Gel was exposed to phosphor-imager screens, which were scanned on a BAS-2500 imager (Fuji).
piRNA cloning
Immunoprecipitations were performed and RNA was isolated using Trizol (Invitrogen). The RNAs were poly(A)-tailed using poly(A) polymerase followed by ligation of a RNA adaptor to the 5′-phosphate of the small RNAs. First-strand cDNA synthesis was performed using an oligo(dT)-linker primer and M-MLV-RNase H- reverse transcriptase. The cDNA was PCR-amplificated for 16 cycles according to the instructions of 454 Live Sciences. Equal amounts of bar-coded cDNA were mixed. The 125–140 bp fraction of the cDNA pool was obtained by fractionation of the cDNA on a preparative 6% polyacrylamide (PAA) gel. The eluted cDNA was finally purified using the Macherey & Nagel NucleoSpin Extract II kit and was sent for 454 GS FLX sequencing (Roche). The following adaptor sequences with NNNN representing the barcodes flank the cDNA inserts—5′-end: 5′-GCCTCCCTCGCGCCATCAGCTNNNNGACCTTGGCTGTCACTCA-3′; 3′-end: 5′-GCCTTGCCAGCCCGCTCAGACGAGACATCGCCCCGCTTTTTTTTTTTTTTTTTTTTTTTTT-3′. Gonads from 5-week-old fish were flash-frozen and small RNA species were isolated using the mirVana miRNA isolation kit. The isolated small RNAs were separated on a denaturing 15% PAA gel and stained with SYBRgreenII. The population of small RNAs with a length of 19–40 bases was obtained by passive elution of the RNAs from the gel. The RNAs were precipitated with ethanol and dissolved in water. The gel-purified RNAs were first poly(A)-tailed using poly(A) polymerase followed by ligation of an RNA adaptor to the 5′-phosphate of the RNAs. First-strand cDNA synthesis was performed using an oligo(dT)-adaptor primer and M-MLV-RNase H- reverse transcriptase. Incubation temperatures were 42 °C for 15 min, ramp to 55 °C followed by 55 °C for 5 min. The resulting cDNAs were then PCR-amplified to about 40 ng/μl using a high-fidelity DNA polymerase. The cDNAs were purified using the Macherey & Nagel NucleoSpin Exctract II kit and subsequently sequenced using the 454 platform (Roche). The following adaptor sequences flank the cDNA inserts—5′-end: 5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′; 3′-end: 5′-CTGCCCCGGGTTCCTCATTCTCTTTTTTTTTTTTTTTTTTTTTTTT-3′.
Sequence analysis
Adaptor sequences were trimmed from generated data using custom scripts. Note that a minor drawback of our cloning strategy is the inability to detect 3′ adenosines. Resulting inserts were mapped to the zebrafish genome (Zv7 assembly, July 2007) using megablast program (Zhang et al, 2000), and genomic annotations of mapped reads were retrieved from Ensembl database (v. 48, Dec 2007) using Perl API provided by Ensembl (Hubbard et al, 2007). Nucleotide frequency graphs were generated by WebLogo software (Crooks et al, 2004). Sequences are available at GEO database under accession number GSE12547.
Quantitative PCR
Total RNA was isolated from juvenile gonads using Trizol (Invitrogen). RNA was treated with DNase and again Trizol-isolated. Oligo dT-primed cDNA was used for quantitative PCR.
Quantitative PCR was conducted on a Biorad ICycler system with SYBR Green. Normalized enrichment values were calculated with a standard formula. Sequences of primers are Vasa_F 5′-GGTCGTGGAAAGATTGGCCTG-3′, Vasa_R 5′-CAGCAGCCATTCTTTGAATATCTTC-3′, GypsyDR2_F 5′-GAAATCACCTGTGCATTTAC-3′, GypsyDR2_R 5′-ATGCAGACATTGGGTAAAGC-3′, EnSpmN1_F 5′-GATTGGCCATTGTGTTCACATGC, EnSpmN1_R 5′-GCTGTGACTGTCATAGGTTTACC-3′, Ngaro_F 5′-GGAGCGATCGAGACCTACC, Ngaro_R 5′-CAATCATATCACGTGCTCCTCTCG-3′, Polinton_F 5′-CCTGACAATGTTGTCAGCCTG-3′, Polinton_R 5′-CATGAAAGCTAAGGGTATAACTCTG-3′, DIRS1a_F 5′-GGGTGCGTCACGCTTGC-3′, DIRS1a_R 5′-GTAACCTCGAACGTTCCCC-3′, L1-5_F 5′-GCACAAAGGACAAATTCACTGGAC-3′, L1-5_R 5′-GTCCACGTTTAGTATTACAGTTGC-3′, LTR2_F 5′-GGTGTCGTTAGAATGCCCTTGAC-3′, LTR2_R 5′-GGTTATACCTGTGGGTCACGTG-3′.
Supplementary Material
Supplementary Figures
Supplementary Table 1
Acknowledgments
We thank Dr Kelly Smith for critical reading of the manuscript, Professor Edwin Cuppen and Ewart de Bruijn for assistance in screening for zebrafish mutants, Professor Mary C Mullins for discussion and help in analysing ovary sections. This work was supported by the European Union Sixth Framework Program Integrated Project SIROCCO (Grant LSHG-CT-2006-037900) and a VIDI fellowship from the Netherlands Organisation for Scientific Research (NWO; RFK).
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Bourc'his D (2008) Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev 22: 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH (2004) Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H (2000) piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H (2002) miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Stout JL, Slade AJ, Moens CB (2004) A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol 77: 91–112 [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H (2006a) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H (2006b) MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci USA 103: 13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A Slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K et al. (2007) Ensembl 2007. Nucleic Acids Res 35 (Database issue): D610–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA (2005) Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res 33: 2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H (2007) Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280 [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Yamaguchi S, Carthew RW (2002) RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev 16: 1884–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Etkin LD (2004) The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol 59: 1–36 [DOI] [PubMed] [Google Scholar]
- Krovel AV, Olsen LC (2002) Expression of a vas∷EGFP transgene in primordial germ cells of the zebrafish. Mech Dev 116: 141–150 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T (2004) Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE (2006) Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC (1997) A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476 [DOI] [PubMed] [Google Scholar]
- Nonomura KI, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N (2007) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA (2002) RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell 9: 315–327 [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC (2004) Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A (2005) SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310: 1327–1330 [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschêne G, Bucheton A, Pélisson A (2004) Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y (2008) Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 9: 129–140 [DOI] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V (2006) Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev 20: 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman K, Wallace RA, Sarka A, Qi X (1993) Stages of oocyte development in the zebrafish Brachydanio rerio. J Morphol 152: 203–224 [DOI] [PubMed] [Google Scholar]
- Uchida D, Yamashita M, Kitano T, Iguchi T (2002) Oocyte apoptosis during the transition from ovary-like tissue to testes during sex differentiation of juvenile zebrafish. J Exp Biol 205 (Part 6): 711–718 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA (2004) The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol 1: 54–58 [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, Raz E (2003) Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol 13: 1429–1434 [DOI] [PubMed] [Google Scholar]
- Westerfield M (1993) The Zebrafish Book. University of Oregon Press: Eugene [Google Scholar]
- Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E (2003) Efficient target-selected mutagenesis in zebrafish. Genome Res 13: 2700–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, Hopkins N (1997) Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 124: 3157–3165 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Haley B (2005) Ribo-gnome: the big world of small RNAs. Science 309: 1519–1524 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table 1