Abstract
The Giardia lamblia cyst wall is required for survival outside the host and infection. Three cyst wall protein (cwp) genes identified to date are highly up-regulated during encystation. However, little is known of the molecular mechanisms governing their gene regulation. Messenger RNAs containing premature stop codons are rapidly degraded by a nonsense-mediated mRNA decay (NMD) system to avoid production of non-functional proteins. In addition to RNA surveillance, NMD also regulates thousands of naturally occurring transcripts through a variety of mechanisms. It is interesting to know the NMD pathway in the primitive eukaryotes. Previously, we have found that the giardial homologue of a conserved NMD factor, UPF1, may be functionally conserved and involved in NMD and in preventing nonsense suppression. In this study, we tested the hypothesis that NMD factors can regulate some naturally occurring transcripts in G. lamblia. We found that overexpression of UPF1 resulted in a significant decrease of the levels of CWP1 and cyst formation and of the endogenous cwp1-3, and myb2 mRNA levels and stability. This indicates that NMD could contribute to the regulation of the cwp1-3 and myb2 transcripts, which are key to G. lamblia differentiation into cyst. Interestingly, we also found that UPF1 may be involved in regulation of eight other endogenous genes, including up-regulation of the translation elongation factor gene, whose product increases translation which is required for NMD. Our results indicate that NMD factor could contribute to the regulation of not only nonsense containing mRNAs, but also mRNAs of the key encystation-induced genes and other endogenous genes in the early-diverging eukaryote, G. lamblia.
Introduction
Giardia lamblia is a major cause of outbreaks of waterborne diarrheal disease worldwide, which contributes greatly to malnutrition and malabsorption in children [1]–[3]. Like Entamoeba histolytica and other intestinal protozoan parasites, G. lamblia undergoes differentiation from a pathogenic trophozoite form into a resistant infectious cyst form [3], [4]. Cyst can survive in the hostile environment and infect a new host because they have a resistant extracellular wall.
Because of the importance of the cyst wall, a lot of researches are focusing on identifying and understanding its key components, cyst wall proteins (CWPs) [5]–[7]. Interestingly, three cwp genes identified to date are highly up-regulated during encystation [5]–[7]. There is little understanding of the molecular mechanisms governing their transcriptional regulation. A Myb family transcription factor (Myb2) is encystation-induced and is involved in coordinating upregulation of the cwp1-3 genes [8]. Two GARP family transcription factors may be involved in transcriptional regulation of many different genes including the encystation-induced cwp1 gene and constitutive ran gene [9]. An ARID family transcription factor can bind to specific AT-rich Inr sequences and function as an important transactivator in the regulation of the cwp1 gene [10]. However, little is known about regulation of mRNA stability of the cwp genes during giardial growth and differentiation.
In late-branching eukaryotes, either a frameshift or a nonsense mutation often leads to rapid degradation of the gene's mRNA by a nonsense-mediated mRNA decay (NMD) pathway [11]–[13]. This surveillance system protects cells from the production of non-functional proteins by eliminating mutant mRNAs. The NMD pathway is present in yeast, plants, Caenorhabditis elegans and mammals [11]–[13]. NMD factors such as up-frameshift 1 (UPF1), UPF2 and UPF3 have been identified in yeast, C. elegans, mice and humans [14]–[19]. They have been shown to interact together and form a complex [20], [21]. Mutations in upf genes stabilize mRNAs with nonsense mutations [22], [23]. UPF1 is one of the most conserved NMD factors [11]–[13]. NMD is a translation-dependent event since its mechanism depends on the recognition of the nonsense mutations by the translational machinery [13]. Studies have shown that NMD factors including UPF1 enhance translation termination at a nonsense codon through interaction with the termination release factors [20], [24]–[26].
In addition to RNA surveillance, NMD factors also function in regulating the abundance of some naturally occurring mRNAs [22], [27]. Hundreds or thousands of wild-type NMD targets have been identified in yeast and humans [28], [29]. Most of them are up-regulated, some of them are down-regulated in upf null mutant [29].
G. lamblia is of biological interest in understanding the progress of eukaryotic evolution [30]–[33]. It has fewer cellular components for DNA synthesis, transcription and RNA processing, possibly due to their divergence or their functional redundancy with other proteins in some pathways [33]. Blast searches of the G. lamblia genome databases identified matches for UPF1, which is the most conserved NMD factor but no matches for UPF2 and UPF3 and some other NMD factors [34]. Interestingly, our previous results showed that: i) the levels of the nonsense transcripts were lower in G. lamblia; ii) the aminoglycoside G418 had an inhibiting effect on NMD in G. lamblia, similar to the effect of aminoglycosides on inhibiting NMD in late-branching eukaryotes; iii) Giardial UPF1 functioned in reducing the levels of nonsense-containing transcripts and in enhancing fidelity of translation termination [34]. Therefore, the NMD phenomenon could be present in G. lamblia. However, G. lamblia does not have some of the components of the NMD pathway and the reduction levels of the nonsense transcripts observed in G. lamblia are lower than those in late-branching eukaryotes, suggesting that the NMD system in G. lamblia may be less-functional.
Previously, we have found that the expression of the upf1 gene was induced in cells expressing the luciferase gene with a nonsense mutation [34]. In this study, we found that the nonsense mutation triggered a decrease in cwp1 and cwp2 mRNA levels and there was a reverse correlation between the expression levels of the cwp1/2 and upf1 mRNA. We also found that overexpression of UPF1 reduced the levels of CWP1 and cyst formation and reduced the mRNA levels and stability of the cwp1, cwp2, cwp3, and myb2 genes. In addition, we found that the expression of other five genes was increased and that of other three genes was decreased by the UPF1 overexpression. For example, the translation elongation factor mRNA was increased by the UPF1 overexpression. This could be a new example of an NMD target whose product increases translation which is required for NMD. Our findings provide new insights into regulation of the giardial cyst wall protein genes and other endogenous genes.
