Abstract
In 1988 McCusker and Haber generated a series of mutants which are resistant to the minimum inhibitory concentration of the protein synthesis inhibitor cycloheximide. These cycloheximide-resistant, temperature-sensitive (crl) mutants, in addition, exhibited other pleiotropic phenotypes, e.g., incorrect response to starvation, hypersensitivity against amino acid analogues, and other protein synthesis inhibitors. Temperature sensitivity of one of these mutants, crl3–2, had been found to be suppressed by a mutation, SCL1–1, which resided in an α-type subunit of the 20S proteasome. We cloned the CRL3 gene by complementation and found CRL3 to be identical to the SUG1/CIM3 gene coding for a subunit of the 19S cap complex of the 26S proteasome. Another mutation, crl21, revealed to be allelic with the 20S proteasomal gene PRE3. crl3–2 and crl21 mutant cells show significant defects in proteasome-dependent proteolysis, whereas the SCL1–1 suppressor mutation causes partial restoration of crl3–2-induced proteolytic defects. Notably, cycloheximide resistance was also detected for other proteolytically deficient proteasome mutants (pre1–1, pre2–1, pre3–1, pre4–1). Moreover, proteasomal genes were found within genomic sequences of 9 of 13 chromosomal loci to which crl mutations had been mapped. We therefore assume that most if not all crl mutations reside in the proteasome and that phenotypes found are a result of defective protein degradation.
INTRODUCTION
Cycloheximide is a potent inhibitor of the ribosome. The drug blocks elongation of the nascent polypeptide chain during translation. When cells of the yeast Saccharomyces cerevisiae are exposed to high levels of cycloheximide, they stop both protein synthesis and growth. Application of the minimum inhibitory concentration of cyloheximide causes cells to stop growth, but in contrast, under these conditions, protein synthesis is not completely inhibited (Shilo et al., 1979). Transition from the G1 to S-phase is a critical step in the cell cycle of S. cerevisiae. When cells have passed this step, they have to complete the next cycle of cell division. Under nutrient-limiting conditions, e.g., when cells reach stationary phase or when they starve for a single amino acid, they stop growth and rest as single unbudded cells. Such a G1 arrest also occurs when protein synthesis is restrained by exposure to the minimum inhibitory level of cycloheximide (McCusker and Haber, 1988b).
In 1988 McCusker and Haber (1988a) isolated a series of temperature-sensitive mutants which were resistant to the minimum inhibitory concentration of cycloheximide. These crl (cycloheximide-resistant ts lethal) mutants resided in 22 complementation groups. Besides cyloheximide resistance and temperature sensitivity, they showed many additional phenotypes, e.g., hypersensitivity against amino acid analogues but also other protein synthesis inhibitors. crl mutants also exhibited a failure to undergo a defined G1 arrest when grown into stationary phase or when starving for a single amino acid (McCusker and Haber, 1988b).
For one of these mutants, crl3–2 an extragenic suppressor mutation, SCL1–1, had been found which resided in an α-type subunit of the 20S proteasome (Balzi et al., 1989). Proteasomes are essential, highly sophisticated protease complexes that are responsible for selective proteolysis in the cytoplasm and nucleus of the eukaroytic cell (Coux et al., 1996; Hilt and Wolf, 1996). Most proteins degraded by this system are tagged for destruction by addition of multiubiquitin chains. Ubiquitin/proteasome-dependent degradation is a significant step in many different regulatory pathways of the cell. Proteasomes are implicated in stress response by removing abnormal proteins (Heinemeyer et al., 1991, 1993; Hilt et al., 1993). They are involved in adaptation of metabolism by degrading enzymes, e.g., fructose-1,6-bisphosphatase (FBPase) (Schork et al., 1995), ornithine decarboxylase (Murakami et al., 1992) as well as transcriptional regulators like Gcn4, which controls expression of metabolic enzymes (Kornitzer et al., 1994). They are also linked to cell differentiation as had been shown for the proteolytically unstable yeast MATα2 transcriptional repressor protein (Chen et al., 1993; Richter-Ruoff et al., 1994). Studies with proteasome mutants indicated that proteasomes play vital functions in the cell division cycle (for reviews, see Deshaies, 1995; Hilt and Wolf, 1996; King et al., 1996). Proteasome-mediated proteolytic processes are required for termination of cell cycle phases (e.g., by degradation of cyclins (Seufert et al., 1995; Yaglom et al., 1995) but also initiate entry into distinct cell cycle transitions (e.g., by degrading cyclin-dependent kinase inhibitors and other cell cycle inhibitors).
The 20S proteasome is a hollow cylindrically shaped proteolytic complex which consists of a stack of four rings, each containing seven subunits. In eukaryotes, seven different β-type subunits are used to form each of the two inner rings, whereas the outer rings contain seven different α-type subunits each. In agreement with this structure 14 genes coding for 20S proteasome subunits are found in the yeast genome. Yeast 20S proteasomes exhibit three different proteolytic activities cleaving chromogenic peptides at the C terminus of hydrophobic, acidic, or basic amino acids, respectively (Heinemeyer et al., 1991). The corresponding active sites reside within certain β-type subunits. Pre2 confers the chymotrypsin-like activity, Pre3 the peptidyl-glutamyl-peptide-hydrolyzing (PGPH) activity, and Pup1 the trypsin-like activity (Heinemeyer et al., 1997). X-ray structure analysis of the 20S proteasome from Thermoplasma acidophilum (Löwe et al., 1995) and recently of the eukaryotic 20S proteasome from S. cerevisiae uncovered that the active sites were located inside the central channel of the complex (Groll et al., 1997). The 20S proteasome had been found to constitute the proteolytic core module of a larger protease complex, the 26S proteasome. Two additional regulatory 19S cap complexes are attached to the ends of the 20S cylinder to build up this 26S complex (Peters, 1994). The 26S proteasome degrades ubiquitinated proteins in an ATP-dependent reaction, whereas the 20S proteasome is unable to do so. The 19S cap complexes are most probably responsible for recognition, unfolding, and translocation of substrate proteins to the proteolytically active sites of the 20S core. Subunits of the 19S regulatory cap complex can be divided into two classes: one containing ATPase motifs and the others not. Several non-ATPase subunits have been characterized from mammalian cells and have also been cloned in the budding yeast. The yeast 19S cap subunit Mcb1 (van Nocker et al., 1996), like its human homologue S5a (Deveraux et al., 1994), is able to bind multiubiquitin chains. Other genes coding for non-ATPase subunits from budding yeast include SEN3 (S1 subunit) (De Marini et al., 1995), NIN1 (S14 subunit) (Kominami and Toh-e, 1994), and SUN2 (S3 subunit). At least six genes [YTA1/TBP1, YTA3/CIM5, SUG1/CIM3, YTA2/YNT1, YTA5/YHS4, and SUG2 (Ghislain et al., 1993; Rubin et al., 1996; Russell et al., 1996; Schnall et al., 1994; Swaffield et al., 1995)] have been found to code for subunits of the 19S cap complex, which due to common sequence features are all classified to be ATPases of the AAA superfamily.
Interestingly, sug1 mutations had initially been detected to function as suppressors of mutations, gal4 (Swaffield et al., 1992) and cdc68 (Xu et al., 1995), which cause transcriptional activation defects. Furthermore, biochemical studies indicated Sug1 to be a component of the RNase II holoenzyme complex (Kim et al., 1994). However, biochemical evidence appeared that Sug1 is an integral component of the 26S proteasome (Rubin et al., 1996).
