Abstract
A critical seasonal event for anadromous Chinook salmon (Oncorhynchus tshawytscha) is the time at which adults migrate from the ocean to breed in freshwater. We investigated whether allelic variation at the circadian rhythm genes, OtsClock1a and OtsClock1b, underlies genetic control of migration timing among 42 populations in North America. We identified eight length variants of the functionally important polyglutamine repeat motif (PolyQ) of OtsClock1b while OtsClock1a PolyQ was highly conserved. We found evidence of a latitudinal cline in average allele length and frequency of the two most common OtsClock1b alleles. The shorter 335 bp allele increases in frequency with decreasing latitude while the longer 359 bp allele increases in frequency at higher latitudes. Comparison to 13 microsatellite loci showed that 335 and 359 bp deviate significantly from neutral expectations. Furthermore, a hierarchical gene diversity analysis based on OtsClock1b PolyQ variation revealed that run timing explains 40.9 per cent of the overall genetic variance among populations. By contrast, an analysis based on 13 microsatellite loci showed that run timing explains only 13.2 per cent of the overall genetic variance. Our findings suggest that length polymorphisms in OtsClock1b PolyQ may be maintained by selection and reflect an adaptation to ecological factors correlated with latitude, such as the seasonally changing day length.
Keywords: Chinook salmon, Clock gene, latitudinal cline, migration timing, PolyQ
1. Introduction
Many organisms use day length (photoperiod) as an environmental cue to regulate seasonal changes in behaviour, growth, development, reproduction, dormancy and migration (Pittendrigh 1981; Vaz Nunes & Saunders 1999; Bradshaw & Holzapfel 2001). In plants and animals, the daily molecular oscillator, known as the circadian clock, senses changes in photoperiod and mediates a diverse number of photoperiodic responses such as flowering time in long- and short-day plants (Arabidopsis, Kardailsky et al. 1999; Oryza, Yano et al. 2000) as well as hormone secretion in mammals (Siberian hamsters, reviewed in Goldman 2001). Most recently, the circadian rhythm gene, timeless, has been shown to affect the incident of diapause, a seasonal event in Drosophila (Tauber et al. 2007).
A critical seasonal event for anadromous fish such as Chinook salmon (Oncorhynchus tshawytscha) is the time at which adults migrate from the ocean to their natal rivers to breed. Intra-annual variability in both water flow and temperature limits access to discrete spawning habitats within these rivers, and consequently, locally adapted migratory populations have evolved low inter-annual variability in their seasonal return time. Thus, the timing of this event is primarily an adaptation to long-term average conditions rather than a proximate response to current conditions. Photoperiod is a stable, long-term environmental cue that fishes could use to coordinate their population-specific migratory runs with these seasonally varying conditions (Quinn & Adams 1996). Previous studies have demonstrated that photoperiod is a key environmental cue for maturation timing in salmonid fish (Beacham & Murray 1988) and because this trait is strongly correlated to migration timing (Quinn et al. 2000), it is reasonable to hypothesize that photoperiod is also an important cue for anadromous salmon.
In North America, there is a considerable amount of diversity in the timing of Chinook salmon migration. To facilitate management of these locally adapted populations, fishery biologists categorize individuals as belonging to one of four seasonal runs based on the peak freshwater return time of the population. In a population genetic study of 118 Chinook salmon runs based on neutral markers, Waples et al. (2004) found that distinct seasonal runs within a geographical region were genetically more similar to each other than to populations with a similar run time from different geographical regions. These results indicate that run timing has evolved multiple times in this species. In the interior Columbia River Basin, however, Waples et al. (2004) and others have concluded that run timing differences reflect a much older divergence of two major lineages.
One notable trend throughout the geographical range of Chinook salmon is an increase in run time diversity with decreasing latitude. For instance, primarily summer run Chinook salmon populations occupy the northern rivers of Alaska and Canada while autumn, spring and summer run populations inhabit regions to the south with multiple run time populations coexisting in the same river. The Sacramento River in California, for example, supports three seasonal runs: autumn, spring and the only remaining winter run population. Thus, there appears to be a latitudinal cline in run time diversity of Chinook salmon populations along the west coast of North America.
We previously characterized two copies of the circadian rhythm gene, Clock, (OtsClock1a and OtsClock1b) as potential candidate loci for migration time in Chinook salmon (O'Malley & Banks 2008). The Clock gene, which codes for one of the most essential proteins of the circadian oscillator (Lowrey & Takahashi 2004), has been characterized in several model organisms including mouse (King et al. 1997), Drosophila (Allada et al. 1998) and zebrafish (Whitmore et al. 1998). Heterodimerization of CLOCK and a second protein, BMAL, produces a transcription-activating complex that regulates the expression of two additional circadian genes, Period and Cryptochrome (Reppert & Weaver 2002). A critical domain of CLOCK, which affects the transcription-activating potential of this protein, is the carboxyl-terminal polyglutamine repeat motif (PolyQ; Darlington et al. 1998). Expansion or contraction in the number of glutamine repeats in this region directly affects the corresponding gene product and thereby influences the circadian phenotype (Gekakis et al. 1998).
In an earlier investigation, we studied seasonal migratory runs of Chinook salmon from two systems and showed that length variation in the OtsClock1b PolyQ domain provides evidence for potentially adaptive genetic differentiation (O'Malley et al. 2007). Furthermore, tests for selective neutrality revealed that OtsClock1b deviated from neutral expectations in both systems indicating that this candidate gene is likely under selection. Thus, these results suggest that OtsClock1b may influence migration timing of Chinook salmon in these two systems.
Here we screened for length polymorphisms in OtsClock1a and OtsClock1b PolyQ domains among 42 runs of Chinook salmon and investigated whether there was any association between variation in allele frequency and migration time across a broad latitudinal gradient along the west coast of North America. We then compared these results to data from microsatellite loci presumed to be selectively neutral.