Results
NMD can be monitored by a constitutive promoter system
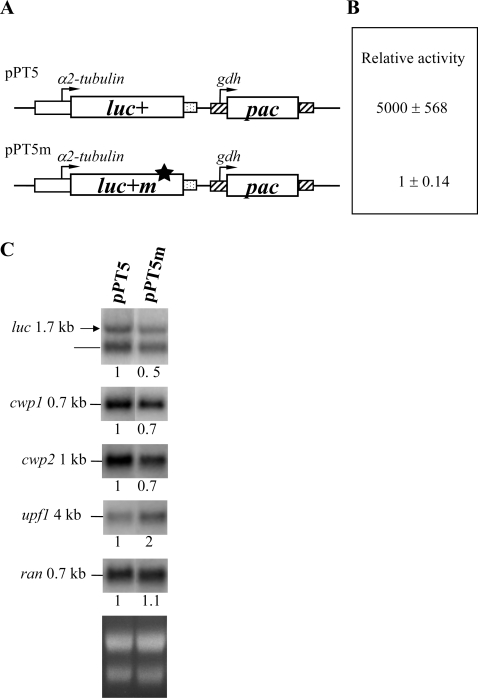
NMD is related to the presence of a premature stop codon or not [11]–[13]. We have tested NMD effect using a luciferase gene with or without a premature stop codon under the control of the encystation-induced cwp1 promoter [34]. We found that NMD could be present in G. lamblia because the results showed that the mRNA produced from the luciferase gene with a stop codon under the control of the cwp1 promoter was decayed compared with wild type luciferase mRNA [34]. We further used a constitutive promoter to test the NMD. We prepared puromycin-based constructs pPT5 and pPT5m in which the wild type luciferase gene (luc+) and the luciferase gene with a stop codon (luc+m) are controlled by the α2-tubulin promoter, respectively (Fig. 1A), and stably transfected them into G. lamblia. The level of luc+m activity in the pPT5m cell line was reduced by approximately 5,000-fold, relative to that of the wild-type luc+ in the pPT5 cell line (Fig. 1B), indicating that luc+m could be non-functional. This result is similar to the data we previously reported for the luc+m under the control of the cwp1 promoter [34]. The level of luciferase mRNA was lower by ∼50% (P<0.05) in the pPT5m transfectants compared with that in the pPT5 transfectants (Fig. 1C, lanes 1 and 2), indicating that the presence of a nonsense mutation in luc+m triggered a decrease in mRNA levels (NMD). Therefore, NMD can be monitored by a constitutive promoter system. The results from the α2-tubulin and the cwp1 promoters [34] similarly indicate that NMD could be present in G. lamblia. As a control, the ran mRNA levels did not change in the pPT5m transfectants compared with the pPT5 transfectants (Fig. 1C, lanes 1 and 2).
Figure 1. Down-regulation of the cwp genes in cells expressing the luciferase gene with a nonsense mutation.
(A) Diagrams of the pPT5 and pPT5m plasmids. The pac gene (open box) is under the control of the 5′- and 3′-flanking regions of the gdh gene (striated box). The luciferase reporter gene (luc+, open box) is under the control of the 5′-flanking region of the α2-tubulin (open box) and the 3′-flanking region of the ran gene (dotted box), respectively. The arrows show the directions of gene transcription. Plasmids pPT5 and pPT5m encode a wild-type luciferase gene (luc+) with 550 amino acids and a luciferase mutant (luc+m) in which Leu 411 was mutated to a termination codon (marked by a star) and Asp 500 was mutated to Asn, respectively. (B) Nonsense mutation leads to a decrease of luciferase activity. After stable transfection with the pPT5 and pPT5m constructs, luciferase activity was measured in vegetative cells. The activity of pPT5 transfectants relative to pPT5m transfectants is presented. Values are shown as mean±standard error. (C) Down-regulation of the luciferase gene with a nonsense mutation. Total RNA blots made from vegetative transfectants were hybridized with specific gene probes as indicated (upper panels). Ribosomal RNA loading controls are in the bottom panel. Representative results are shown. Two different luciferase transcripts, a 1.7 kb full-length transcript (shown by an arrow) and a 1.2 kb transcript (shown by a line), were detected in the pPT5 and pPT5m transfectants. The numbers show the relative activity, which reflects expression relative to that in controls.
Reverse correlation between the expression level of the cwp1/2 and upf1 mRNA
Previously we found that the nonsense mutation triggered an increase in upf1 mRNA levels [34]. In this study, we also found that the levels of the upf1 mRNA increased by ∼2-fold (P<0.05) in the pPT5m cell line compared with the pPT5 cell line (Fig. 1C). Interestingly, the levels of the cwp1 and cwp2 mRNA were lower by ∼30% (P<0.05) in the pPT5m cell line compared with the pPT5 cell line (Fig. 1C). Similarly decreased cwp1 or cwp2 mRNA levels were also detected in the pPW1m cell line that expressed the luciferase gene with a premature stop codon under the control of the cwp1 promoter (data not shown) [34].
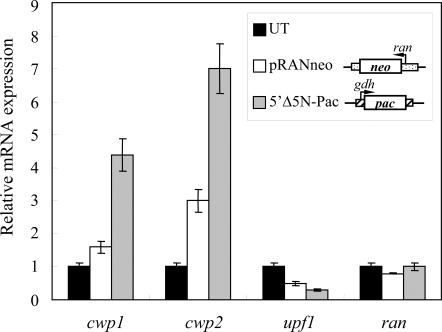
Previously we found that the levels of the upf1 mRNA decreased with increasing levels of the cwp1 and cwp2 mRNA during encystation [34]. We also demonstrated that stable transfection systems can increase the levels of the cwp1, cwp2 and cwp3 transcripts during vegetative growth [35]. We further asked whether stable transfection influenced the upf1 gene expression. We found that the level of the upf1 mRNA decreased by ∼50–70% (P<0.05) in cells stably transfected with the pRANneo or 5′Δ5N-Pac, which contain the neomycin or puromycin selective marker, relative to untransfected cells (Fig. 2). As reported previously, the levels of the cwp1/2 mRNA increased significantly and the levels of the ran and ribosomal mRNA did not change significantly in the transfected cell lines (Fig. 2) [35].
Figure 2. Reverse correlation between the expression levels of the cwp1/2 and upf1 mRNA in stable cell lines.
Diagrams of the pRANneo and 5′Δ5N-Pac plasmids for stable transfection are shown in inset. The neo or pac gene (open box) is under the control of the 5′- and 3′-flanking regions of the ran (dotted box) or gdh gene (striated box). Total RNA blots made from vegetative untransfected cells (UT), pRANneo or 5′Δ5N-Pac transfectants were hybridized with specific probes as indicated. Hybridization signals were imaged and quantified as indicated in Materials and Methods. The results are expressed as relative expression level over untransfected control. Values are shown as mean±standard error.
