Summary
Proteus mirabilis alternates between motile and adherent forms. MrpJ, a transcriptional regulator previously reported to repress motility, is encoded at the 3' end of the mrp fimbrial operon in P. mirabilis. Sequencing of the P. mirabilis genome revealed 14 additional paralogs of mrpJ, ten of which are associated with fimbrial operons. Twelve of these genes, when overexpressed, repressed motility; several distinct patterns of swarming motility were noted. Expression of 10 of the 14 mrpJ paralogs repressed flagellin (FlaA) synthesis. Alignment of the predicted amino acid sequences of MrpJ and its fourteen paralogs revealed a conserved consensus motif (SQQQFSRYE) within the helix-turn-helix domain. Site-directed mutagenesis of these residues coupled with linker insertion mutagenesis of MrpJ confirmed the importance of this domain for repression of motility. Gel shift assays demonstrated that MrpJ and another paralog UcaJ bind directly to the promoter region of the flagellar master regulator flhDC. Thus, P. mirabilis appears to use a related mechanism to inhibit motility during the production of at least ten of its predicted fimbriae.
Keywords: fimbria, flagella, motility, Proteus mirabilis, swarm
Introduction
Motility and attachment represent opposing aspects of the life cycles of many bacterial species, yet both contribute to bacterial fitness and virulence in the host. As might be expected, both systems are subject to complex regulatxion. Bacterial motility is mediated by the flagellum, and attachment by fimbriae (Finlay and Falkow, 1997). However, only a few regulatory networks have been found that simultaneously increase fimbrial expression while decreasing flagellum-mediated motility, or vice versa. These include the bvgAS system of Bordetella species (Akerley et al., 1995), the messenger cyclic di-GMP (Simm et al., 2004), fimZ expression in Salmonella enterica serovar Typhimurium (Clegg and Hughes, 2002), and motility feedback into the ToxR regulatory system in Vibrio cholerae (Gardel and Mekalanos, 1996). Constitutive expression of type 1 fimbriae in uropathogenic Escherichia coli has also been shown to decrease swimming motility (Bryan et al., 2006; Lane et al., 2007). Another strategy for decreasing motility while fimbriae are expressed is mediated by mrpJ, a gene located at the end of the MR/P fimbrial operon which when overexpressed, decreases flagellum-mediated motility in Proteus mirabilis (Li et al., 2001).
P. mirabilis, a member of the Enterobacteriaceae and an opportunistic pathogen of the urinary tract, frequently infects patients with indwelling urinary catheters (reviewed in (Coker et al., 2000)). This organism has long been studied for its ability to swarm across solid surfaces (reviewed in (Rather, 2005)), during which short, vegetative swimmer cells convert to very long (up to 100 μm), hyperflagellated swarmer cells. These swarmer cells also exhibit reduced expression of fimbriae (Jansen et al., 2003; Mobley et al., 1996), suggesting possible coordinate expression between flagella and fimbriae. Prior to the sequencing of the P. mirabilis genome, expression of five different fimbriae had been documented in P. mirabilis: mannose-resistant Proteus-like (MR/P) (Adegbola et al., 1983; Bahrani et al., 1994), uroepithelial cell adhesin (UCA, also known as nonagglutinating fimbriae or NAF) (Cook et al., 1995; Wray et al., 1986), Proteus mirabilis fimbria (PMF) (Adegbola et al., 1983; Massad et al., 1994b), ambient temperature fimbria (ATF) (Massad et al., 1994a), and P. mirabilis P-like pili (PMP) (Bijlsma et al., 1995). The MR/P fimbria is encoded by an operon containing nine genes (Bahrani et al., 1994). The last gene in the mrp operon is mrpJ, which encodes a helix-turn-helix XRE-family transcriptional regulator (Li et al., 2001).
Since our initial report of MrpJ control of both swimming and swarming motility and its functional homolog papX in E. coli (Li et al., 2001), mrpJ homologs have been found associated with fimbrial operons in Xenorhabdus nematophila (He et al., 2004) and Photorhabdus temperata (Meslet-Cladiere et al., 2004), although the phenotype of these mrpJ homologs on motility has not yet been established. However, the recent completion of the P. mirabilis genome (Pearson et al., 2008) predicted that this organism has fourteen additional mrpJ paralogs, most of which (10 of 14) are also associated with fimbrial operons. This report details the role of all fifteen P. mirabilis mrpJ-like genes in both swimming and swarming motility, and presents data on the mechanism of MrpJ action.
Results
P. mirabilis HI4320 has 14 additional mrpJ homologs
The recent annotation of the P. mirabilis genome (Pearson et al., 2008) revealed 17 potential chaperone-usher fimbrial operons. Surprisingly, 10 of these 17 operons contain an mrpJ paralog either at the beginning (5 paralogs) or the end (5 paralogs) of the operon (Table II). One operon, fimbria10, has two mrpJ paralogs. A further scan of the genome located four additional mrpJ paralogs that do not appear to be associated with fimbrial genes (denoted as orphans in Table II).