Here we demonstrate 1) that crl3–2 is allelic with SUG1/CIM3 and 2) that another crl mutation, crl21, is allelic with the 20S proteasomal β-type gene PRE3. Both crl3 and crl21 mutants are defective in proteasomal protein degradation. We also show that the SCL1–1 mutation which suppresses temperature sensitivity caused by the crl3–2 mutation leads to partial restoration of proteasomal activity. Other mutations in various 20S proteasome subunits (Pre1, Pre2, Pre3, Pre4) which diminish proteolytic activity of the complex also cause cycloheximide resistance and temperature sensitivity. Moreover, closely linked to 9 of 12 chromosomal positions to which other crl mutations had been mapped (McCusker and Haber, 1988a) genes coding for subunits of the 26S proteasome were found. We therefore propose that the various phenotypes found in crl mutants are a result of defects in the proteasome degradation pathway.
MATERIALS AND METHODS
Media
Cells were grown either on YPD (1% yeast extract, 2% peptone, 2% glucose), YP ethanol (1% yeast extract, 2% peptone, 2% ethanol), or on minimal medium (0.67% Difco yeast nitrogen base without amino acids) containing 2% glucose or 2% ethanol as a carbon source and supplements (20 μg/ml uracil, 20 μg/ml histidine, 30 μg/ml leucine, 20 μg/ml adenine). Synthetic complete (CM) medium was prepared as outlined in the study by Sherman (1991). Selection for uracil auxotrophic cells was performed on mineral medium containing 50 μg/ml uracil and 0.1% wt/vol 5-fluoroorotic acid (5-FOA). Cycloheximide resistance was determined using either YPD or mineral medium and supplements with a cycloheximide concentration between 0.5 and 10 μg/ml. Sporulation medium contained 1% potassium acetate, 0.1% yeast extract, and 0.05% glucose. Radiolabeling was done in pulse medium (0.17% Difco yeast nitrogen base without amino acid and ammonium sulfate, 0.5% proline, 100 μM ammonium sulfate, 2% ethanol, and required supplements).
General Methods
For mating of yeast cells, sporulation, tetrad dissection, plasmid segregation and separation, as well as the gap repair method, standard protocols were followed (Ausubel et al., 1990; Sherman et al., 1986). Yeast transformations were performed with the improved lithium acetate method of Gietz et al. (1992) using single-stranded salmon sperm DNA as carrier. Growth and manipulation of Escherichia coli strain DH5α were carried out using standard methods described in the studies of Ausubel et al. (1990) and Sambrook et al. (1989).
DNA was sequenced with the dideoxy chain termination method (Sanger et al., 1977) using gene-derived oligonucleotides as primers. SDS-PAGE was performed using 8 to 15% separating gels.
Plasmids
Plasmid pUM20 was generated by inserting a 2.6-kb fragment containing the entire coding region of the scl1+ gene as well as 891 bp and 989 bp from the 5′ and 3′ region, respectively, into the MCS of plasmid pDP83 (URA3/CEN14). Plasmid pUM70/76 was prepared via the gap repair method (Rothstein, 1991): A 1.85-kb MscI/BstEII fragment was excised from pUM20 containing the scl1+ gene, leaving 213-bp and 509-bp flanking sequences, respectively. Strain Y55–1161 was transformed with the gapped pUM20 and repaired plasmids were recovered and sequenced. Plasmids pUM71 (PSCL1-1::scl1+) and pUM72 (Pscl1 + ::SCL1–1) were generated from plasmids pUM20 and pUM70/76. The mutated SCL1–1 open reading frame (ORF) was removed from plasmid pUM70/76 by AvaI/SacI digestion. The gap obtained was filled with a 1692-bp fragment containing the scl1+ ORF which had been excised from plasmid pUM20 by AvaI/SacI digestion. A similar strategy was used for pUM72: This time a 1692-bp AvaI/SacI fragment containing the mutated SCL1–1 ORF was excised from pUM70/76 and cloned into AvaI/SacI sites of plasmid pUM20 after excision of the scl1+ sequence. To clone the crl3–2 mutant allele, a 1.6-kb fragment including the crl3–2 ORF, was amplified by polymerase chain reaction (PCR). Chromosomal DNA isolated from strain Y55–1162 was used as template. PCR primers used were 5′-GATATTCTAGATCCAGC and 5′-CGAATTCGGTACCGTTATATCCTG. Subcloning of the amplified fragment into the vector pWH5 (Rose and Broach, 1991; Wright et al., 1986) yielded plasmid pUM101 which was used for sequencing the crl3–2 mutant allele. For cloning of the crl21 mutant allele, a 1.4-kb fragment reaching from the NruI/SnaBI sites located in the 5′ and 3′ regions of the PRE3(crl21) ORF, respectively, was amplified by PCR. Chromosomal DNA isolated from the Y55crl21 strain was used as template. PCR primers used were 5′-AAAGGATCCGATACGTAGATACAGTCACCA and 5′-AAGGATCCCATACTTACCCTGCTCGCGA. Subcloning of the PCR-amplified fragment into the yeast shuttle vector pRS315 (LEU2 CEN6) (Sikorski and Hieter, 1989; Rose and Broach, 1991) using primer-derived BamHI sites yielded plasmid pRG26.
Olson Blots
Olson blotting offers the possibility to evaluate the position of a distinct gene in the yeast genome. The Olson blots were supplied by Boehringer Mannheim (Indianapolis, IN) and were performed by following a protocol of the manufacturer.
Strains
Strains YUM47, YUM48, YUM60, and YUM61 were derived from Y55–1162 (crl3–2) by transformation with plasmid pUM70/76 (SCL1–1), pUM20 (scl1+), pUM71 (PSC:1–1::scl1+), and pUM72 (Pscl1+ ::SCL1–1), respectively. To generate a crl21 mutant strain isogenic with WCG4a, the diploid strain yRG8, which heterozygously contains a pre3::URA3 deletion, was transformed with the pRG26 (crl21) plasmid. Diploid cells were sporulated and tetrads dissected. Tetrad clones, which due to uracil and leucine prototrophy, were proven to contain the chromosomal pre3::URA3 deletion and plasmid pRG26 (crl21(pre3–6) LEU2) were isolated. To exclude PCR-induced mutations, proteasomal peptide-cleaving activities of the pRG26-dependent pre3–6 (crl21) mutant were determined and compared with the activities of the original strain Y55crl21. Selection on 5-FOA acid agar plates for cells, which due to recombination had replaced the chromosomal pre3::URA3 deletion by the crl21 allele and by this became uracil auxotrophic, yielded the WCG4a isogenic haploid crl21 strain yRG16.
Immunoblotting of Ubiquitin Protein Conjugates
Yeast cells were grown to stationary phase at 30°C, to logarithmic phase at 30°C, or to logarithmic phase at 30°C and thereafter heat stressed by shifting to 37°C for 3 h. Cells were harvested by centrifugation and resuspended in water to yield a 50% (wt/vol) suspension. Samples were heated for 10 min at 95°C, equal amounts of glass beads were added, and cells were broken by vortexing for 30 s six times with intermitted heating (95°C, 1 min). Equal volumes of 4.5% SDS and 2.25 mM EDTA were added, and samples vortexed and heated for 10 min at 95°C. After centrifugation protein concentrations were determined and equal amounts (40 μg) were loaded on SDS gels (10%). After electrophoresis proteins were blotted to nitrocellulose. Filters were blocked by shaking in 5% (wt/vol) milk powder in 100 mM phosphate, 100 mM NaCl (pH 7.5), and 0.1% Tween 20 for 1.5 h and then treated for 1 h with a 1:5000 dilution of anti-ubiquitin protein conjugate serum in coating buffer. Antibodies bound were detected using peroxidase-coupled goat anti-rabbit IgG and an ECL detection system (Amersham Life Sciences, Arlington Heights, IL) following the manufacturer’s instructions.