2. Material and methods
(a) Populations of Chinook salmon
Of the 42 populations included in this study, 40 are part of a standardized DNA collection established by the Chinook Technical Committee (CTC) of the Pacific Salmon Commission (figure 1; Seeb et al. 2007). Individuals from all CTC populations were previously genotyped at 13 microsatellite loci. For this study, we obtained DNA samples of approximately 48 individuals from each of the 40 populations. In June 2004, we collected 20 Chinook salmon liver samples from the remaining two populations, Alaska's Montana Creek and Situk River, and extracted genomic DNA using the DNeasy Tissue Kit (Qiagen).
Figure 1.
Map showing the location of the 42 Chinook salmon populations from California to Alaska.
(b) Genotyping of OtsClock1a and OtsClock1b PolyQ domains
We designed oligonucleotide primers to amplify a 199 bp fragment encompassing the polyglutamine repeat motif of OtsClock1a. The fluorescently labelled sense primer was 5′-GGTTCCTAATGTAGTTCCTGTGCTT-3′ and 5′-GATTTCTCACCTGGACACTGGGCT-3′ the antisense. We used previously designed oligonucleotide primers to amplify a 335 bp fragment of the OtsClock1b PolyQ domain gene (O'Malley et al. 2007).
DNA was amplified in 5 μl reactions using two touchdown PCR profiles: one initial denaturing cycle of 3 min at 94°C, followed by one cycle of 1 min at 94°C, 1 min at 62°C annealing temperature, and 1 min 30 s at 72°C. In subsequent cycles, the annealing temperature was decreased by 2°C until 52°C was reached for OtsClock1a and 56°C for OtsClock1b, followed by 29 more cycles of 1 min at 94°C, 1 min at 52°C/56°C, 1 min 30 s at 72°C, and a final extension of 10 min at 72°C. PCR products were electrophoresed on an Applied Biosystems 3730XL DNA Analyzer and scored as length polymorphisms using GeneMapper software.
(c) Sequencing of OtsClock1a and OtsClock1b PolyQ domain
To determine nucleotide sequence of the Clock PolyQ length variants, we PCR amplified the region from individuals of known genotype (table 1). PCR products were excised from 1.5 per cent agarose gels and purified using QIAquick gel extraction kit (Qiagen). Purified PCR products were then cloned into pCR4-TOPO vector using the TOPO TA Cloning kit for Sequencing (Invitrogen). Plasmid DNA was isolated using Wizard Plus SV Minipreps (Promega) and sequenced using Big Dye Terminator v. 3.1 Cycle Sequencing Ready Reaction. All sequences were generated on an Applied Biosystems 3730XL DNA Analyzer. Multiple sequence alignments were created manually using BioEdit Sequence Alignment Editor (Hall 1997) and automatically using ClustalW (Thompson et al. 1994).
Table 1.
Allele, population, geographical location and run time information of the individuals used to determine nucleotide sequence of the eight novel alleles are listed.
| allele | population | latitude | longitude | run time |
|---|---|---|---|---|
| 293 | McKenzie River, OR | 44.12 | 122.62 | S |
| 329 | Cowlitz River, WA | 46.51 | 122.62 | F |
| 335 | Salmon River, BC | 54.07 | 122.55 | S |
| Cowlitz River, WA | 46.51 | 122.62 | F | |
| McKenzie River, OR | 44.12 | 122.62 | S | |
| 338 | Cowlitz River, WA | 46.51 | 122.62 | F |
| 356 | Salmon River, BC | 54.07 | 122.55 | S |
| 359 | Atnarko River, BC | 52.37 | 126.10 | Su |
| 362 | Imnaha River, OR | 45.73 | 117.86 | S |
| 383 | Chikamin, AK | 55.82 | 130.89 | Su |
| Atnarko River, BC | 52.37 | 126.10 | Su | |
| S. Umpqua River, OR | 43.67 | 124.20 | F |
(d) Data analysis
Calculations of allelic frequencies were performed using Genepop v. 3.3 (Raymond & Rousset 1995). We calculated pairwise estimates of Fst and permuted the data 1000 times using Genetix v. 4.02 (Belkhir 2000). Associations between latitude and allele frequency as well as average allele length were examined using linear regression (SigmaPlot v. 6.0). Average allele length is defined as the sum of all allele lengths of individuals in a given population divided by the total number of individuals in that population. To determine if regression coefficients for the candidate gene alleles differed significantly from neutral expectations, we compared these values to a distribution of regression coefficients (allele frequency on latitude) for 441 alleles (13 microsatellite loci) from the CTC microsatellite baseline dataset.
Partial Mantel tests of association were also used to analyse the patterns of spatial variation by comparing matrices of genetic distance (i.e. Fst, allele frequency, average allele length) to geographical distance (i.e. latitude, longitude). Mantel tests were performed with 10 000 permutations using the software program Passage (Rosenberg 2001).
To partition the overall Fst estimate into genetic variance related to run timing, geographical region and differences among populations, we performed two hierarchical gene diversity analyses (Lewis & Zaykin 2001). The first analysis was based on allelic variation at candidate genes while the second was based on variation at 13 presumed neutral microsatellite loci. To perform the hierarchical analysis, we grouped the 40 populations into geographical regions within one of three run times (table 2). Run time is defined as the peak return time of a given population to freshwater spawning grounds. We primarily used run time designations listed in Waples et al. (2004) and Seeb et al. (2007). To obtain a balanced dataset, the McCloud River winter and Big Qualicum autumn runs were excluded from each analysis. In addition, three populations were excluded from the microsatellite analysis owing to lack of data (Wilson Fall, Siletz Fall and Montana summer).
Table 2.