Table 2.
mrpJ paralogs in P. mirabilis HI4320
| Name | Locus | Fimbrial operon |
|---|---|---|
| mrpJ′ | PMI0261 | MR/P′ |
| mrpJ | PMI0271 | MR/P (mannose resistant/Proteus-like) |
| fim3J | PMI0296 | Fimbria 3 |
| ucaJ | PMI0532 | UCA (uroepithelial cell adhesin) |
| fim5J | PMI1060 | Fimbria 5 |
| fim8J | PMI1470 | Fimbria 8 |
| fim10J1 | PMI2209 | Fimbria 10 |
| fim10J2 | PMI2207 | Fimbria 10 |
| pmpJ | PMI2224 | PMP (P. mirabilis P-like pili) |
| atfJ | PMI2733 | ATF (ambient temperature fimbria) |
| fim14J | PMI3003 | Fimbria 14 |
| PMI0182 | PMI0182 | orphan |
| PMI0982 | PMI0982 | orphan |
| PMI1817 | PMI1817 | orphan |
| PMI3508 | PMI3508 | orphan |
The majority of the mrpJ paralogs repress motility when overexpressed
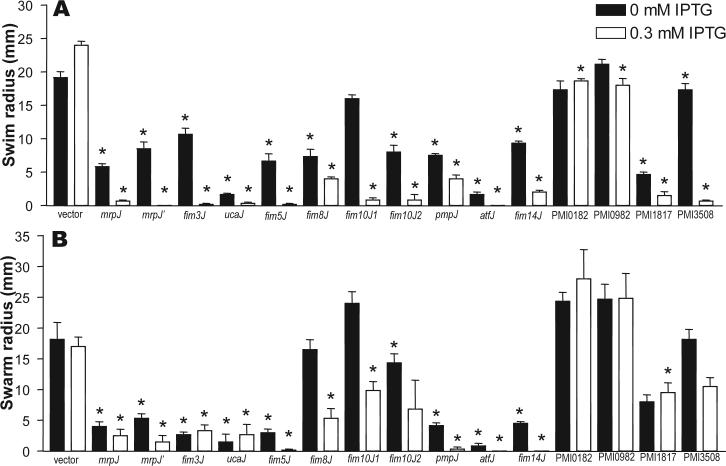
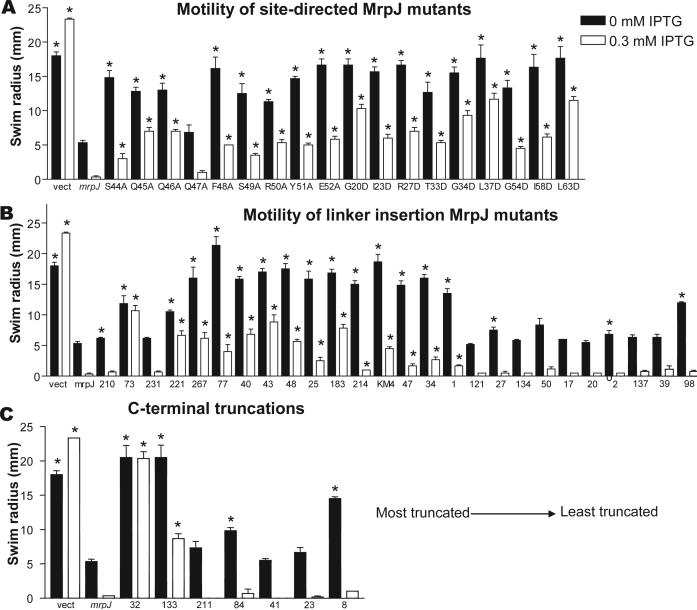
Each of the 14 mrpJ paralogs was cloned into the IPTG-inducible plasmid vector pLX3607. P. mirabilis HI4320 was electroporated with the mrpJ paralog clones, and flagellar motility was examined (Fig. 1). Strain HI4320 overexpressing mrpJ or its paralogs was stabbed into soft agar to measure swimming motility (Fig. 1A). The IPTG-inducible promoter in pLX3607 drives a low level of expression in the absence of IPTG; this level of mrpJ paralog expression was sufficient to significantly reduce swimming motility in 11 of the 14 mrpJ paralog overexpression strains (P<0.05). When expression was induced with 0.3 mM IPTG, 12 of the 14 mrpJ paralog overexpression strains were less motile compared to the induced vector control (P<0.05) (Fig. 1A). Although statistically significant, the slight reduction of motility for the PMI0182 and PMI0982 overexpression strains in the presence of IPTG when compared to the induced vector control is likely not biologically significant.
Figure 1.
Swimming and swarming motility of P. mirabilis when mrpJ or its paralogs are overexpressed. (A) swimming radius of strains stabbed in soft agar; (B) swarming radius of strains spotted on swarm agar. In both panels A and B, black bars represent the swim or swarm radius of uninduced strains, while white bars represent strains that were induced with IPTG. Error bars represent the standard error of the mean. Asterisks indicate P < 0.05 when compared to the vector control.