Overexpression of UPF1 reduced the levels of cyst wall protein 1 and cyst formation
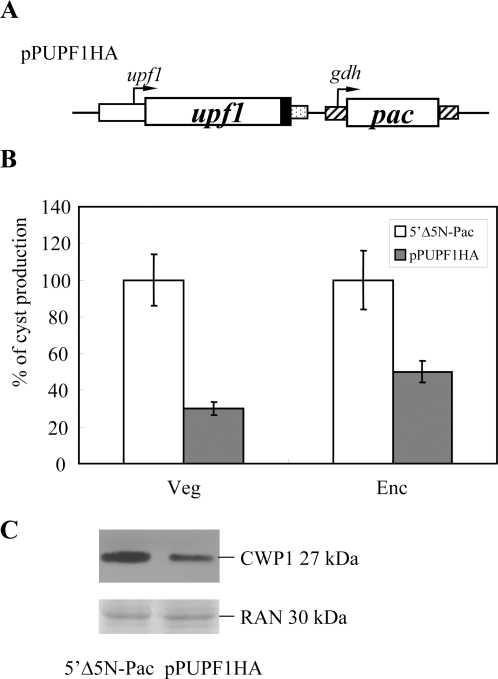
Because of the reverse correlation between the levels of the cwp1/2 and upf1 mRNAs as described above, we further investigated the effect of giardial UPF1 on cyst formation. We stably transfected the UPF1 expression plasmid pPUPF1HA into G. lamblia (Fig. 3A). The UPF1-HA protein was expressed in the stable transfectants as detected in immunofluorescence assays [34]. No significant change in growth rate was observed in the pPUPF1HA cell line (data not shown). We found that the cyst number in the vegetative or encysting pPUPF1HA cell line decreased by ∼30% or ∼50% (P<0.05) relative to the control cell line which only carries the pac gene (5′Δ5N-Pac) (Fig. 3B). We further asked whether the levels of cyst wall protein 1 decreased with the decrease of the cyst formation. As shown by Western blot analysis, UPF1 overexpression also resulted in a reduction of the levels of CWP1 protein (Fig. 3C). As a control, similar levels of intensity of the giardial RAN protein (∼30 kDa) were detected by anti-human RAN antibody (Fig. 3C). The results suggest that UPF1 may function in reducing the level of CWP1 and cyst formation.
Figure 3. Overexpression of UPF1 reduced the levels of CWP1 and cyst formation.
(A) Diagrams of the pPUPF1HA plasmid. The pac gene (open box) expression cassette is the same as in Fig. 2. The upf1 gene is under the control of its own 5′-flanking region (open box) and the 3′-flanking region of the ran gene (dotted box). The filled black box indicates the coding sequence of the HA epitope tag. (B) Overexpression of UPF1 reduced the levels of cyst formation. The 5′Δ5N-Pac and pPUPF1HA stable transfectants were cultured in growth medium to late log/early stationary phase (Veg). Cyst count was performed on these late log/early stationary phase cultures (1.5×106 cells/ml). In another study, the 5′Δ5N-Pac and pPUPF1HA stable transfectants were cultured in encystation medium for 24 h and then subjected to cyst count (Enc). The sum of total cysts is expressed as relative expression level over control. Values are shown as mean±standard error. (C) Overexpression of UPF1 reduced the CWP1 level. The 5′Δ5N-Pac and pPUPF1HA stable transfectants were cultured in encystation medium for 24 h and then subjected to SDS-PAGE and Western blot. The blot was probed by anti-CWP1 antibody. Equal amounts of proteins loaded were confirmed by detection of giardial RAN protein. Representative results are shown.
Overexpression of UPF1 decreased cwp1-3 and myb2 mRNA stability
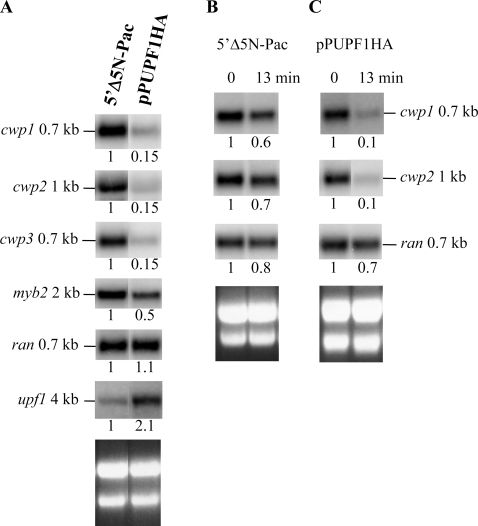
We further investigated whether UPF1 overexpression can influence the expression of cwp and other endogenous genes. We found that the levels of native cwp1, cwp2, cwp3, or myb2 mRNA in the vegetative pPUPF1HA cell line decreased by ∼85% or ∼50% (P<0.05) relative to the control cell line which only carries the pac gene (Fig. 4A). The ran mRNA levels did not change significantly in the pPUPF1HA cell line (Fig. 4A). The upf1 mRNA levels increased significantly (P<0.05) in the pPUPF1HA cell line (Fig. 4A).
Figure 4. Effect of UPF1 overexpression on cwp gene expression.
(A) Overexpression of UPF1 decreased the cwp1-3 and myb2 mRNA levels. Total RNA was harvested from vegetative 5′Δ5N-Pac and pPUPF1HA transfectants. Northern blots were hybridized with specific probes as indicated (upper panels). (B)(C) Overexpression of UPF1 decreased the cwp1 and cwp2 mRNA stability. Total RNA was harvested from either 5′Δ5N-Pac (B) or pPUPF1HA (C) transfectants during vegetative growth. The cells were treated without (0 min) or with 45 µg/ml actinomycin D for 13 min to arrest mRNA synthesis. Northern blot were hybridized with specific gene probes as indicated (upper panels). Ribosomal RNA loading controls are in the bottom panels. Representative results are shown. The numbers show the relative activity, which reflects expression relative to that in controls. The cwp1 and cwp2 signals from Fig. 4B and C were a long exposure to show the difference in the AcD treated and untreated samples.
We further investigated whether the decrease of cwp mRNA levels in the pPUPF1HA transfectants was due to the decrease of mRNA stability. We found that treatment of the pPUPF1HA transfectants with 45 µg/ml of actinomycin D for 13 min decreased mRNA levels of the cwp1 and cwp2 genes by ∼90% (P<0.05) (Fig. 4C). As a control, treatment of the control cell line with actinomycin D for 13 min decreased mRNA levels of the cwp1 and cwp2 genes by 30–40% (P<0.05) (Fig. 4B). Therefore, the cwp1 and cwp2 mRNAs exhibited a half-life of <13 min when the upf1 was overexpressed, while these mRNAs exhibited a half-life of >13 min in the control cell line. The cwp3 and myb2 mRNA stability also decreased significantly in the pPUPF1HA transfectants as compared with that in the control cell line (data not shown). The ran mRNA stability did not change significantly in the pPUPF1HA transfectants as compared with that in the control cell line (Fig. 4C). The results indicate that UPF1 can decrease cwp1-3 and myb2 mRNA stability.