Pulse Chase Experiments and Immunoprecipitation
Wild-type and mutant cells were grown to an A578 of 5 to 6 in mineral medium containing 2% glucose and supplements. After harvesting (5 min, 500 × g) and washing, cells were preincubated for 3.5 h in pulse medium at a density of 3 A578 per ml. Radiolabeling was done by adding [35S]methionine (Amersham, Braunschweig, Germany) to the cell suspension to a final concentration of 50 μCi/ml (pulse). After 1.5 h cells were shifted into mineral medium containing 2% glucose, 5 mM nonradioactive methionine, and 0.5% ammonium sulfate (chase). Samples (3 A578 units) were taken at the times indicated, and the chase period was terminated by addition of trichloroacetic acid (final concentration 5%). Precipitates were washed twice with ethanol (−20°C) and dried. Subsequently, cells were lysed in 100 μl of breaking buffer by glass bead agitation. Immunoprecipitation with antisera specific for FBPase (8 μl) was performed as outlined in the study of Schork et al. (1994a).
RESULTS
Cloning of the CRL3 Gene
The temperature-sensitive crl3–2 mutant strain Y55–1162 (McCusker and Haber, 1988a) was transformed with a YCp50-based yeast genomic library, and transformants were screened for growth at the restrictive temperature (37°C). Strain Y55–1162 showed poor transformability. Therefore, a series of transformations was performed and 48 individual complementing clones were obtained. To verify that growth at 37°C depends on the presence of a YCp50- (URA3) derived library plasmid, clones were plated on 5-FOA medium to select for cells that had lost the plasmid during mitosis. 5-FOA poisons uracil prototrophic cells, hence cells containing the YCp50 (URA3) plasmid do not grow on this medium. Uracil auxotrophic cells that had lost the plasmid were retested for temperature sensitivity. Interestingly, a high rate of plasmid-independent revertants were observed. However, for 29 of these clones growth at 37°C was plasmid dependent. From 18 of these clones plasmids were isolated and four different inserts, about 15-kb long each, were found. All of these inserts contained a common DNA fragment with a length of approximately 2.6 kb, which after subcloning was proven to complement temperature sensitivity of the crl3–2 mutant strain Y55–1162 (Figure 1). The 2.6-kb DNA fragment was isolated and using Olson blots the DNA was mapped close to the centromer of chromosome VII. This localization is in agreement with the previously published position for the crl3–2 mutation (McCusker and Haber, 1988a).
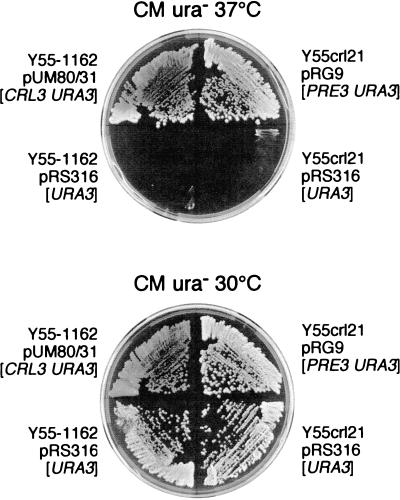
Figure 1.
Plasmid-mediated complementation of crl mutants. Cells were streaked on YPD agar plates and incubated at 37°C and 30°C for 3 d. Plasmids containing the CRL3 (SUG1) or the PRE3 gene complement temperature sensitivity of crl3–2 (left side) and crl21 mutants (right side), respectively. Control strains (bottom) contain the empty plasmid pRS316. After 4 d the strain Y55–1162 (crl3–2 ade1) carrying the CRL3 plasmid (wild type) accumulates significant amounts of a red dye (indicated by the darker color of the colonies, 30°C control plate), which is typically found when starving cells which harbor distinct mutations in the adenine biosynthesis pathway. In contrast, crl3–2 mutant cells generate only tiny amounts of this red product.
Further subcloning of the insert reduced the complementing region to a 1.8-kb fragment. Sequencing uncovered an ORF which showed 100% identity to the previously described SUG1(CIM3/TBPy) gene (Swaffield et al., 1992; Ghislain et al., 1993) which codes for an AAA-ATPase subunit of the proteasomal 19S regulatory cap complex.
Cloning of the crl3–2 Mutant Allele
The crl3–2 mutant allele was cloned from the mutant strain Y55–1162 via PCR. To exclude PCR-induced mutations, five independent fragments were subcloned, sequenced, and compared with the wild-type gene. Ten base pair exchanges within the CRL3 coding region were detected, only one of these led to an amino acid exchange (G41V). The high number of conservative changes found (G566C, C756T, A777G, A834G, G867A, T993C, C1068T, C1128T, C1207T) may reflect some evolutionary distance between the crl3–2 mutant strain Y55–1162 and strains used for sequencing the SUG1/CRL3 wild-type gene (S288C background).
Cloning and Characterization of the SCL1–1 Mutant Allele
An extragenic suppressor mutation SCL1–1, which complemented temperature sensitivity of crl3–2 mutants (McCusker and Haber, 1988a) was found to reside in an α-type subunit of the yeast 20S proteasome (Balzi et al., 1989; Fujiwara et al., 1990; Emori et al., 1991). The SCL1–1 mutant allele was isolated from strain Y55–1161 (crl3–2 SCL1–1) via the plasmid gap repair method (Rothstein, 1991) and sequenced. Six base pair exchanges were detected within the coding region. Five of these were conservative bp exchanges (see legend to Figure 2) and may also be attributed to some evolutionary distance between strains congenic with Y55 and strains with the genetic background of S288C. One base pair exchange led to an amino acid substitution (Y30C) located at a highly conserved short peptide stretch within the H0-helix of 20S proteasomal α-type subunits (Figure 2). Interestingly, bp mutations were also found in the SCL1–1 promoter region (Figure 2). When the crl3–2 mutant strain Y55–1162 was transformed with a SCL1–1-containing plasmid (strain YUM47), the mutant SCL1–1 gene was able to suppress the crl3–2-induced temperature sensitivity also in the presence of a chromosomal copy of the scl1+ wild-type gene (Figure 3). However, in comparison to the crl3–2 SCL1–1 double mutant strain Y55–1161, which only contained a chromosomal SCL1–1 allele, these cells exhibited slightly decreased growth at 37°C (Figure 3), indicating that the suppressor SCL1–1 allele is only semidominant. This finding was confirmed by the fact that expression of a plasmid-derived scl1+ wild-type gene in the crl3–2 SCL1–1 double mutant strain Y55–1161 also caused reduced growth at 37°C. However, in comparison to strain YUM47 (crl3 scl1+ (SCL1–1)) growth appeared to be reduced to a slightly lesser extent (Figure 3). We attribute this difference to a stronger expression of the mutant Scl1–1 protein when it is derived from its chromosomal locus than if it is derived from the plasmid encoded gene.
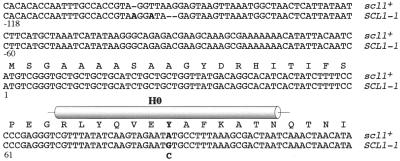
Figure 2.
DNA sequence of the SCL1–1 suppressor allele. DNA and protein sequence of the promoter region and the N terminus of the scl1+ wild-type and SCL1–1 mutant allele. Mutations are indicated by bold letters. Additional conservative bp exchanges found in the SCL1–1 mutant sequence were C280T, G450A, C534T, A540G, and C648T. The position of the H0-helix region is indicated.
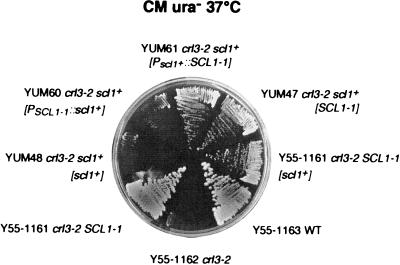
Figure 3.