Hierarchy used to partition the overall Fst into genetic variance related to run timing, region and differences among rivers.
| autumn run | |||||
|---|---|---|---|---|---|
| California | Oregon | Columbia River | |||
| Battle Creek | Umpqua River | Cowlitz River | |||
| Russian River | Siletz River | L. Deschutes River | |||
| Klamath River | Nestucca River | ||||
| Trinity River | Wilson River | ||||
| Rogue River |
| spring run | |||||
|---|---|---|---|---|---|
| California | Oregon | Columbia River | British Columbia | ||
| Butte Creek | Umpqua River | Cowlitz River | Spius River | ||
| Deer Creek | Siletz River | Carson River | Birkenhead River | ||
| Trinity River | Rogue River | McKenzie River | Salmon River |
| summer run | |||||
|---|---|---|---|---|---|
| Columbia/Snake Rivers | S SE Alaska | SE Alaska | N SE Alaska | Interior BC | Coastal BC |
| Methow River | Chikamin River | Andrews Creek | Big Boulder Creek | Clearwater | Klinaklini River |
| Imnaha River | Clear Creek | King Salmon River | Klukshu River | Morkill | Atnarko |
| Secesh River | Keta River | Nakina River | Situk River | ||
| Montana Creek |
3. Results
(a) Conservation of the OtsClock1a PolyQ domain
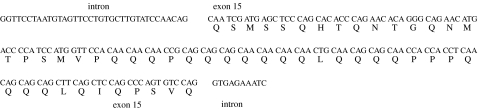
We found no evidence of length polymorphism in the OtsClock1a PolyQ domain among the Chinook salmon populations as all individuals were homozygous for the 199 bp allele. The OtsClock1a PolyQ fragment consists of 52 amino acid residues flanked by 36 and 11 bp of non-coding sequence (figure 2).
Figure 2.
Nucleic and amino acid sequence of OtsClock1a PolyQ locus.
(b) Length variation of the OtsClock1b PolyQ domain
We identified eight length variants of the OtsClock1b PolyQ domain among 42 Chinook salmon populations ranging from California to Alaska. Two length variants, 335 and 359 bp, are most common with frequencies of 0.737 and 0.225, respectively. Two less common variants, 338 and 383 bp, have frequencies of 0.010 and 0.026, respectively. The combined frequency of the remaining four alleles, 293, 329, 356, and 362 bp, is 0.002 (table 3). We identified 16 genotypic classes among the 42 populations. The frequency of the four most common genotypes are: 335/335 (0.56); 335/359 (0.29); 359/359 (0.07); and 335/383 (0.03). The remaining 12 genotypic classes (293/335, 329/329, 329/335, 329/335, 329/359, 335/338, 338/338, 338/359, 335/356, 359/362, 359/383 and 383/383 bp) have a frequency less than or equal to 0.01.
Table 3.
OtsClock1b PolyQ variant frequencies in 42 populations of Chinook salmon (O. tshawytscha). Latitude/longitude (degrees), number of alleles (N),and run time (autumn, A; winter, W; spring, S; summer, Su) are listed for each population.
| population | run time | °latitude | °longitude | N | 293 | 329 | 335 | 338 | 356 | 359 | 362 | 383 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Montana River, AK | Su | 62.06 | 150.04 | 40 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.50 | 0.00 | 0.00 |
| Situk River, AK | Su | 59.45 | 139.57 | 40 | 0.00 | 0.00 | 0.47 | 0.00 | 0.00 | 0.53 | 0.00 | 0.00 |
| Big Boulder Creek, AK | Su | 59.43 | 136.19 | 86 | 0.00 | 0.00 | 0.69 | 0.00 | 0.00 | 0.24 | 0.00 | 0.07 |
| Klukshu River, AK | Su | 59.17 | 138.53 | 82 | 0.00 | 0.00 | 0.41 | 0.00 | 0.00 | 0.59 | 0.00 | 0.00 |
| Nakina River, AK | Su | 58.42 | 133.99 | 88 | 0.00 | 0.00 | 0.51 | 0.00 | 0.00 | 0.40 | 0.00 | 0.09 |
| King Salmon River, AK | Su | 57.68 | 133.17 | 96 | 0.00 | 0.00 | 0.40 | 0.00 | 0.00 | 0.47 | 0.00 | 0.13 |
| Andrews Creek, AK | Su | 56.67 | 132.25 | 86 | 0.00 | 0.00 | 0.46 | 0.00 | 0.00 | 0.42 | 0.00 | 0.12 |
| Clear Creek, AK | Su | 56.08 | 131.07 | 64 | 0.00 | 0.00 | 0.64 | 0.00 | 0.00 | 0.23 | 0.00 | 0.13 |
| Chikamin, AK | Su | 55.82 | 130.89 | 90 | 0.00 | 0.00 | 0.66 | 0.00 | 0.00 | 0.03 | 0.00 | 0.31 |
| Keta River, AK | Su | 55.34 | 130.48 | 80 | 0.00 | 0.00 | 0.82 | 0.00 | 0.00 | 0.09 | 0.00 | 0.09 |