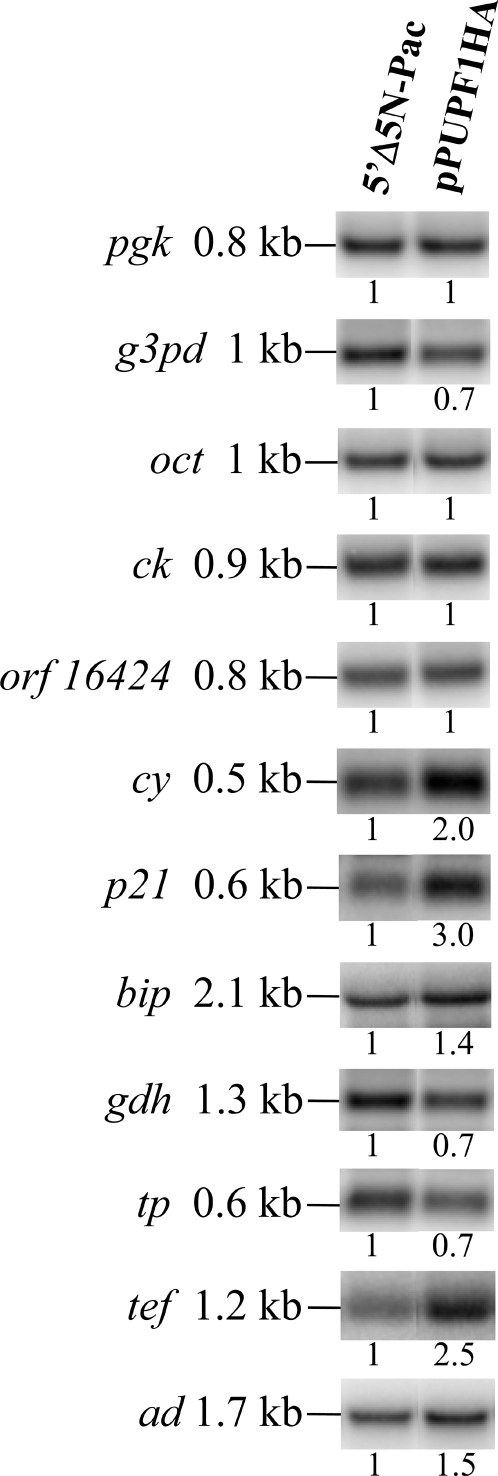
Overexpression of UPF1 changed transcript levels of other endogenous genes
In the previous studies, we have found that the expression of the cwp1, cwp2, myb2 genes was upregulated in stable cell line with drug selection and that the expression of other eight genes was also upregulated [35]. They encodes enzymes involved in anaerobic glycolysis, phosphoglycerate kinase (PGK) and glyceraldehyde-3-phosphate dehydrogenase (G3PD), enzymes for arginine hydrolysis, ornithine carbamoyltransferase (OCT) and carbamate kinase (CK), enzymes involved in protein folding, cyclophilin (CY), co-chaperone-like protein p21, and Bip, and open reading frame (ORF) 16424 with unknown function [1], [35], [36]. We wished to understand the importance of UPF1 for expression of these genes. To achieve this goal, we compared expression of these genes in the UPF1 overexpressed cell line and the control cell line. Of these eight genes, three were upregulated by UPF1 overexpression, including cy, p21, and bip (P<0.05) (Fig. 5). Interestingly, one gene was downregulated by UPF1 overexpression (g3pd, P<0.05) (Fig. 5). The transcript levels of the other four genes, pgk, oct, ck, and orf 16424, were not changed significantly by UPF1 overexpression (Fig. 5). We also found that two other genes with no change in mRNA levels in the stable cell lines, glutamate dehydrogenase (gdh), and thioredoxin peroxidase (tp), were downregulated by UPF1 overexpression (P<0.05) (Fig. 5). In addition, we found two newly identified genes, translation elongation factor (tef, γ subunit of translation elongation factor 1B) [37] and arginine deiminase (ad) [1], were upregulated by UPF1 overexpression (P<0.05) (Fig. 5).
Figure 5. Effect of UPF1 overexpression on endogenous gene expression.
Total RNA blots made from the 5′Δ5N-Pac and pPUPF1HA transfectants were hybridized with specific gene probes as described. Equal RNA loading was confirmed by ethidium bromide staining of ribosomal RNA (data not shown). Representative results are shown. The numbers show the relative activity, which reflects expression relative to that in controls.
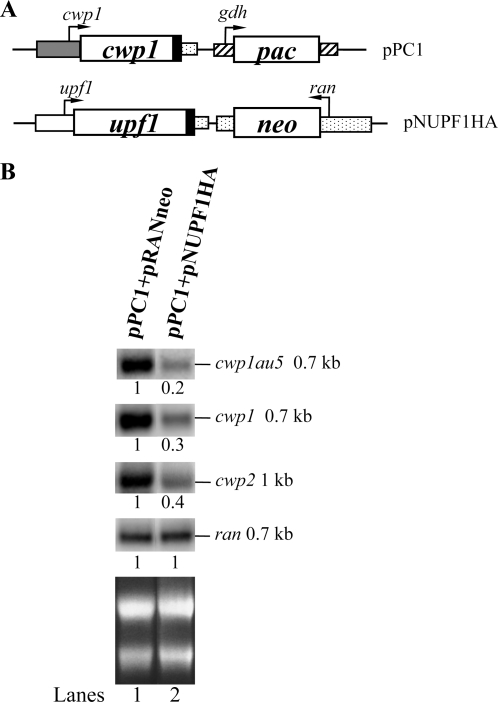
Overexpression of UPF1 decreased mRNA levels of vector-expressed cwp1 gene
We next tested whether overexpression of UPF1 influenced vector-expressed cwp1 gene. We stably transfected the UPF1 expression plasmid pNUPF1HA (Fig. 6A) together with construct pPC1 in which the cwp1 gene is controlled by its own promoter and contains an AU5 epitope tag at their C terminus (Fig. 6A). The CWP1-AU5 protein was expressed in the stable transfectants as detected in immunofluorescence assays and Western blot analysis (data not shown). Northern blot analysis showed that the levels of the cwp1-au5 mRNA decreased by ∼80% (P<0.05) in the pPC1+pNUPF1HA co-transfectants relative to the pPC1+pRANneo control cell line during vegetative growth (Fig. 6B). The levels of the cwp1 (including endogenous cwp1 and vector-derived cwp1-au5) and cwp2 mRNA also decreased by ∼60–70% (P<0.05) in the UPF1 overexpressed cell line, pPC1+pNUPF1HA (Fig. 6B). The results indicate that overexpression of UPF1 not only can decrease the expression of the endogenous cwp genes but also can decrease the expression of the vector-expressed cwp1 gene.
Figure 6. Overexpression of UPF1 decreased mRNA levels of vector-based cwp1 gene.