The mutation in the SCL1–1 structural gene suppresses crl3–2-induced temperature sensitivity. Cells were streaked on synthetic complete medium and incubated at 37°C for 3 d. Control strains which had originally been generated by McCusker and Haber are: the wild-type strain (Y55–1163), the crl3–2 mutant strain (Y55–1162), and the crl3–2 SCL1–1 double mutant strain (Y55–1161). Plasmid-encoded SCL1–1 suppresses crl3–2-induced temperature sensitivity even in the presence of a chromosomal copy of the scl1+ wild-type gene (strain YUM47). In comparison to the original crl3–2 SCL1–1 suppressor strain, Y55–1161 cells show decreased growth rates, demonstrating semidominance of the SCL1–1 allele. Y55–1161 (crl3–2 SCL1–1) cells harboring a plasmid-encoded scl1+ wild-type gene also showed decreased growth but in comparison to YUM47 to a slightly minor extent. YUM61 cells carrying plasmid pUM72 containing the SCL1–1 mutant structural gene driven by the scl1+ promoter were able to grow. YUM60 cells expressing the scl1+ wild-type gene under the control of the mutated SCL1–1 promoter region (plasmid pUM71) did not grow. YUM48 (crl3–2 scl1+) cells also expressing a plasmid-derived Scl1 wild-type protein were not able to grow, demonstrating that an enhanced gene dose does not cause suppression.
The suppressor function of the SCL1–1 allele may be due to the mutation in the structural gene and therefore a result of modified protein structure. Alternatively, also a change of SCL1–1 expression due to the altered promoter sequence may contribute or be solely responsible for suppressor function. To address this question, we tried to test both mutations separately. Plasmids were constructed 1) containing the mutated SCL1–1 promoter sequence attached to the scl1+ ORF (plasmid pUM71) and 2) the mutated SCL1–1 coding region driven by the scl1+ wild-type promoter (plasmid pUM72). These plasmids were transformed into the crl3–2 mutant strain Y55–1162 and cells were tested for growth at 37°C. The mutated SCL1–1 structural gene controlled by the scl1+ wild-type promoter (strain YUM61) was able to complement temperature sensitivity to a rate as had been found for the original SCL1–1 mutant allele, whereas the scl1+ wild-type gene under the mutant SCL1–1 promoter (strain YUM60) was not able to do so (Figure 3). These data clearly demonstrate that suppression is exclusively due to the amino acid exchange within the Scl1–1 protein.
SCL1–1-mediated Suppression Is Due to Compensation of crl3-induced Proteolytic Defects
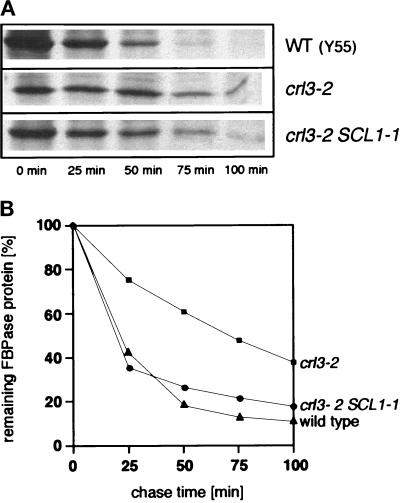
crl3–2 mutants are deficient in proteasome-mediated protein degradation. As had been measured for other proteolytically defective proteasome mutants (Heinemeyer et al., 1991, 1993; Hilt et al., 1993) crl3–2 mutants (Y55–1162) showed accumulation of ubiquitinated proteins (Figure 4). At restrictive but also at permissive temperatures significant amounts of ubiquitinated proteins were found in crl3–2 mutants, whereas in wild-type cells no or only traces of ubiquitinated proteins could be detected under such conditions (Figure 4). Defective proteasomal activity of crl3–2 mutant cells could further be proven by the fact that degradation of a defined proteasomal in vivo substrate protein, FBPase (Schork et al., 1994a, 1995) is significantly reduced in crl3–2 mutant cells (Figure 5). However, when crl3–2 mutant cells contained the SCL1–1 suppressor mutation (strain Y55–1161), a significant restoration of proteasomal activity was observed: In comparison to crl3 mutants, crl3 SCL1–1 double mutant cells accumulated reduced amounts of ubiquitinated proteins under starvation and heat stress conditions (Figure 4). Furthermore, proteolysis of FBPase via the proteasome was restored to nearly wild-type rates in SCL1–1 crl3–2 double mutants (Figure 5). These data made evident that suppression of crl3–2-induced temperature sensitivity by the SCL1–1 mutation is due to restored proteasomal activity.
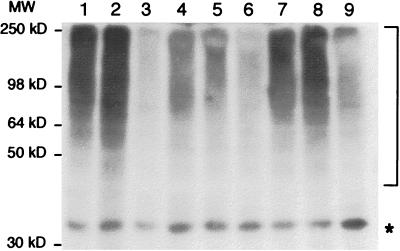
Figure 4.
crl mutants accumulate ubiquitinated proteins. Western blot analysis of crude extracts using anti-ubiquitin protein conjugate serum: wild-type cells, Y55–1163 (lanes 3, 6, and 9), crl3–2 mutant cells Y55–1162 (lanes 2, 5, and 8) and crl3–2 SCL1–1 double mutant cells, Y55–1161 (lanes 1, 4, and 7). Cells were grown at 30°C to stationary phase (lanes 1–3), to logarithmic phase at 30°C (lanes 4–6), or to logarithmic phase at 30°C and thereafter heat stressed by shifting to 37°C for 3 h (lanes 7–9). Ubiquitin protein conjugates are indicated by a bracket. Asterisk, cross-reacting protein. Molecular weights are as indicated.
Figure 5.
Proteolytic stability of FBPase in crl3–2 and crl3–2 SCL1–1 mutants. (A) Pulse chase analysis of FBPase degradation in wild-type (Y55–1163), crl3–2 (Y55–1162), and crl3–2 SCL1–1 (Y55–1161) mutants. Cells were pulse labeled during derepression of FBPase in ethanol containing medium and chased with the addition of glucose followed by extraction, immunoprecipitation, and SDS-PAGE (see MATERIALS AND METHODS). (B) Quantitation of pulse chase analysis of wild-type cells (▴), crl3–2 (▪), and crl3–2 SCL1–1 (•) mutants.
crl21 Is Allelic with the Proteasomal β-Type Gene PRE3
Another crl mutation (crl21) had been mapped close to the centromer of chromosome X (McCusker and Haber, 1988a). The PRE3 gene coding for a β-type subunit of the 20S proteasome was found to be located at the same chromosomal position (Enenkel et al., 1994). To test whether the crl21 mutation is allelic with PRE3, a crl21 mutant strain (Y55crl21) was transformed with a plasmid [pRG9 (URA3)] which contained the PRE3 gene. As shown in Figure 1, this plasmid was able to complement crl21-induced temperature sensitivity of strain Y55crl21.
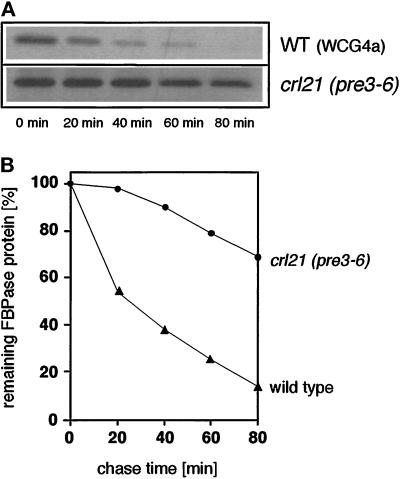
Using PCR the crl21 mutant allele was amplified from chromosomal DNA of strain Y55crl21. The PCR products were subcloned into the vector pRS315 (CEN6 LEU2) yielding plasmid pRG26 and the PRE3 region was sequenced. Besides two conservative bp exchanges (T477A, A650T) within the PRE3 coding sequence, the crl21 allele contained a deletion of four codons encoding L85 to E88. To generate crl21 (pre3–6) mutants with the genetic background of our preferred wild-type strain, WCG4a, the crl21 allele was transferred into strain WCG4a by a one-step gene replacement yielding strain YRG16 (see MATERIALS AND METHODS). As found for the original strain, Y55crl21, also YRG16 showed strongly reduced proteasomal peptide-cleaving activities: As compared with wild-type cells strain YRG16 exhibited 29% of chymotrypsin-like activity, 23% of PGPH activity, and 50% of trypsin-like activity. Moreover, in the original strain Y55crl21 and also in strain YRG16 (Figure 6) significantly reduced rates of degradation of the proteasomal in vivo substrate FBPase (Schork et al., 1994a, 1995) were measured. As compared with wild-type cells, also enhanced accumulation of ubiquitinated protein was detected in YRG16 cells under heat stress conditions. Taken together, these data make evident that the crl21 mutation resides in the 20S proteasome β-type gene PRE3 and that this mutation also causes defects in proteasome-dependent protein degradation.