To measure the effect of mrpJ paralog overexpression strains on swarming across surfaces, cultures of these strains were spotted onto the center of swarm agar plates. Eight of the 14 strains displayed reduced swarming motility compared to the vector control without IPTG induction (P<0.05) (Fig. 1B). When overexpression was induced with IPTG, ten of the 14 strains (i.e., two additional paralogs) were significantly less motile than the vector (P<0.05).
Overexpression of most mrpJ paralogs reduces expression of flagellin in broth culture
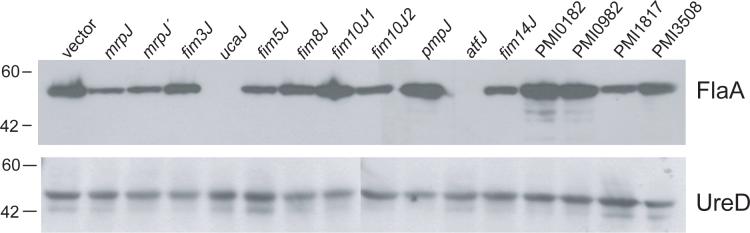
Overexpression of mrpJ causes a reduction in the level of the major flagellin FlaA in P. mirabilis (Li et al., 2001). To test whether this was the case for the other 14 mrpJ paralogs in P. mirabilis HI4320, whole cell lysates of stationary-phase HI4320 expressing each of these paralogs in plasmid pLX3607 were analyzed by Western blot for FlaA expression (Fig. 2). Overexpression of mrpJ paralogs resulted in a wide range of FlaA expression, with most having visibly less FlaA expression. The fim10J1, pmpJ, PMI0182 and PMI0982 overexpression constructs, however, had FlaA expression levels similar to the vector control. The same alterations in FlaA expression were observed when Western blot analysis of the strains shown in Fig. 3 was conducted on mid-logarithmic phase cultures (data not shown).
Figure 2.
Western blot of flagellin expression in P. mirabilis mrpJ or mrpJ paralog overexpression strains. Whole cell lysates of uninduced strains were denatured, electrophoresed on 10% SDS-PAGE, and blotted with anti-FlaA antibody, which recognizes the major subunit of the flagellum. Lysates were also blotted with anti-UreD antibody as a loading control (lower panel). Molecular weight markers are indicated on the left side in kDa.
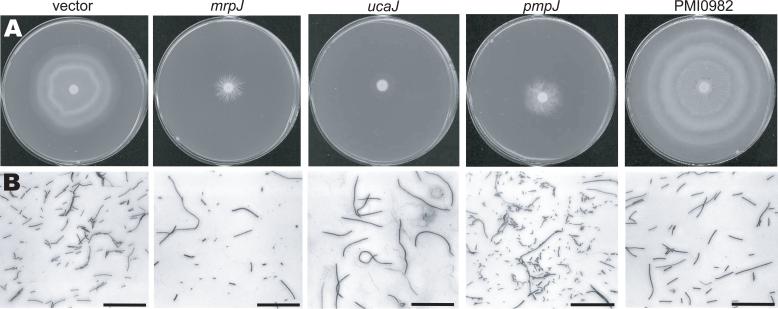
Figure 3.
(A), representative swarming phenotypes of strain HI4320 expressing mrpJ paralogs. (B), Gram stains of bacteria taken from the edge of swarm fronts. The reference bar is 50μm.
Overexpression of some mrpJ paralogs inhibits normal swarm cell differentiation
In addition to reduced overall motility, at least four different swarm patterns were observed in HI4320 overexpressing mrpJ or mrpJ paralogs (Fig. 3). Not only did some strains fail to exhibit the classic “bulls-eye” swarming pattern of P. mirabilis, but radial growth (mrpJ), thin films (ucaJ), or disorganized swarming (pmpJ) was observed. In contrast, overexpression of PMI0982 resulted in swarm rings of larger diameter as compared to the vector control (Fig. 3A).
To investigate whether these strains were not swarming due to an inability to differentiate into swarmer cells, Gram stains were prepared from the edge of swarm fronts and imaged by light microscopy (Fig. 3B). P. mirabilis from the edge of the vector control or the PMI0982 swarm had differentiated into swarmer cells of various lengths. Strains overexpressing mrpJ and pmpJ were found to have mixed populations of normal-appearing swarm cells and much shorter cells. Surprisingly, the strain overexpressing ucaJ was a uniform population of very long swarmer cells (Fig. 3B), although this strain exhibits almost no swarming motility (Fig. 3A).