(A) Diagrams of the pPC1 and pNUPF1HA plasmids. The neo or pac gene (open box) expression cassette is the same as in Fig. 2. In pPC1, the cwp1 gene (open boxes) is flanked by its own 5′-flanking region (gray box) and 3′-flanking region of the ran gene (dotted box) and the filled black box indicates the coding sequence of the AU5 epitope tag. In pNUPF1HA, the upf1 gene is under the control of its own promoter (open box) and the 3′-flanking of the ran gene (dotted box) and the filled black box indicates the coding sequence of the HA epitope tag. (B) Northern blot analysis of au5 tagged cwp1 transcripts in vegetative cells (upper panel). Total RNA blots made from pPC1+pRANneo or pPC1+pNUPF1HA transfectants were hybridized with the au5 specific probe (au5hyb), and specific gene probes as indicated (upper panels). Ribosomal RNA loading controls are in the bottom panel. Representative results are shown. The numbers show the relative activity, which reflects expression relative to that in controls.
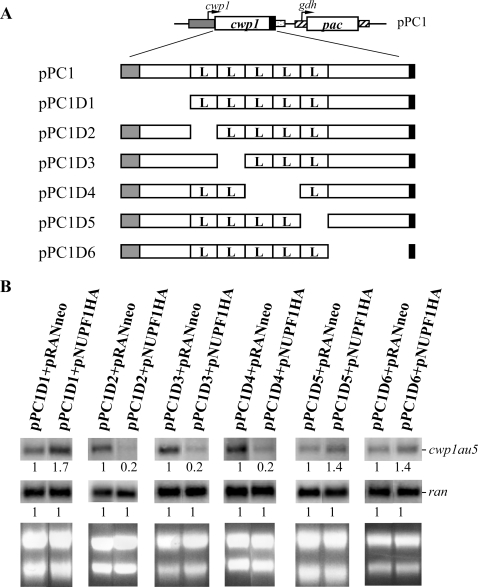
Mapping of the region in the cwp1 gene needed for the UPF1 dependent decay
We further determined the region within the cwp1 mRNA responsible for the UPF1 dependent decay by constructing a series of deletions (Fig. 7A). Deletion of the sequence encoding the first, second or third to fourth LRRs (nucleotides 193–264, construct D2; nucleotides 265–345, construct D3; nucleotides 346–474, construct D4, Fig. 7A) still resulted in a significant decrease of cwp1-au5 mRNA in the pNUPF1HA co-transfectants relative to the control cell line (Fig. 7B). However, deletion of the sequence encoding the region N terminal to the LRRs (nucleotides 4–192, construct D1, Fig. 7A), the fifth LRR (nucleotides 475–552, construct D5, Fig. 7A) or the region C terminal to the LRRs (nucleotides 553–723, construct D6, Fig. 7A) resulted in an increase of the levels of the cwp1-au5 mRNA to ∼1.4 or 1.7-fold (P<0.05) in the pNUPF1HA co-transfectants relative to the control cell line (Fig. 7B). The results indicate that 5′ (nucleotides 4–192) or 3′ (nucleotides 475–723) sequence of the cwp1 gene may contain the sequence responsible for the UPF1 dependent decay.
Figure 7. Mapping of the region in the cwp1 gene needed for the UPF1 dependent decay.
(A) Diagrams of the pPC1 and plasmids for CWP1 deletion mapping. The pac or cwp1 gene (open box) expression cassette is the same as in Fig. 2 and Fig. 6, respectively. The predicted signal peptide is in gray. The LRRs are indicated as open boxes labeled “L”. (B). Analysis of au5 tagged cwp1 transcripts in vegetative cells. The pPC1D1-6+pRANneo and pPC1D1-6+pNUPF1HA transfectants were cultured in encystation medium for 24 h and then subjected to Northern blot analysis. Total RNA blots were hybridized with the au5 specific probe (au5hyb), and ran gene probe (upper panels). Ribosomal RNA loading controls are in the bottom panel. Representative results are shown. The numbers show the relative activity, which reflects expression relative to that in controls.
Discussion
In addition to RNA surveillance, NMD factors also function in regulating the abundance of some naturally occurring mRNAs [23], [28], [38]. Hundreds or thousands of wild-type NMD targets that may be up-regulated or down-regulated by NMD factors have been identified in yeast and humans [28], [29], [39]. Their changes in mRNA abundance may be correlated (direct targets of NMD) or not correlated (indirect targets of NMD) with changes in the mRNA stability [28], [39]. For example, abundance of one subset of wild type mRNAs increased in upf1 null mutant, including PPR1, URA1, URA3, and URA4 mRNAs [28], [40]. However, only PPR1 is the direct target of NMD because only PPR1 mRNA stability increased in upf1 null mutant [28], [40]. PPR1 is a positive transcriptional activator for these URA genes. Altered half-life of this regulatory protein could indirectly influence the abundance of the mRNAs of the downstream targets (URA genes) [28], [40]. An UPF1 dependent destabilizing element (UDE) was mapped to a region located within the 5′-untranslated region and the first 92 bases of the PPR1 coding region [40]. Similarly, one of the UDEs was mapped to a region located within the 5′ (nucleotides 4–192) sequence of the cwp1 gene.
In G. lamblia, we identified the four encystation-induced genes, cwp1, cwp2, cwp3, and myb2, as wild-type targets of NMD. These four genes may be the direct targets of NMD because overexpression of UPF1 led to decreased levels of their mRNA stability. In addition, UDEs were mapped to a region located within the 5′ (nucleotides 4–192) or 3′ (nucleotides 475–723) sequence of the cwp1 gene. It is possible that the cwp2, cwp3 and myb2 genes also have UDEs and that their mRNAs are targeted by UPF1-dependent decay. UPF1 may coordinately down-regulate the encystation-induced cwp1-3 and myb2 genes during vegetative growth. Because Myb2 acts as a positive transcriptional activator for these cwp genes [8], a decrease of the level of the myb2 transcripts that may lead to a decrease of the levels of the Myb2 protein and then results in a decrease of the cwp transcripts. In addition, we have shown that the levels of UPF1 decreased significantly during encystation [34]. During encystation, the up-regulation of these encystation-induced genes may be correlated with the presence of the lower level of UPF1 protein. Interestingly, our results show that the levels of the cwp1 and cwp2 mRNAs were lower in the cells containing luciferase nonsense transcripts. This could be due to the down-regulation of the cwp genes by the presence of the higher level of UPF1 protein upon NMD induction.