Figure 6.
Stabilization of FBPase crl21 (pre3–6) mutants. (A) Pulse chase analysis of FBPase degradation in wild-type (WCG4a) and crl21 (pre3–6) (YRG16) mutants. Cells were pulse labeled during derepression of FBPase in ethanol containing medium and chased with the addition of glucose followed by extraction, immunoprecipitation, and SDS-PAGE (see MATERIALS AND METHODS). (B) Quantitation of pulse chase analysis of wild-type cells (▴) and crl21 (pre3–6) (•) mutants.
Other 20S Proteasome Mutants Also Exhibit Cycloheximide Resistance and Temperature Sensitivity
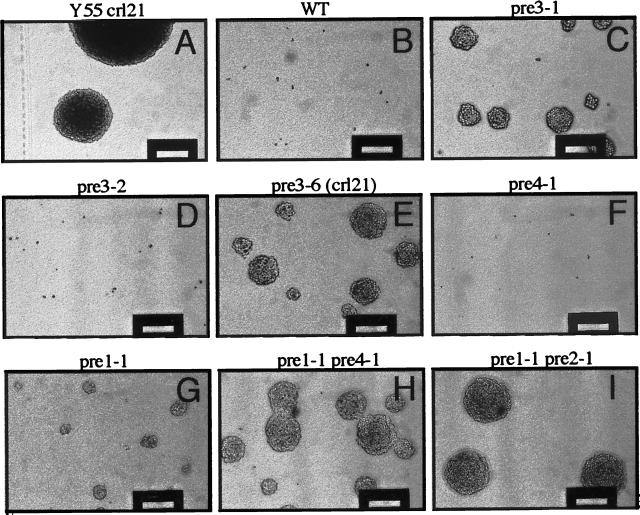
Data obtained with crl3 and crl21 mutants suggested that yeast cells defective in the proteolytic activity of the proteasome are generally resistant to the minimum inhibitory concentration of cycloheximide. To prove this, strains containing various mutations in β-type subunits of the 20S proteasome were tested for growth on agar plates containing 1.5 μg/ml cycloheximide (Figure 7). As expected wild-type cells of strain WCG4a were not able to grow on this medium, whereas the originally isolated crl21 mutant strain Y55crl21 (McCusker and Haber, 1988a) showed normal growth. Interestingly, the mutant strain YRG16 harboring the crl21 (pre3–6) mutation in the genetic background of strain WCG4a showed rather slow growth on cycloheximide medium. However, in contrast to its isogenic wild-type strain (WCG4a), YRG16 mutant cells formed small colonies under these conditions. A similar phenotype was observed for another pre3 mutant allele (pre3–1) which also leads to strong deficiency in proteasome-mediated proteolysis (Gückel, unpublished data). Also pre1–1 mutants carrying a mutation in another 20S proteasome β-type subunit which results in defective chymotrypsin-like activity (Heinemeyer et al., 1991) and stabilization of defined proteasomal substrates (Richter-Ruoff et al., 1992; Heinemeyer et al., 1993) showed significant cycloheximide resistance. For yeast cells with a WCG4a genetic background, the best colony formation on cycloheximide medium was observed for two double mutants strains, pre1–1 pre2–1 (WCG4–1121) and pre1–1 pre4–1 (YHI29/14), which are both strongly defective in proteasome-mediated proteolysis (Richter-Ruoff et al., 1992; Heinemeyer et al., 1993; Hilt et al., 1993; Kornitzer et al., 1994; Schork et al., 1994a). Two other proteasomal mutants containing a different pre3 allele (pre3–2) or a mutation residing in another β-type subunit of the 20S proteasome, pre4–1, were tested. Both mutations had been shown to result in reduced PGPH activity but did not cause any detectable deficiency in protein degradation (Gückel, unpublished data; Hilt et al., 1993). Interestingly, both strains YRG12 (pre3–2) and YHI29/4 (pre4–1) behaved like wild-type cells and were unable to form colonies on medium containing 1.5 μg/ml cycloheximide. These data provide more evidence that resistance against the minimum inhibitory concentration of cycloheximide is a result of proteasomal mutations which cause defects in protein degradation.
Figure 7.
Growth of crl21 and other proteasomal mutants on cycloheximide medium: Cells were plated on YPD agar medium containing 1.5 μg/ml cycloheximide, incubated for 7 d, and colonies photographed using a Zeiss long distance objective (elevation 320×). (A) Original Y55crl21 mutant strain; all other strains are isogenic with WCG4a. (B) YHI29/W wild-type; (C) YRG11 (pre3–1); (D) YRG12 (pre3–2); (E) YRG16 (pre3–6 (crl21)); (F) YHI29/4 (pre4–1); (G) YHI29/1 (pre1–1); (H) YHI29/14 (pre1–1 pre4–1); and (I) WCG4–1121 (pre1–1 pre2–1).
DISCUSSION
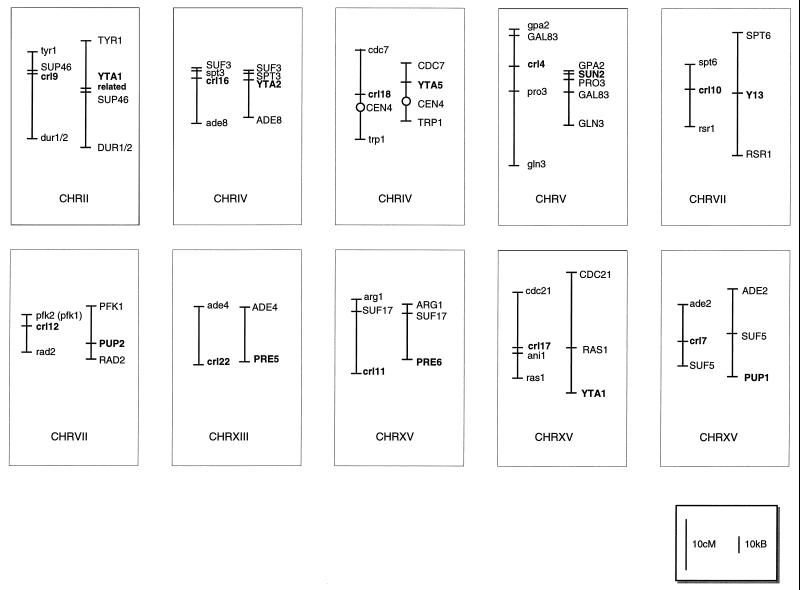
We cloned the CRL3 gene by complementation of temperature sensitivity of the crl3–2 mutant. CRL3 is identical to the SUG1/CIM3 gene which had been shown to code for a 26S proteasome subunit. Additionally, we found that another mutant gene, crl21, is allelic with PRE3 which codes for a β-type subunit of the 20S proteasome. Previously McCusker and Haber (1988b) had found crl13 to be allelic with SUG2, which recently had been proven to code for a 19S cap subunit of the 26S proteasome (Russell et al., 1996). Additionally, we found that certain single and double mutants harboring mutations in β-type subunits of the 20S proteasome which cause strong reduction of proteasomal activity exhibit both cycloheximide resistance and temperature sensitivity. These findings led to the idea that many, if not all, crl mutations isolated (McCusker and Haber, 1988a) may reside in subunits of the proteasome. Therefore, we checked the DNA sequence of chromosomal loci of the other mapped crl mutations (McCusker and Haber, 1988a; Mortimer et al., 1991). In 9 of these 12 cases, genes coding for 26S proteasome subunits were found closely linked to the mapped positions (Figure 8). In six cases the sequence positions of such 26S proteasome genes perfectly matched the mapped positions of crl mutations. In three cases 26S proteasome genes were found very near to the mapped positions (<30 kb away) (McCusker and Haber, 1988a; Mortimer et al., 1991). We conclude that these moderate deviations might be due to mapping errors. In one position a gene was found which codes for a protein with sequence similarity to the 19S cap subunit Yta1.