Overexpression of mrpJ or mrpJ paralogs results in altered fimbrial and flagellar structures
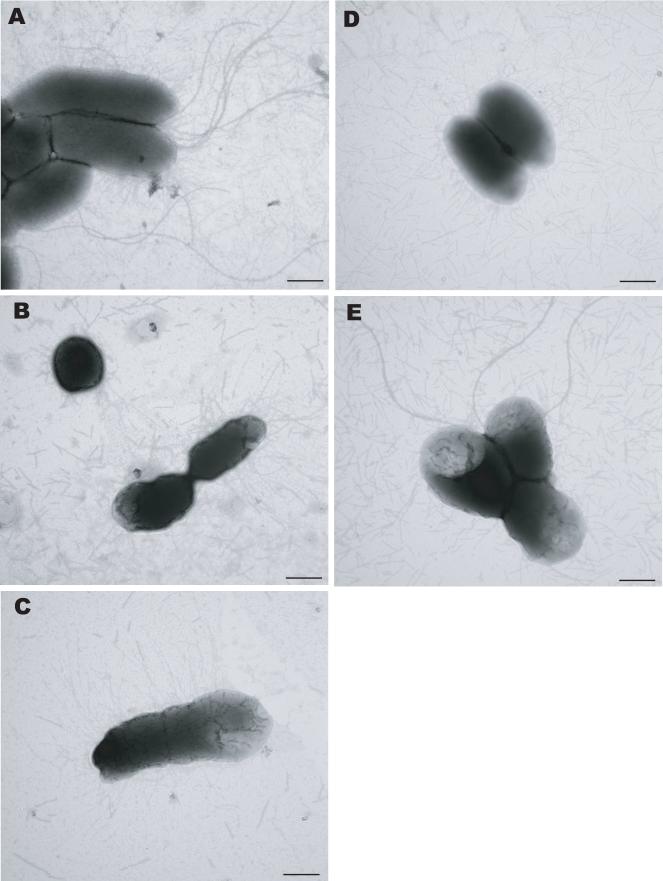
To examine the effect of overexpression of mrpJ or mrpJ paralogs on individual bacteria in a population, overnight cultures of the strains assessed in Fig. 3 were prepared for transmission electron microscopy and representative fields were selected. As expected, overexpression of mrpJ or ucaJ led to a marked decrease in the number of bacteria that possessed flagella (Fig. 4B and 4C). However, although the strain that overexpressed pmpJ had apparently normal levels of FlaA flagellin (Fig. 2), most bacteria in the population lacked a flagellum (Fig. 4D). In contrast, many of the vector control or PMI0982 overexpression bacteria expressed intact flagella (Fig. 4A and 4E).
Figure 4.
Transmission electron microscopy of P. mirabilis strains overexpressing mrpJ paralogs. Flagella are the long, whip-like structures seen in (A) and (E), while fimbriae are the shorter, thinner structures visible in all five panels. (A), empty vector control; (B), mrpJ; (C), ucaJ; (D), pmpJ; (E), PMI0982.
All five of the strains examined by TEM expressed abundant fimbriae (Fig. 4). Many of these fimbriae were seen as sheared fragments in the background. Intriguingly, the appearance of individual fimbriae varied between the different overexpression strains. Specifically, the mrpJ overexpression strain possessed relatively short, thick fimbriae (Fig. 4B) while the fimbriae on the ucaJ overexpression strain (Fig. 4C) were predominantly long, thin, and straight compared to the vector control.
The predicted helix-turn-helix region of MrpJ is required for MrpJ function
To locate the amino acid residues of MrpJ critical for its function, MrpJ and the predicted sequences of the other 14 paralogs were aligned using the CLUSTALW algorithm (Fig. 5A). Three residues are conserved in all 15 proteins; in MrpJ these are R27, Q46, and E52. An additional 20 amino acids are conserved in at least 9 of 15 MrpJ-type proteins encoded by HI4320 (Fig. 5A, orange and green residues in the consensus sequence). A core region of conserved amino acids in MrpJ, SQQQFSRYE (Fig. 5B, green residues), was identified as a likely critical motif within this protein. Each of these MrpJ amino acids was substituted with Ala using site-directed mutagenesis of the mrpJ overexpression plasmid pLX3805. All site-directed mutants had significantly increased swimming motility compared to the mrpJ control (P<0.05) except for the Q47A mutant (Fig. 6A), confirming the importance of these amino acids for the function of MrpJ. The nine additional conserved amino acid residues located outside the core conserved region (Fig. 5B, blue residues) were changed to Asp residues and examined for swimming motility (Fig. 6A). All nine mutants had significantly increased swimming motility compared to the vector control (P<0.05).
Figure 5.