| Salmon River, BC | S | 54.07 | 122.55 | 88 | 0.00 | 0.02 | 0.61 | 0.00 | 0.01 | 0.36 | 0.00 | 0.00 |
| Morkill, BC | Su | 53.60 | 120.70 | 90 | 0.00 | 0.00 | 0.73 | 0.00 | 0.00 | 0.27 | 0.00 | 0.00 |
| Atnarko, BC | Su | 52.37 | 126.10 | 84 | 0.00 | 0.00 | 0.65 | 0.00 | 0.00 | 0.31 | 0.00 | 0.04 |
| Clearwater, BC | Su | 51.55 | 120.20 | 86 | 0.00 | 0.00 | 0.72 | 0.00 | 0.00 | 0.28 | 0.00 | 0.00 |
| Klinaklini River, BC | Su | 51.10 | 125.72 | 80 | 0.00 | 0.00 | 0.80 | 0.00 | 0.00 | 0.17 | 0.00 | 0.03 |
| Birkenhead River, BC | S | 50.32 | 122.60 | 94 | 0.00 | 0.00 | 0.97 | 0.00 | 0.00 | 0.03 | 0.00 | 0.00 |
| Spius River, BC | S | 50.15 | 121.02 | 50 | 0.00 | 0.00 | 0.74 | 0.00 | 0.00 | 0.26 | 0.00 | 0.00 |
| Big Qualicum, BC | F | 49.40 | 124.62 | 94 | 0.00 | 0.00 | 0.91 | 0.00 | 0.00 | 0.09 | 0.00 | 0.00 |
| Carson River, WA | S | 48.48 | 120.19 | 54 | 0.00 | 0.00 | 0.46 | 0.00 | 0.00 | 0.48 | 0.00 | 0.06 |
| Methow River, WA | Su | 48.13 | 120.06 | 58 | 0.00 | 0.00 | 0.45 | 0.03 | 0.00 | 0.52 | 0.00 | 0.00 |
| Cowlitz River, WA | A | 46.51 | 122.62 | 86 | 0.00 | 0.02 | 0.84 | 0.01 | 0.00 | 0.13 | 0.00 | 0.00 |
| Cowlitz River, WA | S | 46.51 | 122.62 | 70 | 0.00 | 0.06 | 0.74 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 |
| Imnaha River, OR | S | 45.73 | 117.86 | 82 | 0.00 | 0.00 | 0.65 | 0.00 | 0.00 | 0.34 | 0.01 | 0.00 |
| Wilson River, OR | A | 45.47 | 123.75 | 72 | 0.00 | 0.00 | 0.80 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 |
| L. Deschutes River, OR | A | 45.28 | 121.02 | 76 | 0.00 | 0.00 | 0.80 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 |
| Secesh River, ID | S | 45.23 | 115.81 | 76 | 0.00 | 0.00 | 0.63 | 0.00 | 0.00 | 0.37 | 0.00 | 0.00 |
| Nestucca River, OR | A | 45.12 | 123.40 | 92 | 0.00 | 0.00 | 0.95 | 0.01 | 0.00 | 0.04 | 0.00 | 0.00 |
| Siletz River, OR | S | 44.92 | 124.02 | 64 | 0.00 | 0.00 | 0.94 | 0.02 | 0.00 | 0.05 | 0.00 | 0.00 |
| Siletz River, OR | A | 44.92 | 124.02 | 60 | 0.00 | 0.00 | 0.93 | 0.00 | 0.00 | 0.07 | 0.00 | 0.00 |
| McKenzie River, OR | S | 44.12 | 122.62 | 86 | 0.01 | 0.00 | 0.66 | 0.00 | 0.00 | 0.33 | 0.00 | 0.00 |
| S. Umpqua River, OR | A | 43.67 | 124.20 | 94 | 0.00 | 0.00 | 0.82 | 0.00 | 0.00 | 0.16 | 0.00 | 0.02 |
| N. Umpqua River, OR | S | 43.67 | 124.20 | 88 | 0.00 | 0.00 | 0.78 | 0.00 | 0.00 | 0.22 | 0.00 | 0.00 |
| Rogue River, OR | S | 42.65 | 122.68 | 46 | 0.00 | 0.00 | 0.83 | 0.02 | 0.00 | 0.15 | 0.00 | 0.00 |
| Rogue River, OR | A | 42.42 | 123.45 | 48 | 0.00 | 0.00 | 0.83 | 0.02 | 0.00 | 0.15 | 0.00 | 0.00 |
| Klamath River, CA | A | 41.27 | 123.78 | 90 | 0.00 | 0.00 | 0.97 | 0.00 | 0.00 | 0.03 | 0.00 | 0.00 |
| McCloud River, CA | W | 41.09 | 122.12 | 188 | 0.00 | 0.00 | 0.81 | 0.07 | 0.00 | 0.12 | 0.00 | 0.00 |
| Trinity River, CA | A | 40.72 | 122.80 | 94 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Trinity River, CA | S | 40.72 | 122.80 | 82 | 0.00 | 0.00 | 0.99 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 |
| Sacramento, CA | A | 40.42 | 122.03 | 182 | 0.00 | 0.00 | 0.73 | 0.06 | 0.00 | 0.21 | 0.00 | 0.00 |
| Deer Creek, CA | S | 40.17 | 121.60 | 156 | 0.00 | 0.00 | 0.72 | 0.01 | 0.00 | 0.27 | 0.00 | 0.00 |
| Butte Creek, CA | S | 39.97 | 121.58 | 36 | 0.00 | 0.00 | 0.72 | 0.00 | 0.00 | 0.28 | 0.00 | 0.00 |
| Russian River, CA | A | 38.52 | 122.98 | 64 | 0.00 | 0.00 | 0.84 | 0.02 | 0.00 | 0.14 | 0.00 | 0.00 |
| total (N) | — | — | — | 3462 | 0.0002(1) | 0.002(8) | 0.737(2550) | 0.010(33) | 0.0002(1) | 0.225(779) | 0.0002(1) | 0.026(89) |
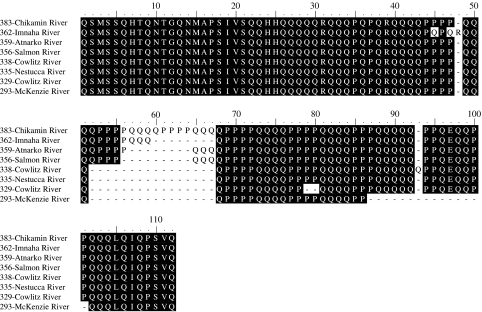
Length variation of the eight OtsClock1b PolyQ alleles (80–110aa) is primarily characterized by the insertion and deletions consisting of both glutamine (Q) and proline (P) repeats (figure 3). Two exceptions include allele 293 bp, which lacks one glutamic acid and allele 362 bp, which contains one additional arginine residue and two unique and non-conservative amino acid changes (P–Q).
Figure 3.