Figure 8.
Chromosomal map positions of crl mutations and corresponding loci in the yeast genomic sequence: Left maps comprise chromosomal positions of crl mutations as had been determined by genetic mapping, respectively. Right maps comprise corresponding loci as found in the yeast genomic sequence (Source: MIPS database http://www.mips.biochem.mpg.de/). Bars indicate genetic distances (cM) and sequence distances (kb), respectively. crl mutations and predicted corresponding proteasomal genes are indicated by bold letters. Proteasomal genes are characterized as: YTA2, 19S AAA protein; YTA5, 19S AAA protein; SUN2, 19S non-ATPase; Y13, 20S α-type; PUP2, 20S α-type; PRE5, 20S α-type; PRE6, 20S α-type; YTA1, 19S AAA protein; and PUP1, 20S β-type. YTA1 related: not indicated as 19S cap subunit, homologous to YTA1.
The crl3–2 mutation caused significant defects in proteasome-dependent degradation. This has been proven by accumulation of ubiquitinated proteins as well as stabilization of FBPase, a selective substrate of the ubiquitin proteasome pathway (Schork et al., 1994a,b, 1995). The SCL1–1 mutation which functions as an extragenic suppressor of the temperature-sensitive phenotype of crl3 mutants resides in an α-type subunit of the 20S proteasome (McCusker and Haber, 1988a; Balzi et al., 1989). We found that the SCL1–1 mutation leads to significant restoration of proteasomal activity. These data provide additional evidence that Crl3 is a subunit of the proteasomal 19S cap complex. Cloning of the SCL1–1 mutant allele showed that the suppressor mutation is due to an exchange of tyrosine 30 against cysteine. Tyr 30 is entirely conserved in all 20S proteasome α-type subunits known so far. It locates within a highly conserved region near the N terminus of the Scl1 protein (Figure 2). As derived from the 20S proteasome x-ray structure the mutation resides central to the H0-helix of the Scl1 protein. This domain, which locates at the top or bottom of the 20S proteasome cylinder, had been indicated to be responsible for interaction between the 20S core and 19S cap complexes. It had been suggested that the ATPase subunits of the 19S cap complexes form a hetero-oligomeric ring, possibly at the interface to the 20S complex (Lupas et al., 1995). Our results indicate that Crl3 and Scl1 interact physically. One may imagine that the crl3–2 mutation may diminish formation of 26S proteasome complexes by disturbing the 20S core–19S cap interaction. The SCL1–1 mutation would then suppress by reconstituting assembly of the 26S proteasome. An alternative explanation for the suppressor phenotype might be that the crl3–2 mutation disturbs Crl3 activity. Then the SCL1–1 mutation may suppress by restoring Crl3 activity through a neigbouring effect.
We conclude that phenotypes found for crl mutants are due to a defective proteasome. Our data indicate that crl mutations reside in different types of 19S cap as well as 20S core subunits. We therefore assume that these mutations do not impair a specific function of the proteasome but cause a general defect in proteasomal protein degradation. McCusker and Haber (1988a) calculated that there are approximately 50 genes which can mutate to confer a crl phenotype. This number of possible crl genes is in good agreement with the predicted number of genes which could be mutated to cause a defect in proteasomal protein degradation: Mutations will be expected to reside in 14 different 20S proteasomal genes and in at least 20 genes coding for 19S cap subunits. In addition, some crl mutations may reside in defined components of the ubiquitination system.
Proteasome-dependent proteolysis is crucial for many different regulatory pathways (Hilt and Wolf, 1996). Therefore one should expect that proteasomal mutations cause multiple phenotypic effects. Indeed, besides cycloheximide resistance and temperature sensitivity, crl mutants exhibited many additional phenotypes, e.g., incorrect response to starvation conditions, sensitivity against amino acid analogues, hypersensitivity against certain protein synthesis inhibitors, tightening of leaky auxotrophic mutations, and suppression of hygromycin B-suppressible mutations (McCusker and Haber, 1988b).
McCusker and Haber (1988a,b) suggested that crl mutations may cause defects in protein translation. They proposed that crl mutations may reside in ribosomal subunits, but could not easily imagine how mutations in so many different ribosomal proteins could cause such similar phenotypes (McCusker and Haber, 1988b). As a second explanation crl mutations were thought to reside in one or more pathways which may influence ribosome function as for example in pathways responsible for modification of rRNA, ribosomal proteins, or other translation factors (McCusker and Haber, 1988b). However, with the finding that crl mutations reside in structural genes of the proteasome and cause defects in proteasome-dependent protein degradation, many of the observed phenotypes can now be explained without assuming that these mutations directly affect ribosome function or enhance translational misreading. Sensitivity against amino acid analogues and elevated temperatures was found as typical stress phenotypes in mutants deficient in proteasomal activities as well as mutants defective in certain ubiquitination pathways (Seufert and Jentsch, 1990; Heinemeyer et al., 1991; Hilt et al., 1993). These stress conditions caused proteasome mutants to accumulate ubiquitinated proteins (Heinemeyer et al., 1991; Hilt et al., 1993 and see also Figure 4). These findings provide strong evidence that such mutants insufficiently remove abnormal proteins. Rates of formation of abnormal proteins induced by application of amino acid analogues or heat stress most probably do not differ in wild-type and crl mutant cells. Therefore, both phenotypes, sensitivity against amino acid analogues, and heat stress are well explained as a result of insufficient response to such stress conditions. Due to the fact that screening for cycloheximide-resistant and temperature-sensitive mutations yielded so many complementation groups, it had been expected that tightly linked pathways may be affected to induce these phenotypes. However, due to the finding that crl mutations reside in the proteasome, one may imagine that relating to cellular function quite distant pathways may be disturbed to induce these two phenotypes.
Data presented in this work show that cycloheximide resistance depends on defects in protein degradation. Proteasomal mutations (pre3–2 and pre4–1), which only affect peptide-cleaving activities but do not influence degradation of substrate proteins (Hilt et al., 1993; Gückel, unpublished data), did not show any cycloheximide resistance. In contrast, proteasomal mutations (pre1–1, pre1–1 pre2, pre1–1 pre4–1), which due to stabilization of different substrate proteins (Richter-Ruoff et al., 1992; Seufert and Jentsch, 1992; Kornitzer et al., 1994; Richter-Ruoff et al., 1994; Yaglom et al., 1995) had been proven to be defective in protein degradation, caused cycloheximide resistance. Moreover, a clear dependence between strength of cycloheximide resistance and reduction in proteasome-dependent degradation of substrate proteins was observed. Interestingly, proteasomal mutants isogenic with strain WCG4a in contrast to the original crl strains with Y55 background (McCusker and Haber, 1988a) exhibited extremely slow growth. This was even visible when the crl21 mutation was transferred from Y55 to WCG4a background. Strains with Y55 background seem to be quite evolutionary distant from WCG4a. This had been indicated by various conservative bp exchanges found within CRL3/SUG1 scl1+/PRS2 and CRL21/PRE3 loci when sequenced from Y55-derived strains. We therefore suggest that decreased growth rates of mutants with WCG4a strain background are due to some unknown genetic alterations.