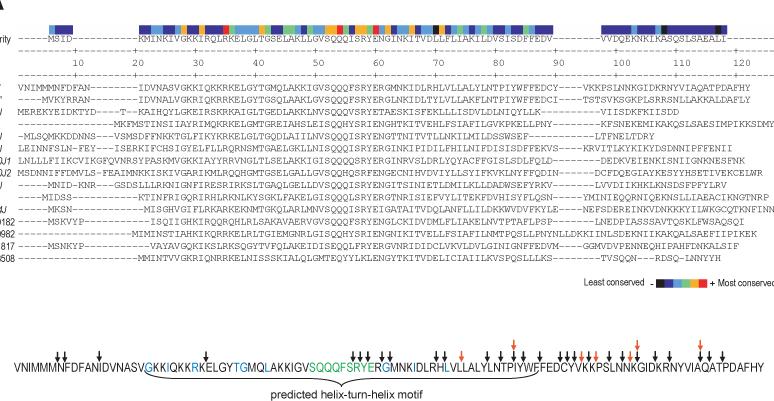
Identification and mutation of conserved amino acid residues of MrpJ. (A) Alignment of MrpJ and the fourteen P. mirabilis MrpJ paralogs. The CLUSTALW algorithm was used to align the amino acid sequences of MrpJ with its paralogs. The consensus sequence is shown in the first line. The degree of amino acid conservation is shown at the top, with highly conserved amino acids indicated in red or orange. (B) Location of mutations in MrpJ. Site-directed mutations in amino acids conserved amongst MrpJ and its paralogs are highlighted in green (Ala substitution) and blue (Asp substitution). The locations of 5-aa in-frame insertions are indicated by black arrows, while 5-aa insertions that result in premature stop codons are indicated by red arrows. The predicted helix-turn-helix domain of MrpJ is indicated with a bracket.
Figure 6.
Swimming motility of HI4320ΔmrpJ overexpressing mutated forms of mrpJ. MrpJ mutants were stabbed into soft agar and the radius of motility was measured. (A) amino acids conserved in MrpJ and its paralogs were substituted with Ala or Asp. (B) transposon mutagenesis was used to introduce random 5-aa insertions in MrpJ. Mutations are listed in order from N-terminal to C-terminal. (C) random insertions that resulted in premature stop codons in mrpJ. Mutants are listed from most to least truncated. In all panels, black bars represent swarm or swim radii of uninduced strains, while white bars represent strains that were induced with IPTG. Asterisks (*) indicate P<0.05 when compared to the mrpJ control. Error bars represent the standard error of the mean.
To further localize regions of MrpJ necessary for function, 15-nt linkers were inserted randomly into mrpJ. Twenty-eight unique in-frame insertion mutants (Fig. 5B, black arrows; see also supplementary figure S1) were measured for swimming motility (Fig. 6B). Mutations within a region corresponding to the predicted helix-turn-helix domain of MrpJ (Fig. 5B) had the greatest effect on swimming motility (Fig. 6B, mutant 221 to mutant 1). Seven mrpJ mutants with insertions that result in premature stop codons (Fig. 5B, red arrows) were also examined for swimming motility (Fig. 6C).
Deletion of the C-terminal 27 amino acids of 110-amino acid MrpJ (Fig. 6C and S1, mutant 211) had no effect on MrpJ function. Deletion of the C-terminal 37 amino acids of MrpJ (Fig. 6C and S1, mutant 133) resulted in a partial loss of MrpJ function (Fig. 6C, compare uninduced and induced expression of mutant 133); only when the C-terminal 45 amino acids (41% of MrpJ) were removed (Fig. 6C and S1, mutant 32) was the MrpJ protein unable to function.
MrpJ binds the promoter region of flhDC
FlhD2C2 is the master regulator of the flagellar operon (Macnab, 1996). To determine whether MrpJ controls motility by regulating FlhD2C2, the promoter region of flhDC was amplified by PCR, labeled, and used as a target in electrophoretic mobility shift assays (Fig. 7A). Addition of MrpJ-His6 to the flhDC promoter (Fig. 7A, lanes 3−7) resulted in a retardation of the mobility of the DNA compared to the promoter alone. In contrast, there was no mobility shift when MrpJ-His6 was added to the flhD coding sequence (Fig. 7A, lanes 1−2).
Figure 7.