Multiple sequence alignment of the eight OtsClock1b length variants. Allele length (base pairs) and source population is listed for each sequence.
(c) Latitudinal distribution of OtsClock1b PolyQ alleles compared to microsatellites
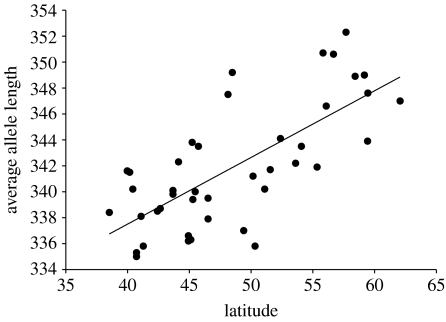
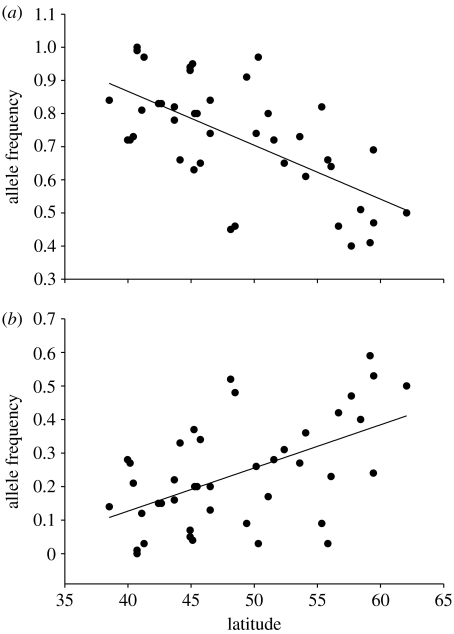
We found a significant association between average allele length of OtsClock1b PolyQ variants and latitude using linear regression (R2=0.516, p<0.0001, regression coefficient=0.514; figure 4). Examination of the two most common OtsClock1b alleles (335 and 359 bp) revealed a significant association between frequency and latitude albeit in the opposite direction. The short 335 bp allele increases in frequency at lower latitudes (R2=0.407, p<0.0001, regression coefficient=−0.016) while the long 359 bp allele increases in frequency at higher latitudes (R2=0.285, p=0.0003, regression coefficient=0.013) (figure 5a,b).
Figure 4.
Plot of latitude in degrees (x-axis) against average allele length. (R2=0.516, p=0.0001; y=316.9+0.51x).
Figure 5.
Plot of latitude in degrees (x-axis) against frequencies of alleles for (a) 335 bp (R2=0.407, p=0.0001; y=1.52+−0.016x) and (b) 359 bp (y-axis) (R2=0.285, p=0.0003; y=−0.39+0.012x).
To test for departure from neutral expectations, we calculated the regression coefficient of allele frequency on latitude for each of the 441 microsatellite alleles and compared these values to those for the two candidate alleles. The regression coefficients for 335 bp (−0.016) and 359 bp (0.012) were exceeded by less than 1 per cent of the microsatellites.
To examine the spatial distribution of OtsClock1b alleles further, we performed partial Mantel tests of association and compared allele frequency (335 and 359 bp), average allele length and pairwise Fst estimates to latitude and longitude. We found a significant positive association between all three measures of genetic distance (allele frequency, average length and Fst estimates) and latitude (constant=longitude; table 4). By contrast, we found no significant association between the three genetic distance measures and longitude (constant=latitude) indicating that the major directional component of the observed clinal patterns is north–south (table 4).
Table 4.
Partial Mantel tests of association between OtsClock1B PolyQ genetic distance and geographical distance. (p<0.05 are shown in italic.)
| genetic matrices | distance matrices | |||
|---|---|---|---|---|
| constant | r | p | ||
| Fst | latitude | longitude | 0.2397 | 0.0005 |
| longitude | latitude | 0.0478 | 0.6315 | |
| allele frequency 335 | latitude | longitude | 0.2879 | 0.0003 |
| longitude | latitude | 0.0188 | 0.8569 | |
| allele frequency 359 | latitude | longitude | 0.1783 | 0.0037 |
| longitude | latitude | 0.1492 | 0.0672 | |
| average allele length | latitude | longitude | 0.4032 | 0.0001 |
| longitude | latitude | −0.0989 | 0.2434 | |
We discovered an analogous pattern of geographical distribution for the two less common length variants (338 and 383 bp). The short 338 bp allele is only present in 12 populations located south of 48° latitude while the long 383 bp allele is present in 9 populations located north of 48° latitude. One exception to this distinct geographical separation occurs in the Umpqua River, where two individuals have a 383 bp allele. In summary, the overall trend in OtsClock1b allelic variation among North American Chinook salmon populations is the prevalence of long OtsClock1b PolyQ alleles in northern populations (359 and 383 bp) and short PolyQ alleles (335 and 338 bp) in southern populations.
(d) Genetic structure based on OtsClock1b PolyQ alleles compared to microsatellites
The overall Fst estimate among the 42 populations based on OtsClock1b was 0.142 while the overall Fst estimate based on microsatellite loci was 0.073. We found no significant association between pairwise Fst estimates based on OtsClock1b allele frequency and pairwise Fst estimates based on microsatellite allele frequencies (Mantel test: r=0.037, p=0.409).
A hierarchical gene diversity analysis of OtsClock1b PolyQ variation showed that genetic variance related to run timing was 0.056, indicating that run timing explained 40.9 per cent of the overall genetic differences among populations. Genetic variance related to differences among populations was 0.065 (47.4%) while the remaining 0.016 (11.7%) resulted from differences among regions. By contrast, a hierarchical gene diversity analysis of 13 microsatellite loci revealed that genetic variance related to run timing was 0.010, indicating that run timing only explained 13.2 per cent of the overall genetic differences among populations. Genetic variance related to differences among populations was 0.057 (75.0%) while the remaining 0.0096 (11.8%) resulted from differences among regions.