It is interesting to know how NMD factors recognize their targets. In yeast, it has been thought that NMD promotes rapid decay of the nonsense-containing mRNA through interaction of a RNA-binding protein(s) with specific RNA elements [12]. Heterogeneous nuclear ribonucleoprotein 1 (HRP1) may be a marker protein that binds to the downstream element of the nonsense mutation and interacts with NMD factors [12]. On the other hand, wild-type mRNAs without premature stop codon were also regulated by a UPF1-dependent mechanism [38], [41]. Four natural targets for an RNA-binding protein, Stau1, were identified in humans, including ADP ribosylation factor 1, c-JUN, SERPINE1, IL7R, and GAP43 mRNAs. Stau1 can bind to the 3′-untranslated region of these targets' mRNAs for Stau1-mediated mRNA decay, which depends on translation and recruitment of the NMD factor UPF1 [38], [41]. Short untranslated regions are typical of giardial transcripts [3]. Interestingly, one of the UDEs was mapped to a region located within the 3′ (nucleotides 475–723) sequence of the cwp1 gene. It is possible that G. lamblia may have similar RNA binding proteins as marker proteins for abnormal or natural NMD targets. Blast searches of the G. lamblia genome databases identified a match for HRP1, which has five RNA binding domains [34]. However, it is very different from the yeast HRP1, which has two RNA binding domains. Further studies will be required to identify the RNA binding proteins and elucidate their roles in NMD.
In addition to encystation-induced genes, we also found that UPF1 may be involved in regulating transcripts of many different genes. Overexpression of UPF1 led to enhanced levels of five of twelve genes, including cy, p21, bip, tef and ad. Three genes were downregulated, including g3pd, gdh, and tp. The other four genes tested were not changed, including pgk, oct, ck, and orf 16424. The affected genes encode proteins involved in protein translation (TEF) [37], protein folding (CY, p21, and Bip) [35], cell metabolism (AD, G3PD, GDH, and TP) [1], [36]. They may be direct or indirect targets of NMD and this requires further investigation.
NMD may increase translation accuracy because NMD factors including UPF1-3 could enhance translation termination at a nonsense codon through interaction with the termination release factors in yeast [20], [24]–[26]. NMD requires translation since its mechanism depends on the recognition of the nonsense mutations by the translational machinery [13]. NMD factors including UPF1-3 also function in stimulating translation in human [42]. In addition, NMD factors including UPF1-3 may increase translation through down-regulation of Ebs1p, which is a global inhibitor of translation in yeast [43]. This occurs without a change in the EBS1 mRNA stability, indicating that EBS1 is an indirect target of NMD and it may be targeted by NMD-regulated transcription factors. In this study, we also found that UPF1 may be involved in upregulating transcripts of translation elongation factor (tef, γ subunit of translation elongation factor 1B) [37]. This suggests that the giardial NMD factors may function in enhancing translation through upregulation of tef. Therefore, we provided a new example of an NMD target whose product increases translation initiation which is required for NMD.
Our results indicate that NMD can affect some endogenous genes involved in differentiation, metabolism, and protein translation and folding in the early-diverging protozoan G. lamblia. Our findings also lead to greater understanding of the regulation of mRNA stability of the genes involved in cyst wall pathway and provide a model to investigate the mechanism of cell differentiation.
Materials and Methods
G. lamblia culture
Trophozoites of G. lamblia WB (ATCC 30957) clone C6 were cultured in modified TYI-S33 medium [44] and encysted as previously described [7]. Cyst count was performed on vegetative cultures as previously described [35]. Cyst count was also performed on 24 h encysting cultures. In experiments exposing G. lamblia vegetative trophozoites to actinomycin D, trophozoites were cultured in medium 45 µg/ml actinomycin D (in PBS) for indicated time in the legends of Figures 4B and C.
RNA extraction and Northern blot analysis
Total RNA was extracted from G. lamblia clones C6 at the indicated differentiation stages in the legends of Figures 1, and 3– 7 using TRIzol reagent (Invitrogen). For Northern blot analysis, 10 µg total RNA was fractionated and transferred to charged Nylon membranes (Biodyne B membrane, Pall). Full-length coding region probes of luciferase, cwp1 (GenBank accession no. U09330), cwp2 (GenBank accession no. U28965), cwp3 (GenBank accession no. AY061927), myb2 (GenBank accession no. AY082882), ran (GenBank accession no. U02589), upf1 (GenBank accession no. DQ861427), phosphoglycerate kinase (pgk, GenBank accession no. for genomic DNA XM_762975), glyceraldehyde-3-phosphate dehydrogenase (g3pd, GenBank accession no. for genomic DNA M88062), ornithine carbamoyltransferase (oct, GenBank accession no. for genomic DNA XM_765341), carbamate kinase (ck, GenBank accession no. for genomic DNA XM_765099), orf 16424 (GenBank accession no. for genomic DNA XM_764168; orf 16424 in G. lamblia genome database, http://www.giardiadb.org/giardiadb/)(Morrison et al., 2007), cyclophilin (cy, GenBank accession no. for genomic DNA XM_774688), p21 (GenBank accession no. for genomic DNA XM_762782, for protein XP_767875), bip (GenBank accession no. for genomic DNA XM_766560, for protein XP_771653), glutamate dehydrogenase (gdh, GenBank accession no. for genomic DNA XM_773614), thioredoxin peroxidase (tp, GenBank accession no. for genomic DNA XM_774576)[36], γ subunit of translation elongation factor 1B (tef, GenBank accession no. for genomic DNA XP_778603), and arginine deiminase (ad, GenBank accession no. for genomic DNA U49236) genes were prepared by PCR amplification of genomic DNA using primers lucF (ATGGAAGACGCCAAAAAC) and lucR (TTACACGGCGATCTTTCC), cwp1F (ATGATGCTCGCTCTCCTT) and cwp1R (TCAAGGCGGGGTGAGGCA), cwp2F (ATGATCGCAGCCCTTGTT) and cwp2R (TCACCTTCTGCGGACAAT), cwp3F (ATGTTTTCTCTGCTTCTT) and cwp3R (TTATCTGTAGTAGGGCGG), myb2F (ATGTTACCGGTACCTTCT) and myb2R (TCAGGGTAGCTTCTCACG), ranF (ATGTCTGACCCAATCAGC) and ranR (TCAATCATCGTCGGGAAG), upf1F (ATGGAGCCTTGTGCATTG) and upf1R (CTATGCCTTAGGAATTAC), pgkF (ATGTCCTTAGCGAAGCTCTCC) and pgkR (CTTCTTGTCAGACAGTCTGAT), g3pdF (ATGCCTATTCGCCTCGGAATC) and g3pdR (GCAGCCCTTGGACCCGACGTA), octF (ATGCCGTTCAAGCAGACCCGC) and octR (CTCCATCTTGCAGTCATGCAA), ckF (ATGTCGGCAGGGAAAACGGTT) and ckR (ATCCTTGATGATGCGGGTCCC), 16424F (CACCATGAGTAGAACGCCAAAC) and 16424R (GTAGCGACGATTACCGGA), cyF (ATGAACTCTCCAGTTTCTGAC) and cyR (CTGGAGCACGCCACAGTCGGC), p21F (ATGCACCATCCGACGATCTA) and p21R (CTCCTCTGCCTTCTCTTCGCC), bipF (ATGACGTCTAGTCACGTTAA) and bipR (GAGTTCATCTTTTTCTGCAT), gdhF (ATGCCTGCCCAGACGATCGAG) and gdhR (CACGCAGCCCTGCTCGATCAT), tpF (ATGCCCGTCCCCATCCCCGGC) and tpR (CTTCTTGAACGTCTTGGAGAA), tefF (ATGCAGATCACAGGCAGTCAG) and tefR (CTAGTGCCAAGTCTCCCCATC), adF (ATGACTGACTTCTCCAAGGAT) and adR (TCACTTGATATCGACGCAGAT), respectively. Radiolabeled probes were prepared using the Rediprime II kit (GE Healthcare). An oligonucleotide probe complementary to the AU5 tag coding sequence and its flanking region (au5hyb, GAATTCTCACTTGAGGTAGAAATCGGTAGGCGGGGTGAGG, AU5 tag coding sequence is underlined) was end-labeled using [γ-32P] ATP and T4 polynucleotide kinase. The membranes were hybridized and washed as previously described [45]. Equal loading was confirmed by reprobing the Northern blots with radiolabeled ribosomal DNA. The ribosomal DNA fragment for large subunit ribosomal RNA (X05397) was amplified by PCR using primers RIBOF (GGCCTGCCCCTCGCCCGC) and RIBOR (CCCCTCAGTCCTCCGGGG) and a genomic DNA template. Radiolabeled ribosomal DNA probes were prepared as described above. For detection, the blots were exposed to a storage phosphor screen and the radioactive signals were quantitated using a Typhoon Trio™ Variable Mode Imager (GE Healthcare). Two independently generated stably transfected lines were made from each construct and each of these cell lines was assayed three separate times. The results are expressed as relative expression level over control. Student's t-tests were used to determine statistical significance of differences between samples.