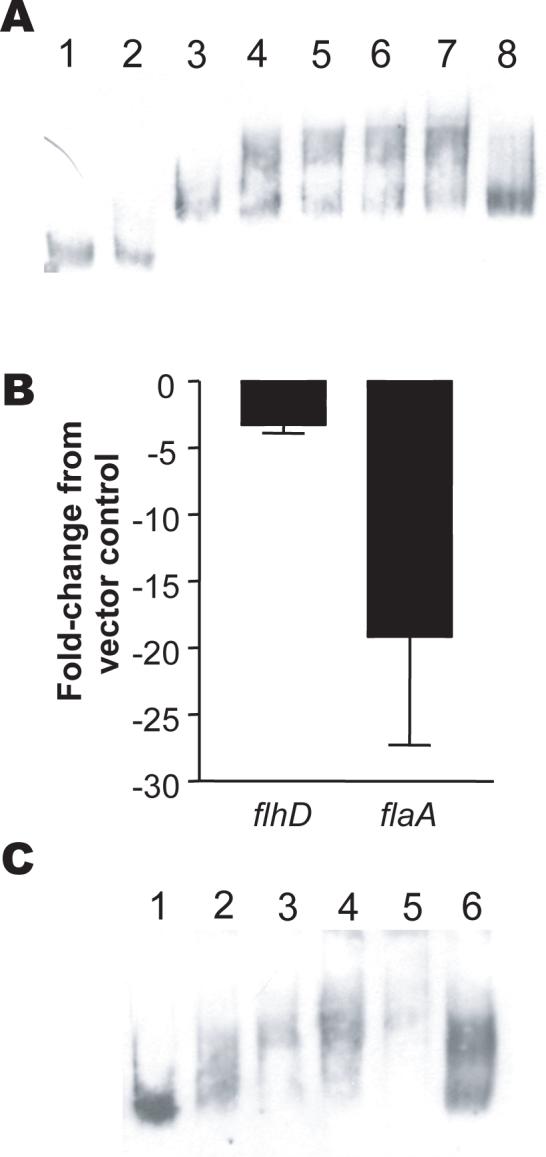
MrpJ directly regulates flhDC expression. (A) Gel shift of the flhDC promoter by MrpJ-His6. MrpJ-His6 was added to either the DIG-labeled P. mirabilis flhD coding sequence or the putative flhDC promoter sequence; DNA-protein interactions were detected by retardation of electrophoretic mobility of DNA relative to unbound DNA in a native polyacrylamide gel. Lanes: 1, flhD coding sequence DNA (0.4ng) only; 2, flhD DNA plus 320ng MrpJ-His6; 3, flhD promoter sequence DNA (0.4ng) only; 4, DNA plus 40ng MrpJ-His6; 5, DNA plus 80ng MrpJ-His6; 6, DNA plus 160ng MrpJ-His6; 7, DNA plus 320ng MrpJ-His6; 8, DNA plus 160ng MrpJ-His6 plus 200-fold excess unlabeled DNA. (B) effect of mrpJ overexpression on flagellar master regulator flhD and flagellin flaA transcripts. qRT-PCR was used to compare transcript levels of flhD and flaA in HI4320 overexpressing mrpJ with the HI4320 vector control. The Y-axis shows the fold-change of flhD or flaA transcripts in the mrpJ overexpression strain compared to the vector control. The data are from three independent experiments. Error bars represent the standard error of the mean. (C) Gel shift of the flhDC promoter by UcaJ-His6. UcaJ-His6 was added to the DIG-labeled P. mirabilis putative flhDC promoter sequence. Lanes: 1, flhD promoter sequence DNA (0.4ng) only; 2, DNA plus 150ng UcaJ-His6; 3, DNA plus 300ng UcaJ-His6; 4, DNA plus 600ng UcaJ-His6; 5, DNA plus 1200ng UcaJ-His6; 6, DNA plus 600ng UcaJ-His6 plus 200-fold excess unlabeled DNA.
To confirm that MrpJ regulates flhDC, quantitative real-time PCR was used to measure flhD and flaA (flagellin) transcript levels in the presence or absence of the mrpJ overexpression plasmid pLX3805 (Fig. 7B). Indeed, overexpression of mrpJ led to an average 3.3-fold decrease in flhD transcript and an average 19.1-fold decrease in flaA transcript (Fig. 7B).
To test whether other MrpJ paralogs might bind the flhDC promoter, UcaJ-His6 was purified and added to the flhDC promoter (Fig. 7C). As expected, addition of increasing amounts of UcaJ-His6 to the flhDC promoter led to a corresponding decrease in DNA mobility compared to the promoter alone.
Discussion
The fact that P. mirabilis HI4320 possesses 15 copies of mrpJ demonstrates that this bacterium is committed to tightly regulating expression of flagella for motility versus fimbriae for adherence. MrpJ and its homologs are transcriptional regulators of the bacterial XRE (xenobiotic response element) family, defined by the presence of a DNA-binding helix-turn-helix domain (HTH). Alignment of the predicted protein sequences of all 15 mrpJ-type genes in P. mirabilis revealed that the conserved amino acid consensus sequence motif within these proteins is consistently located within the HTH domain. Mutation of individual conserved amino acid residues, coupled with data from linker insertion mutagenesis of mrpJ, confirmed the critical contribution of the HTH domain in MrpJ function. This family of regulators likely mediates repression of motility by direct binding to the flagellar master regulator FlhD2C2 resulting in repression of its transcription and the downregulation of flagellar synthesis. When any of the ten fimbrial operons that carry an active MrpJ paralog are expressed, motility will be dramatically downregulated. Thus the opposing forces of motility and adherence are minimized by the action of MrpJ paralogs.