4. Discussion
(a) Characterization of duplicated Clock PolyQ domains
The PolyQ region corresponds to the transactivation domain of the CLOCK protein. Studies have demonstrated that length variation in this glutamine-rich region can affect the binding affinity of this transcription factor and thereby alter the circadian phenotype (Darlington et al. 1998). This study is one of a limited number investigating repeat length expansion of the Clock PolyQ domain in natural populations (birds: Johnson et al. 2007; Drosophila: Saleem et al. 2001, Weeks et al. 2006; naked mole rat: Avivi et al. 2001). We identified eight length variants of OtsClock1b PolyQ while the OtsClock1a domain was highly conserved. We previously discovered that a 1200 bp fragment located downstream of the OtsClock1a PolyQ shows a 91 per cent sequence identity to the Atlantic salmon Transferrin gene (O'Malley & Banks 2008). Considering that this fragment is not present in OtsClock1b, it is possible that strong purifying selection may inhibit repeated expansion of the OtsClock1a PolyQ domain.
(b) Latitudinal cline of OtsClock1b maintained by selection
DeBruyne et al. (2006) discovered that Clock plays a central role in the light input pathway of the circadian timing mechanism. We found evidence for a latitudinal cline in average allele length as well as frequency of the two most common alleles; the longer variant increasing in frequency in northern latitudes while the shorter variant increases in frequency in southern latitudes. Comparing the frequency distribution of these two OtsClock1b PolyQ alleles with presumably selectively neutral microsatellite alleles strongly suggests that this latitudinal cline is maintained by selection. We also found an analogous geographical pattern in the frequency distribution of the two less common alleles with the longer variant present primarily in northern populations (greater than 48°) while the shorter variant was only present in southern populations (less than 48°). We hypothesize that the observed clinal variation in OtsClock1b PolyQ alleles could reflect an adaptation to photoperiodic parameters correlated with latitude as the seasonal variation in day length is more pronounced in Alaska compared to California. A recent study by Johnson et al. (2007) detected a similar latitudinal cline in the avian Clock PolyQ domain (longer alleles in north, shorter alleles in south), which the authors propose may be indicative of local adaptation to latitudinal gradients in the seasonal rate of change of photoperiod.
Pittendrigh et al. (1991) first observed that the amplitude of the circadian pacemaker declines as the duration of the entraining light pulse (photoperiod) is increased, ultimately producing a latitudinal cline in nature. More recently, Vitaterna et al. (2006) demonstrated that a mutation in the Clock PolyQ domain modifies the amplitude of the pacemaker which, in turn, can have significant effects on the entrainment behaviour of organisms to light and other resetting stimuli. For Chinook salmon, the duration of the daily photoperiod throughout the breeding season is steadily increased as one moves north. Natural length variation in the OtsClock1b PolyQ may also modify the circadian pacemaker amplitude such that selection for the conservation of pacemaker amplitude during the breeding season may produce the observed latitudinal cline in OtsClock1b allele frequency.
Other ecological factors correlated with latitude may also contribute to the clinal variation in OtsClock1b PolyQ alleles as has been demonstrated for the threonine–glycine-encoding (Thr–Gly) repeat region of the Drosophila circadian rhythm gene, PERIOD (Costa et al. 1991; Castiglione-Morelli et al. 1995; Sawyer et al. 1997; Rosato & Kyriacou 2001; Sawyer et al. 2006). Similar to our discoveries for OtsClock1b, two alleles make up approximately 90 per cent of the natural variation in the Drosophila PERIOD gene, (Thr–Gly)20 and (Thr–Gly)17, with the longer allelic variant predominating in the northern Europe and the shorter variant predominating in the southern region (Costa et al. 1992). This clinal variation of the Drosophila Period gene is thought to be maintained by climate-related selection, as temperature compensation of the circadian clock differs among the Thr–Gly variants (Sawyer et al. 1997). (Thr–Gly)17 variants show a 24 hour cycle at higher temperatures but at a shorter period as temperatures are decreased. By contrast, the (Thr–Gly)20 variants are not sensitive to changes in temperature and show, on average, a period slightly shorter than 24 hours. Therefore, the two major (Thr–Gly) variants appear to be adapted to the thermal environments in which they predominate, (Thr–Gly)17 in southern and (Thr–Gly)20 in northern Europe.
Temperature is a critical environmental variable particularly during the freshwater stages of Chinook salmon life history. Seasonal fluctuations in temperature can create thermal barriers to salmon migrating to freshwater spawning habitats (Richter & Kolmes 2005). Consequently, populations have adapted to long-term average conditions for specific rivers throughout their geographical range. It is plausible that climate-related selection may contribute to the latitudinal cline in OtsClock1b PolyQ variants as has been documented for the Drosophila Period gene. However, the relationship between the Clock gene and temperature has yet to be explored in any model organism.
The primary aim of this study was to investigate whether there was any association between variation in allele frequency and migration timing among Chinook salmon populations along the west coast of North America. Considering the clinal variation in run time diversity, one would predict increasing gene diversity in the southern populations. We, however, found no evidence for this trend. In fact, California's Trinity autumn and Trinity spring populations were essentially fixed for the 335 bp allele (frequency=1.00 and 0.99, respectively).
(c) Contrasting population genetics based on OtsClock1b compared to microsatellites
Pairwise Fst values based on OtsClock1b were not significantly associated with those based on microsatellites indicating that they do not reflect similar patterns of population structure and/or history. Thus, while we cannot rule out the possibility that the spatial distribution of OtsClock1b allelic variants is influenced by historical separations, our findings provide evidence for clinal variation probably maintained by selection.
The hierarchical gene diversity analysis of eight OtsClock1b PolyQ variants revealed that almost half of the overall genetic variance is related to run timing (40.9%) with the remaining variation attributed to differences among regions and populations (11.7 and 47.4%, respectively). These results contrast sharply with those obtained for assumed neutral microsatellite loci, which indicate that run timing explains only 13.2 per cent of the overall genetic variance. Our microsatellite findings are in accord with those reported by Waples et al. (2004) where only 10.2 per cent of the overall genetic variance among 118 Chinook salmon populations was explained by run timing based on allozymes.