Plasmid construction
All constructs were verified by DNA sequencing with a BigDye Terminator 3.1 DNA Sequencing kit and an ABI 3100 DNA Analyser (Applied Biosystems). Plasmid 5′Δ5N-Pac was a gift from Dr. Steven Singer and Dr. Theodore Nash [46]. Plasmids pRANneo, pPW1, pPW1m, pPC1, pPUPF1HA, and pNUPF1HA have been described previously [7], [34], [35], [47]. A NheI/ClaI fragment containing the luciferase gene, 32-bp ran promoter and two copies of a 19-bp tet operator sequence from pPop2N [34] was replaced by the NheI/ClaI excised luciferase gene and α2-tubulin promoter from pNT5, resulting in pPT5. An NcoI/ClaI fragment containing the wild type luciferase gene from pPT5 [34] was replaced by the NcoI/ClaI excised mutated luciferase gene from pPW1m, resulting in pPT5m. For constructing pPC1D1, a PCR with oligonucleotide cwp1D1NF (GGCGCCATGGATGCCCTGGATCTTTCGGACATG) and ran3C (GCGGATCGATGTAACGAACCGCTAGAAG) generated a 0.8-kb product that was digested with NcoI and ClaI. Another PCR with primers T3 (ATTAACCCTCACTAAAG) and cwp1D1NR (GGCGCCATGGCCCTGATATTTTATTTCTGTG) generated a 0.3-kb PCR product that was digested with NcoI and NheI and cloned into NheI/ClaI digested pPop2N with the 0.8-kb NcoI/ClaI fragment. The resulting pPC1D1 contains a cwp1 gene lacking the coding sequence for the predicted signal peptide sequence and the sequence N terminal to LRRs (nucleotides 4–192). For constructing pPC1D2, a PCR with oligonucleotide cwp1D2BF (GGCGGGATCCACCCTTTACTTGAGCAACAAC) and ran3C generated a 0.6-kb product that was digested with BamHI and ClaI. Another PCR with primers T3 and cwp1D2BR (GGCGGGATCCGATAACGTAGTTATTCGAGGC) generated a 0.4-kb PCR product that was digested with BamHI and NheI and cloned into NheI/ClaI digested pPop2N with the 0.6-kb BamHI/ClaI fragment. The resulting pPC1D2 contains a cwp1 gene lacking the coding sequence for the first LRR (nucleotides 193–264). Similar strategy was used to constructs pPC1D3, pPC1D4, pPC1D5, and pPC1D6, which contain cwp1 gene with deletion of the coding sequence for the second LRR (nucleotides 265–345), third to fourth LRR (nucleotides 346–474), fifth LRR (nucleotides 475–552), and the sequence C terminal to LRRs (nucleotides 553–723), respectively.
Transfection, luciferase assay, and Western blot analysis
Cells transfected with pN series plasmids were selected with G418 as described [47]. Stable transfectants were maintained at 150 µg/ml G418. Cells transfected with pP series plasmids containing the pac gene were selected and maintained with 54 µg/ml puromycin. For co-transfection assays (see Figs. 6 and 7), G. lamblia cells were first transfected with pP series plasmids and selected in 54 µg/ml puromycin. The stable transfectants were transfected with pN series plasmids and then the cells were doubly selected in both 150 µg/ml G418 and 54 µg/ml puromycin. After stable transfection with specific constructs, luciferase activity was determined in vegetative cells at late log/stationary phase (1.5×106 cells/ml) or in 24 h encysting cells as described [45] and was measured with an Optocomp I luminometer (MGM Instruments). Two independently generated stably transfected lines were made from each construct and each of these lines was assayed three separate times. Western blots were probed with anti-CWP1 antibody (1/10,000) [48] or anti-human RAN antibody (1/5,000) (Santa Cruz Biotechnology), and detected with peroxidase-conjugated goat anti-mouse IgG (Pierce, 1/5,000) and enhanced chemiluminescence (GE Healthcare).