The presence of fifteen copies of mrpJ-type genes in P. mirabilis, in retrospect, likely explains why the original mrpJ deletion strain had no apparent phenotype (Li et al., 2001). However, it is possible that not all the mrpJ paralogs function the same way, or that they regulate overlapping but not identical pathways. This possibility is suggested by the unique swarming patterns (Fig. 3) and different fimbrial appearances (Fig. 4) of mrpJ paralog overexpression strains. Additionally, the two mrpJ paralogs that had very little effect on motility, PMI0182 and PMI0982, may either be nonfunctional or regulate other pathways beside motility. Examination of the predicted protein sequences of PMI0182 and PMI0982 reveals an arginine residue that is conserved in these two proteins, yet is not found in the other 13 MrpJ proteins (Fig. 5A, residue 39 of the consensus sequence). Although all mrpJ paralog clones grow at the same rate as the vector control when uninduced, it should be noted that many strains, including the mrpJ overexpression strain, have a significant growth lag when induced with 0.3 mM IPTG (data not shown). Yet, Western blot analysis demonstrates that levels of the major flagellin FlaA are clearly reduced in most mrpJ overexpression strains, even when overexpression was not induced. Therefore, reduced swimming motility radii in induced strains are likely a combined result of reduced flagellar expression and slower growth.
The full mechanism of MrpJ regulation remains unknown. Because MrpJ decreases the amount of flagella produced by P. mirabilis, we predicted that MrpJ binds the promoter of the flagellar master regulator, flhDC. However, Li et al. (Li et al., 2001) reported that His-tagged MrpJ does not bind these sequences. There are several possible explanations for our current finding that MrpJ actually does bind the flhDC promoter. Primarily, in this study, a larger fragment of the region upstream of flhDC was assessed for MrpJ binding (550nt compared to 400nt). It is also possible that the shift was not detected due to poorer resolution or less sensitive detection; in the current study, chemiluminescent detection was used on fragments separated on a polyacrylamide gel, while the previous report used ethidium bromide detection on fragments separated with an agarose gel. There is a large distance (2757 nt) between flhD and the next gene (PMI1673, a hypothetical gene), providing possible sites for multiple regulators to bind this region. Although MrpJ and most of its paralogs act to reduce expression of flhDC, the different contributions of each paralog to swarming motility suggest that MrpJ likely also acts on other pathways. It is worth noting that the role of the C-terminus of MrpJ remains undetermined, particularly as it was possible to delete the C-terminal quarter of MrpJ and still repress motility. A future area of focus will be using unbiased methods to locate other genes that are regulated by MrpJ.
Little is currently known about regulation of the 17 different potential fimbriae encoded by P. mirabilis. MR/P fimbriae are preferentially expressed in vitro during static culture at 37°C and are expressed during urinary tract infection in mice (Bahrani et al., 1991). Expression of MR/P is controlled by an invertible element in the mrp promoter (Zhao et al., 1997) which is in turn regulated by the MrpI recombinase (Li et al., 2002). UCA/NAF fimbriae have been isolated from P. mirabilis cultured in minimal medium (Wray et al., 1986) as well as P. mirabilis grown on LB agar plates grown at 37°C (Tolson et al., 1995), but expression was not detected at 23°C (Tolson et al., 1995). Both MR/P and UCA/NAF were also found to vary in expression within a swarming colony of P. mirabilis (Latta et al., 1999). The ATF fimbriae are preferentially expressed during static culture at 23°C in rich medium but not at 42°C, culture in minimal medium, or when grown on solid medium (Massad et al., 1994a). Mannose-resistant/Klebsiella-like (MR/K) fimbriae, which are likely PMF fimbriae (Bahrani et al., 1993) are known to be produced during static culture (Adegbola et al., 1983); however, because MR/K fimbriae were only characterized by erythrocyte binding (i.e., hemagglutination) and not by protein or nucleotide sequence analysis, it is not certain that the fimbriae analyzed were truly PMF. PMP expression has been confirmed by N-terminal sequencing of purified P. mirabilis fimbrial protein (Bijlsma et al., 1995), although the culture condition used was not specified. The remaining 12 potential fimbriae encoded by P. mirabilis (Pearson et al., 2008) remain uncharacterized. Multiple types of P. mirabilis fimbriae can be expressed simultaneously (Adegbola et al., 1983; Bahrani and Mobley, 1993; Massad et al., 1994a; Tolson et al., 1995). It is intriguing to consider whether MrpJ or its paralogs may contribute to regulation of fimbrial expression in addition to regulation of flagella, particularly in light of the different fimbrial appearances exhibited by mrpJ or ucaJ overexpression strains (Fig. 4).
Although mrpJ homologs are found, in a few instances, associated with fimbrial operons in other species (He et al., 2004; Li et al., 2001; Meslet-Cladiere et al., 2004), this unusual method of motility regulation has been embraced by P. mirabilis to modulate expression of its surface appendages. This organism is notorious for its ability to swarm over surfaces, morphing alternately between swarmer cells with abundant flagella and fimbriated swimmer cells expressing only a few flagella. Notably, swarm cells have reduced expression of fimbriae (Mobley et al., 1996). Additionally, this organism possesses the machinery to make up to 17 distinct chaperone-usher fimbriae (Pearson et al., 2008). Indeed, ten fimbrial operons include an mrpJ paralog, emphasizing the idea that when P. mirabilis is adherent, it seeks to downregulate motility. The exact mechanisms for mrpJ control of motility, and swarm cell downregulation of fimbriae, will make interesting future directions of study.