We discovered several examples where temporally divergent runs within a river have similar OtsClock1b allele frequencies (i.e. Trinity, Rogue, Umpqua and Siletz Rivers). Therefore, the large percentage of genetic variance attributed to run timing is likely a reflection of the strong latitudinal cline in allele frequency as primarily summer runs inhabit the northern range of this species where average allele length is large. To test this, we excluded the summer run populations and reanalysed data from 20 autumn and spring run populations in Oregon, Washington and California and found that the overall genetic variance related to run timing decreased to 20 per cent while the differences among populations increased to 79 per cent. Thus, other environmental factors or life-history traits correlated with latitude probably confound the analysis.
(d) Summary
Duplicated Clock genes in Chinook salmon show distinct patterns of length variation in the functionally significant PolyQ domain. In contrast to the highly conserved PolyQ of OtsClock1a, we identified eight length variants of the OtsClock1b PolyQ domain. Based on the OtsClock1b PolyQ variation, we found evidence for a latitudinal cline in average allele length and frequency as well as a large component of genetic variance explained by run timing. These results contrast sharply with those obtained for presumed selectively neutral microsatellite markers suggesting that the observed variation may be maintained by selection and reflect an adaptation to ecological factors correlated with latitude such as photoperiod.
Acknowledgments
This research was supported by the California Department of Water Resources and the Mamie M. Markham Research grant. We are grateful to M. Ford, M. Camara and three anonymous reviewers for their comments that considerably improved the manuscript.
References
- Allada R, White N.E, So W.V, Hall J.C, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. doi:10.1016/S0092-8674(00)81440-3 [DOI] [PubMed] [Google Scholar]
- Avivi A, Albrecht U, Oster H, Joel A, Beiles A, Nevo A.M. Biological clock in total darkness: the Clock/MOP3 circadian system of the blind subterranean mole rat. Proc. Natl Acad. Sci. USA. 2001;98:13 751–13 756. doi: 10.1073/pnas.181484498. doi:10.1073/pnas.181484498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beacham T.D, Murray C.B. Influence of photoperiod and temperature on timing of sexual maturity of pink salmon (Oncorhynchus gorbucha) Can. J. Zool. 1988;66:1729–1732. [Google Scholar]
- Belkhir, K. 2000 GENETIX, logiciel sous Windows pour la genetique des populations. Montpelier, France: Laboratoire Genome et Populations, CNRS UPR 9060, Universite de Montpellier II.
- Bradshaw W.E, Holzapfel C.M. Phenotypic evolution and the genetic architecture underlying photoperiodic time measurement. J. Insect Physiol. 2001;47:809–820. doi:10.1016/S0022-1910(01)00054-3 [Google Scholar]
- Castiglione-Morelli M.A, Guantieri V, Villani V, Kyriacou C.P, Costa R, Tamburro A.M. Conformational study of the Thr–Gly repeat in the Drosophila clock protein, PERIOD. Proc. R. Soc. B. 1995;260:155–163. doi: 10.1098/rspb.1995.0073. doi:10.1098/rspb.1995.0073 [DOI] [PubMed] [Google Scholar]
- Costa R, Peixoto A.A, Thackeray J.R, Dalgleish R, Kyriacou C.P. Length polymorphism in the Threonine–Glycine-encoding repeat region of the Period gene in Drosophila. J. Mol. Evol. 1991;32:238–246. doi: 10.1007/BF02342746. doi:10.1007/BF02342746 [DOI] [PubMed] [Google Scholar]
- Costa R, Peixoto A.A, Barbujani G, Kyriacou C.P. A latitudinal cline in a Drosophila clock gene. Proc. R. Soc. B. 1992;250:43–49. doi: 10.1098/rspb.1992.0128. doi:10.1098/rspb.1992.0128 [DOI] [PubMed] [Google Scholar]
- Darlington T.K, Wager-Smith K, Ceriani M.F, Staknis D, Gekakis N, Steeves T.D.L, Weitz C.J, Takahashi J.S, Kay S.A. Closing the circadian loop: clock-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. doi:10.1126/science.280.5369.1599 [DOI] [PubMed] [Google Scholar]
- DeBruyne J.P, Noton E, Lambert C.M, Maywood E.S, Weaver D.R, Reppert S.M. A Clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. doi:10.1016/j.neuron.2006.03.041 [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen H.B, Davis F.C, Wilsbacher L.D, King D.P, Takahashi J.S, Weitz C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. doi:10.1126/science.280.5369.1564 [DOI] [PubMed] [Google Scholar]
- Goldman B.D. Mammalian photoperiodic system: formal properties and neuroendocrine mechanisms of photoperiodic time measurement. J. Biol. Rhythms. 2001;16:283–301. doi: 10.1177/074873001129001980. doi:10.1177/074873001129001980 [DOI] [PubMed] [Google Scholar]
- Hall T. Ibis Therapeutics; Carlsbad, CA: 1997. BioEdit sequence alignment editor. [Google Scholar]
- Johnson A, et al. Avian Clock gene polymorphism: evidence for a latitudinal cline in allele frequencies. Mol. Ecol. 2007;12:4867–4880. doi: 10.1111/j.1365-294X.2007.03552.x. doi:10.1111/j.1365-294X.2007.03552.x [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla V.K, Ahn J.H, Dagenais N, Christensen S.K, Nguyen J.T, Chory J, Harrison M.J, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. doi:10.1126/science.286.5446.1962 [DOI] [PubMed] [Google Scholar]
- King D.P, et al. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. doi:10.1016/S0092-8674(00)80245-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. O. & Zaykin D. 2001 Genetic data analysis: computer program for the analysis of allelic data V. 1.0 (d16c). See http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php