How can cycloheximide resistance of proteolysis-defective proteasome mutants be explained? When protein synthesis is restricted due to inhibition with cycloheximide or due to limitation of nutrients, the start of a new cell cycle is prevented and cells stop growth during the G1 phase (Hartwell and Unger, 1977; McCusker and Haber, 1988b). Thus, cells must possess a sensor which enables them to monitor the rate of protein synthesis. We hypothesize that this sensor consists of a protein which is permanently synthesized and in parallel degraded via the proteasome. Application of a minimum inhibitory concentration of cycloheximide will partially block protein synthesis. As in wild-type cells, proteolysis works at normal rates. These conditions will lead to a decrease in the intracellular concentration of the hypothetical sensor protein. Finally, the sensor will drop beyond a critical threshold level which may then be the signal causing cells to stop growth. However, the sensor protein is stabilized when introducing a proteasomal mutation which leads to defects in proteolysis. Thus, even if synthesis is impaired the sensor may reach cellular concentrations which enable cells to continue proliferation. This model would also explain lack of a specific G1 arrest of crl mutants when starving for nutrients. Such conditions may also lead to reduction of protein synthesis and therefore decreased levels of the hypothetical sensor protein. However, due to proteolytic stabilization in proteasome mutants, the sensor may not decline beyond the critical threshold concentration and cells may undergo the G1-S transition. Because crl mutant cells showed hypersensitivity and not resistance against other protein synthesis inhibitors such as anisomycin, hygromycin B, and G418 (McCusker and Haber, 1988b) McCusker and Haber (1988a) suggested that crl mutations may not generally allow initiation of the cell cycle under conditions of limited protein synthesis. This suggestion seems to be contradictory to our proposed model. However, one may imagine that a distinct balancing between reduction of protein synthesis and decrease of protein degradation may be a prerequisite for resistance to occur. Appropriate reduction of protein synthesis may only be reached when using defined concentrations of cycloheximide but not when applying other kinds of inhibitors. Some evidence for this explanation is provided by the fact that for another protein synthesis inhibitor, cryptopleurine, eight crl mutants were found to be resistant (McCusker and Haber, 1988b). Moreover, as it is known for hygromycin B, such inhibitors may strongly induce translational misreading. Therefore, when using such inhibitors, the ability of crl mutants to divide under protein synthesis-limiting conditions may be superimposed by a growth defect caused by the inability of these mutant cells to adequately respond to the formation of abnormal proteins. crl mutants share several phenotypes with gcn (general control nonderepressing) mutants (McCusker and Haber, 1988b): they are hypersensitive against the histidine biosynthesis inhibitor 3-aminotriazol and they also tighten leaky auxotrophic mutations. Therefore, like gcn mutants, crl mutants are thought to fail in derepression of amino acid and purine biosynthetic pathways. We obtained additional evidence for a failure in derepression of biosynthetic pathways when crl3–2 mutant cells, which additionally contained an ade1 mutation, were grown to stationary phase. Due to the block in adenine biosynthesis under such derepression conditions, ade1 CRL3 “wild-type” cells (strain Y55–1162 containing a CRL3 plasmid; Figure 1) accumulate a red dye. However, most probably due to the failure in derepression of the adenine synthesis pathway, accumulation of this red dye is significantly diminished in crl3 mutant cells (strain Y55–1162 containing the control plasmid pRS316; Figure 1). Interestingly, a clear link between the proteasome and Gcn4 was uncovered (Kornitzer et al., 1994). This transcriptional activator for general amino acid and purine synthesis control is degraded via the ubiquitin proteasome pathway. As a response to starvation, this Gcn4 inactivation process is slowed down (Kornitzer et al., 1994). Therefore, one may expect that proteolytic stabilization of Gcn4 in proteasome mutants results in enhanced and not reduced derepression of biosynthetic pathways. We speculate that proteasomal degradation defects may cause some failure in the starvation response which may superimpose the gain of Gcn4 function.
Interestingly, crl mutations have been found to suppress a limited spectrum of hygromycin B-suppressible amino acid auxotrophic mutations (McCusker and Haber, 1988b). These hygromycin B-suppressible auxotrophic mutations are nonsense mutations that cause expression of truncated proteins. In certain cases, due to their structural defects, these products may be rapidly removed via proteasomal degradation. However, if such truncated proteins exhibit some residual enzymatic activity, suppression observed may be explained by proteolytic stabilization occurring in proteasome mutants.
Most phenotypes found in crl mutants can be explained by a distinct effect: for instance, insufficient degradation of false proteins or stabilization of some defined regulatory protein. However, defects in proteasome function may cause disturbance in many different cellular pathways which themselves may be interconnected. Therefore, some phenotypes found in proteasomal mutants may be a consequence of complex dissarrangements occurring in various cellular pathways.
Table 1.
S. cerevisiae strains used
| Strain | Genotype | Origin |
|---|---|---|
| Y55-1161a | HO crl3-2 SCL1-1 ade1-1 ura3-1 gal3 MAL1 SUC1 | McCusker and Haber, 1988a |
| Y55-1162a | HO crl3-2 ade1-1 ura3-1 gal3 MAL1 SUC1 | McCusker and Haber, 1988a |
| Y55-1163a | HO ade1-1 ura3-1 gal3 MAL1 SUC1 | McCusker and Haber, 1988a |
| YUM47a | HO crl3-2 ade1-1 ura3-1 gal3 MAL1 SUC1 [SCL1-1URA3 CEN14] | Derivative of Y55-1162, this work |
| YUM48a | HO crl3-2 ade1-1 ura3-1 gal3 MAL1 SUC1 [scl1+ URA3 CEN14] | Derivative of Y55-1162, this work |
| YUM60a | HO crl3-2 ade1-1 ura3-1 gal3 MAL1 SUC1 [PSCL1-1::scl1+ URA3 CEN14] | Derivative of Y55-1162, this work |
| YUM61a | HO crl3-2 ade1-1 ura3-1 gal3 MAL1 SUC1 [Pscl1+::SCL1-1 URA3 CEN14] | Derivative of Y55-1162, this work |
| Y55crl21a | HO crl21 ura3-1 gal3 MAL1 SUC1 | McCusker and Haber, 1988a |
| WCG4a | MATa ura3 leu2-3,112 his3-11,15 Cans Gal+ | Heinemeyer et al., 1993 |
| YRG8b | MATa/MATα PRE3/pre3Δ::URA3 ura3/ura3 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 Cans/Cans Gal+/Gal+ | Gückel, Hilt |
| YRG11b | MATa pre3-1 ura3 leu2-3,112 his3-11,15 Cans Gal+ | Gückel, Hilt |
| YRG12b | MATa pre3-2 ura3 leu2-3,112 his3-11,15 Cans Gal+ | Gückel, Hilt |
| YRG16b | MATa pre3-6 (crl21) ura3 leu2-3,112 his3-11,15 Cans Gal+ | This work |
| YHI29Wb | MATa ura3 leu2-3,112 his3-11,15 Cans Gal+ | This work |
| YHI29/1b | MATα pre1-1 ura3 leu2-3,112 his3-11,15 Cans Gal+ | This work |
| YHI29/4b | MATα pre4-1ura3 leu2-3,112 his3-11,15 Cans Gal+ | This work |
| YHI29/14b | MATa pre1-1 pre4-l ura3 leu2-3,112 his3-11,15 Cans Gal+ | This work |
| WCG4-1121b | MATa pre1-1 pre2-1ura3 leu2-3,112 his3-11,15 Cans Gal+ | Heinemeyer et al., 1993 |
These strains are congenic with strain Y55.