Acknowledgments
We thank the researchers and administrators of the G. lamblia Genome database. We also thank Dr. Ming-Shyue Lee, Dr. Tsai-Kun Li, and Dr. Chien-Kuo Lee for helpful comments, and Ms Yi-Li Liu and I-Ching Huang and Mr. Chao-Cheng Cho for technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the National Science Council (NSC 96-2320-B-002-040-MY3) and the National Health Research Institutes (NHRI-EX96-9510NC and NHRI-EX97-9510NC) in Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wolfe MS. Giardiasis. Clin Microbiol Rev. 1992;5:93–100. doi: 10.1128/cmr.5.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall MM, Naumovitz D, Ortega Y, Sterling CR. Waterborne protozoan pathogens. Clin Microbiol Rev. 1997;10:67–85. doi: 10.1128/cmr.10.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam RD. Biology of Giardia lamblia. Clin Microbiol Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ondarza RN. Drug targets from human pathogenic amoebas: Entamoeba histolytica, Acanthamoeba polyphaga and Naegleria fowleri. Infect Disord Drug Targets. 2007;7:266–280. doi: 10.2174/187152607782110059. [DOI] [PubMed] [Google Scholar]
- 5.Lujan HD, Mowatt MR, Conrad JT, Bowers B, Nash TE. Identification of a novel Giardia lamblia cyst wall protein with leucine-rich repeats. Implications for secretory granule formation and protein assembly into the cyst wall. J Biol Chem. 1995;270:29307–29313. doi: 10.1074/jbc.270.49.29307. [DOI] [PubMed] [Google Scholar]
- 6.Mowatt MR, Lujan HD, Cotton DB, Bower B, Yee J, et al. Developmentally regulated expression of a Giardia lamblia cyst wall protein gene. Mol Microbiol. 1995;15:955–963. doi: 10.1111/j.1365-2958.1995.tb02364.x. [DOI] [PubMed] [Google Scholar]
- 7.Sun CH, McCaffery JM, Reiner DS, Gillin FD. Mining the Giardia lamblia genome for new cyst wall proteins. J Biol Chem. 2003;278:21701–21708. doi: 10.1074/jbc.M302023200. [DOI] [PubMed] [Google Scholar]
- 8.Sun CH, Palm D, McArthur AG, Svard SG, Gillin FD. A novel Myb-related protein involved in transcriptional activation of encystation genes in Giardia lamblia. Mol Microbiol. 2002;46:971–984. doi: 10.1046/j.1365-2958.2002.03233.x. [DOI] [PubMed] [Google Scholar]
- 9.Sun CH, Su LH, Gillin FD. Novel plant-GARP-like transcription factors in Giardia lamblia. Mol Biochem Parasitol. 2006;146:45–57. doi: 10.1016/j.molbiopara.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Wang CH, Su LH, Sun CH. A novel ARID/Bright-like protein involved in transcriptional activation of cyst wall protein 1 gene in Giardia lamblia. J Biol Chem. 2007;282:8905–8914. doi: 10.1074/jbc.M611170200. [DOI] [PubMed] [Google Scholar]
- 11.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez CI, Bhattacharya A, Wang W, Peltz SW. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene. 2001;274:15–25. doi: 10.1016/s0378-1119(01)00552-2. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson MF. A new function for nonsense-mediated mRNA-decay factors. Trends in Genetics. 2005;21:143–148. doi: 10.1016/j.tig.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ, Jr, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Applequist SE, Selg M, Raman C, Jack HM. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 18.Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol. 2000;20:8944–8957. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;117:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 23.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–4603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, González CI. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:3390–3400. doi: 10.1128/MCB.26.9.3390-3400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahlseid JN, Puziss J, Shirley RL, Atkin AL, Hieter P, et al. Accumulation of mRNA coding for the ctf13p kinetochore subunit of Saccharomyces cerevisiae depends on the same factors that promote rapid decay of nonsense mRNAs. Genetics. 1998;150:1019–1035. doi: 10.1093/genetics/150.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 30.Sogin ML, Gunderson JH, Elwood HJ, Alonso RA, Peattie DA. Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science. 1989;243:75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto T, Nakamura Y, Nakamura F, Shirakura T, Adachi J, et al. Protein phylogeny gives a robust estimation for early divergences of eukaryotes: phylogenetic place of a mitochondria-lacking protozoan, Giardia lamblia. Mol Biol Evol. 1994;11:65–71. doi: 10.1093/oxfordjournals.molbev.a040093. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto T, Nakamura Y, Kamaishi T, Nakamura F, Adachi J, et al. Phylogenetic place of mitochondrion-lacking protozoan, Giardia lamblia, inferred from amino acid sequences of elongation factor 2. Mol Biol Evol. 1995;12:782–793. doi: 10.1093/oxfordjournals.molbev.a040256. [DOI] [PubMed] [Google Scholar]
- 33.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 34.Chen YH, Su LH, Sun CH. Incomplete nonsense-mediated mRNA decay in Giardia lamblia. Int J Parasitol. 2008 doi: 10.1016/j.ijpara.2008.02.006. doi:10.1016/j.ijpara.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Su LH, Lee GA, Huang YC, Chen YH, Sun CH. Neomycin and puromycin affect gene expression in Giardia lamblia stable transfection. Mol Biochem Parasitol. 2007;156:124–135. doi: 10.1016/j.molbiopara.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Wang CC. A likely molecular basis of the susceptibility of Giardia lamblia towards oxygen. Mol Microbiol. 2006;59:202–211. doi: 10.1111/j.1365-2958.2005.04896.x. [DOI] [PubMed] [Google Scholar]
- 37.Sanders J, Brandsma M, Janssen GM, Dijk J, Möller W. Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1 beta gamma delta in the endoplasmic reticulum. J Cell Sci. 1996;109:1113–1117. doi: 10.1242/jcs.109.5.1113. [DOI] [PubMed] [Google Scholar]
- 38.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 39.Taylor R, Kebaara BW, Nazarenus T, Jones A, Yamanaka R. Gene set coregulated by the Saccharomyces cerevisiae nonsense-mediated mRNA decay pathway. Eukaryot Cell. 2005;4:2066–2077. doi: 10.1128/EC.4.12.2066-2077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kebaara B, Nazarenus T, Taylor R, Forch A, Atkin AL. The Upf-dependent decay of wild-type PPR1 mRNA depends on its 5′-UTR and first 92 ORF nucleotides. Nucleic Acids Res. 2003;31:3157–3165. doi: 10.1093/nar/gkg430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ford AS, Guan Q, Neeno-Eckwall E, Culbertson MR. Ebs1p, a negative regulator of gene expression controlled by the Upf proteins in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:301–312. doi: 10.1128/EC.5.2.301-312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983;77:487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- 45.Knodler LA, Svard SG, Silberman JD, Davids BJ, Gillin FD. Developmental gene regulation in Giardia lamblia: first evidence for an encystation-specific promoter and differential 5′ mRNA processing. Mol Microbiol. 1999;34:327–340. doi: 10.1046/j.1365-2958.1999.01602.x. [DOI] [PubMed] [Google Scholar]
- 46.Singer SM, Yee J, Nash TE. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol Biochem Parasitol. 1998;92:59–69. doi: 10.1016/s0166-6851(97)00225-9. [DOI] [PubMed] [Google Scholar]
- 47.Sun CH, Chou CF, Tai JH. Stable DNA transfection of the primitive protozoan pathogen Giardia lamblia. Mol Biochem Parasitol. 1998;92:123–132. doi: 10.1016/s0166-6851(97)00239-9. [DOI] [PubMed] [Google Scholar]
- 48.Huang YC, Su LH, Lee GA, Chiu PW, Cho CC, Wu JY, Sun CH. Regulation of cyst wall protein promoters by Myb2 in Giardia lamblia. J Biol Chem. 2008 doi: 10.1074/jbc.M805023200. Sep 2008; doi:10.1074/jbc.M805023200. [DOI] [PMC free article] [PubMed] [Google Scholar]