- Lowrey P.L, Takahashi J.S. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genom. Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. doi:10.1146/annurev.genom.5.061903.175925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley K.G, Banks M.A. Duplicated Clock genes with unique polyglutamine domains provide evidence for nonhomologous recombination in Chinook salmon (Oncorhynchus tshawytscha) Genetica. 2008;132:87–94. doi: 10.1007/s10709-007-9151-8. doi:10.1007/s10709-007-9151-8 [DOI] [PubMed] [Google Scholar]
- O'Malley K.G, Camara M.D, Banks M.A. Candidate loci reveal genetic differentiation between temporally divergent migratory runs of Chinook salmon (Oncorhynchus tshawytscha) Mol. Ecol. 2007;16:4930–4941. doi: 10.1111/j.1365-294X.2007.03565.x. doi:10.1111/j.1365-294X.2007.03565.x [DOI] [PubMed] [Google Scholar]
- Pittendrigh C.S. Circadian organization and the photoperiodic phenomena. In: Follet B.K, Follet D.E, editors. Biological clocks in seasonal reproductive cycles. John Wright; Bristol, UK: 1981. pp. 1–35. [Google Scholar]
- Pittendrigh C.S, Kyner W.T, Takamura T. The amplitude of circadian oscillations: temperature dependence, latitudinal clines, and the photoperiodic time measurement. J. Biol. Rhythms. 1991;6:299–313. doi: 10.1177/074873049100600402. doi:10.1177/074873049100600402 [DOI] [PubMed] [Google Scholar]
- Quinn T.P, Adams D.J. Environmental changes affecting the migratory timing of American shad and sockeye salmon. Ecology. 1996;77:1151–1162. doi:10.2307/2265584 [Google Scholar]
- Quinn T.P, Unwin M.J, Kinnison M.T. Evolution of temporal isolation in the wild: genetic divergence in timing of migration and breeding in introduced Chinook salmon populations. Evolution. 2000;54:1372–1385. doi: 10.1111/j.0014-3820.2000.tb00569.x. doi:10.1554/0014-3820(2000)057[1372:EOTIIT]2.0.Co;2 [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. doi:10.2307/2410454 [DOI] [PubMed] [Google Scholar]
- Reppert S.M, Weaver D.J. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. doi:10.1038/nature00965 [DOI] [PubMed] [Google Scholar]
- Richter A, Kolmes S.A. Maximum temperature limits for Chinook, coho, and chum salmon, and steelhead trout in the Pacific Northwest. Rev. Fish. Sci. 2005;13:23–49. doi:10.1080/10641260590885861 [Google Scholar]
- Rosato E, Kyriacou C.P. Flies, clocks and evolution. Phil. Trans. R. Soc. B. 2001;356:1769–1778. doi: 10.1098/rstb.2001.0961. doi:10.1098/rstb.2001.0961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. S. 2001 Passage Pattern analysis, spatial statistics, and geographic exegesis V. 1.0. Tempe, AZ: Department of Biology, Arizona State University.
- Saleem Q, Anand A, Jain S, Brahmachari S.K. The polyglutamine motif is highly conserved at the Clock locus in various organisms and is not polymorphic in humans. Hum. Genet. 2001;109:136–142. doi: 10.1007/s004390100550. doi:10.1007/s004390100550 [DOI] [PubMed] [Google Scholar]
- Sawyer L.A, Hennessy J.M, Peixoto A.A, Rosato E, Parkinson H, Costa R, Kyriacou C.P. Natural variation in a Drosophila clock gene and temperature compensation. Science. 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. doi:10.1126/science.278.5346.2117 [DOI] [PubMed] [Google Scholar]
- Sawyer L.A, Sandrelli F, Pasetto C, Peixoto A.A, Rosato E, Costa R, Kyriacou C.P. The Period gene Thr–Gly polymorphism in Australian and African Drosophila melanogaster populations: implications for selection. Genetics. 2006;174:465–480. doi: 10.1534/genetics.106.058792. doi:10.1534/genetics.106.058792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeb L.W, et al. Development of a standardized DNA database for Chinook salmon. Fisheries. 2007;32:540–552. doi:10.1577/1548-8446(2007)32[540:DOASDD]2.0.CO;2 [Google Scholar]
- Tauber E, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–1897. doi: 10.1126/science.1138412. doi:10.1126/science.1138412 [DOI] [PubMed] [Google Scholar]
- Thompson J.D, Higgins D.G, Gibson T.J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz Nunes M, Saunders D. Photoperiodic time measurement in insects: a review of clock models. J. Biol. Rhythms. 1999;14:84–104. doi: 10.1177/074873049901400202. doi:10.1177/074873099129000470 [DOI] [PubMed] [Google Scholar]
- Vitaterna M.H, Ko C.H, Chang A, Buhr E.D, Fruechte E.M, Schook A, Antoch M.P, Turek F.W, Takahashi J.S. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase–response curve amplitude. Proc. Natl Acad. Sci. USA. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. doi:10.1073/pnas.0603601103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R.S, Teel D.J, Meyers J.M, Marshall A.R. Life-history divergence in Chinook salmon: historic contingency and parallel evolution. Evolution. 2004;58:386–403. doi:10.1554/03-323 [PubMed] [Google Scholar]
- Weeks A.R, McKechnie S.W, Hoffmann A.A. In search of clinal variation in the period and clock timing genes in Australian Drosophila melanogaster populations. J. Evol. Biol. 2006;19:551–557. doi: 10.1111/j.1420-9101.2005.01013.x. doi:10.1111/j.1420-9101.2005.01013.x [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes N.S, Strahle U, Sassone-Corsi P. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat. Neurosci. 1998;1:701–707. doi: 10.1038/3703. doi:10.1038/3703 [DOI] [PubMed] [Google Scholar]
- Yano M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. doi:10.1105/tpc.12.12.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]