These strains are isogenic with WCG4a Heinemeyer et al., 1993.
ACKNOWLEDGMENTS
The authors thank J.E. Haber for providing strains. We also thank C. Mann for friendly reception and support of UMG during his stay in Gif-sur-Yvette. Thanks also to W. Heinemeyer for reading the manuscript. We thank the Deutsche Akademische Austauschdienst, Bonn, for providing a Programme de Coopération Scientifique grant for scientific exchange. This work was supported by the German Israeli Foundation, Jerusalem.
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Greene Publishing Association Wiley; 1990. [Google Scholar]
- Balzi E, Chen WN, Capieaux E, McCusker JH, Haber JE, Goffeau A. The suppressor gene scl1+ of Saccharomyces cerevisiae is essential for growth (published erratum appears in Gene 89, 151, 1990) Gene. 1989;83:271–279. doi: 10.1016/0378-1119(89)90113-3. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- De Marini DJ, Papa FR, Swaminathan S, Ursic D, Rasmussen TP, Culbertson MR, Hochstrasser M. The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent protein degradation in vivo. Mol Cell Biol. 1995;15:6311–6321. doi: 10.1128/mcb.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. Make it or break it: the role of ubiquitin-dependent proteolysis in cellular regulation. Trends Cell Biol. 1995;5:428–434. doi: 10.1016/s0962-8924(00)89102-3. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Emori Y, Tsukahara T, Kawasaki H, Ishiura S, Sugita H, Suzuki K. Molecular cloning and functional analysis of three subunits of yeast proteasome. Mol Cell Biol. 1991;11:344–353. doi: 10.1128/mcb.11.1.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C, Lehmann H, Kipper J, Guckel R, Hilt W, Wolf DH. PRE3, highly homologous to the human major histocompatibility complex- linked LMP2 (RING12) gene, codes for a yeast proteasome subunit necessary for the peptidylglutamyl-peptide hydrolyzing activity. FEBS Lett. 1994;341:193–196. doi: 10.1016/0014-5793(94)80455-9. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Orino E, et al. Proteasomes are essential for yeast proliferation. cDNA cloning and gene disruption of two major subunits. J Biol Chem. 1990;265:16604–16613. [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jeans A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Unger MW. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol. 1977;75:422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272:25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity - Mutations link the proteasome to stress-dependent and ubiquitin-dependent proteolysis. J Biol Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- Hilt W, Wolf DH. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- Kim YJ, Björklund S, Li Y, Sayrem MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters J-M, Kirschner MW. How proteolysis drives the cell cycles. Science. 1996;274:1652–1658. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kominami KI, Toh-e A. Characterizing of the NIN1 gene product of Saccharomyces cerevisiae. Exp Cell Res. 1994;211:203–211. doi: 10.1006/excr.1994.1079. [DOI] [PubMed] [Google Scholar]
- Kornitzer D, Raboy B, Kulka RG, Fink GR. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Stock D, Jap B, Zwick P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at a 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Lupas A, Zwickl P, Wenzel T, Seemüller E, Baumeister W. Structure and function of the 20S proteasome and of its regulatory complexes. Cold Spring Harbor Symp Quant Biol. 1995;60:515–524. doi: 10.1101/sqb.1995.060.01.055. [DOI] [PubMed] [Google Scholar]
- McCusker JH, Haber JE. Cycloheximide-resistant temperature-sensitive lethal mutations of Saccharomyces cerevisiae. Genetics. 1988a;119:303–315. doi: 10.1093/genetics/119.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker JH, Haber JE. crl mutants of Saccharomyces cerevisiae resemble both mutants affecting general control of amino acid biosynthesis and omnipotent translational suppressor mutants. Genetics. 1988b;119:317–327. doi: 10.1093/genetics/119.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Schild D, Contopoulou CR, Kans JA. Genetic and physical maps of Saccharomyces cerevisiae. Methods Enzymol, 1991;194:827–863. doi: 10.1016/0076-6879(91)94060-p. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Peters JM. Proteasomes: protein degradation machines of the cell. Trends Biochem Sci. 1994;19:377–382. doi: 10.1016/0968-0004(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Richter-Ruoff B, Heinemeyer W, Wolf DH. The proteasome/multicatalytic-multifunctional proteinase. In vivo function in the ubiquitin-dependent N-end rule pathway of protein degradation in eukaryotes. FEBS Lett. 1992;302:192–196. doi: 10.1016/0014-5793(92)80438-m. [DOI] [PubMed] [Google Scholar]
- Richter-Ruoff B, Wolf DH, Hochstrasser M. Degradation of the yeast MAT alpha 2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 1994;354:50–52. doi: 10.1016/0014-5793(94)01085-4. [DOI] [PubMed] [Google Scholar]
- Rose MD, Broach JR. Cloning genes by complementation in yeast. Methods Enzymol, 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement and allelle rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rubin DM, Coux O, Wefes I, Hengartner C, Young RA, Goldberg AL, Finley D. Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature. 1996;379:655–657. doi: 10.1038/379655a0. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Sathyanarayana UG, Johnston SA. Isolation and characterization of SUG2. A novel ATPase family component of the yeast proteasome. J Biol Chem. 1996;271:32810–32817. doi: 10.1074/jbc.271.51.32810. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall R, Mannhaupt G, Stucka R, Tauer R, Ehnle S, Schwarzlose C, Vetter I, Feldmann H. Identification of a set of yeast genes coding for a novel family of putative ATPases with high similarity to constituents of the 26S protease complex. Yeast. 1994;10:1141–1155. doi: 10.1002/yea.320100903. [DOI] [PubMed] [Google Scholar]
- Schork SM, Bee G, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase in yeast is mediated by the proteasome. FEBS Lett. 1994a;349:270–274. doi: 10.1016/0014-5793(94)00668-7. [DOI] [PubMed] [Google Scholar]
- Schork SM, Bee G, Thumm M, Wolf DH. Site of catabolite inactivation. Nature. 1994b;369:283–284. doi: 10.1038/369283a0. [DOI] [PubMed] [Google Scholar]
- Schork SM, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W, Jentsch S. In vivo function of the proteasome in the ubiquitin pathway. EMBO J. 1992;11:3077–3080. doi: 10.1002/j.1460-2075.1992.tb05379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:19–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hick JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Shilo B, Riddle VG, Pardee AB. Protein turnover and cell-cycle initiation in yeast. Exp Cell Res. 1979;123:221–227. doi: 10.1016/0014-4827(79)90462-2. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffield JC, Bromberg JF, Johnston SA. Alterations in a yeast protein resembling HIV Tat-binding protein relieve requirement for an acidic activation domain in GAL4. Nature. 1992;357:698–700. doi: 10.1038/357698a0. [DOI] [PubMed] [Google Scholar]
- Swaffield JS, Melcher K, Johnston SA. A highly conserved ATPase protein as a mediator between acidic activation domains and the TATA-binding protein. Nature. 1995;374:88–91. doi: 10.1038/374088a0. [DOI] [PubMed] [Google Scholar]
- van Nocker S, Deveraux Q, Rechsteiner M, Vierstra RD. Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc Natl Acad Sci USA. 1996;93:856–860. doi: 10.1073/pnas.93.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Maundrell K, Heyer WD, Beach D, Nurse P. Vectors for the construction of gene banks and the integration of cloned genes in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Plasmid. 1986;15:156–158. doi: 10.1016/0147-619x(86)90051-x. [DOI] [PubMed] [Google Scholar]
- Xu Q, Singer RA, Johnston GC. Sug1 modulates yeast transcription activation by Cdc68. Mol Cell Biol. 1995;15:6025–6035. doi: 10.1128/mcb.15.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom J, Linskens MHK, Sadis S, Rubin DM, Futcher B, Finley D. p34CDC28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]