Experimental Procedures
Bacterial strains and culture conditions
P. mirabilis HI4320 was isolated from the urine of an elderly, long-term catheterized woman (Mobley and Warren, 1987). E. coli DH5α was used as the host strain for cloning of mrpJ paralogs and mrpJ mutagenesis experiments. All strains and mutants were cultured at 37°C in nonswarming LB broth (per liter: 10g tryptone, 5g yeast extract, 0.5g NaCl) or on LB medium solidified with 1.5% agar. Antibiotic supplementation of chloramphenicol (20 μg/ml), ampicillin (100 μg/ml), or kanamycin (25 μg/ml) was provided as necessary.
Cloning of mrpJ paralogs
The IPTG-inducible plasmid pLX3607 and mrpJ overexpression plasmid pLX3805 have been described (Li et al., 2001). The fourteen additional mrpJ paralogs in the HI4320 genome were amplified by PCR (primers in Supplementary Table I) and cloned into the NcoI and HindIII sites of pLX3607 (Table I), with the exception of fim3J, which was cloned into the NcoI and BamHI sites of pLX3607. After all sequences were confirmed by nucleotide sequence analysis, wild-type P. mirabilis HI4320 was electroporated with each of the plasmids.
Table 1.
Plasmids used in this study.
| Name | Description | Source |
|---|---|---|
| pLX3607 | IPTG-inducible cloning vector; ampR | (Li et al., 2001) |
| pLX3805 | pLX3607 with mrpJ in the NcoI-HindIII sites | (Li et al., 2001) |
| pMP123 | pLX3607 with mrpJ′ in the NcoI-HindIII sites | This study |
| pMP124 | pLX3607 with fim3J in the NcoI-HindIII sites | This study |
| pMP125 | pLX3607 with ucaJ in the NcoI-HindIII sites | This study |
| pMP126 | pLX3607 with fim5J in the NcoI-HindIII sites | This study |
| pMP128 | pLX3607 with pmpJ in the NcoI-HindIII sites | This study |
| pMP129 | pLX3607 with atfJ in the NcoI-HindIII sites | This study |
| pMP131 | pLX3607 with fim10J2 in the NcoI-HindIII sites | This study |
| pMP165 | pLX3607 with fim14J in the NcoI-HindIII sites | This study |
| pMP183 | pLX3607 with fim8J in the NcoI-HindIII sites | This study |
| pMP184 | pLX3607 with fim10J1 in the NcoI-HindIII sites | This study |
| pMP185 | pLX3607 with PMI0182 in the NcoI-HindIII sites | This study |
| pMP186 | pLX3607 with PMI0982 in the NcoI-HindIII sites | This study |
| pMP187 | pLX3607 with PMI1817 in the NcoI-HindIII sites | This study |
| pMP188 | pLX3607 with PMI3508 in the NcoI-HindIII sites | This study |
| pLX2501 | pLX3607 with mrpJ in the NcoI-BglII sites; His-tag | (Li et al., 2001) |
| pMP201 | pBAD/Myc-HisA with ucaJ in the NcoI-HindIII sites; His-tag | This study |
Swimming and swarming motility assays
Swimming motility was assessed by stabbing a swim plate (1% tryptone, 0.5% NaCl, 0.25% agar) with a late logarithmic-phase bacterial culture (OD600 = 1.0). Care was taken to not stab through to the bottom of the plate, to prevent twitching motility. Swim plates were incubated for 19 h at 30°C before the radius of motility was measured. Swarming motility was assessed by spotting 5 μl late logarithmic-phase bacterial culture (OD600 = 1.0) onto the center of a swarm plate (1% tryptone, 1% NaCl, 0.5% yeast extract, 1.5% agar). Swarm plates were incubated inverted at 30°C for 17 h before the radius of motility was measured. Ampicillin (100 μg/ml) was added to plates as necessary to maintain overexpression plasmids. All results represent the mean of three independent experiments. The paired t test was used to compare motility radii with either the vector or mrpJ controls; one-tailed P-values < 0.05 were considered significant.
Western blot analysis
Overnight, aerated cultures of HI4320 with overexpression plasmids containing mrpJ or its paralogs were adjusted to OD600 = 1.0. Alternatively cultures were grown to mid-logarithmic phase (OD600 = 0.6). Cultures were not induced with IPTG. A 1 ml sample of each culture was collected by centrifugation (6000 × g, 8 min). The bacterial cell pellet was suspended in 100 μl 2X Laemmli sample buffer, boiled for 10 min, and separated by SDS-PAGE. Western blot analysis was conducted using anti-FlaA antibody (Mobley et al., 1996) or anti-UreD antibody (Heimer and Mobley, 2001) as previously described and peroxidase-conjugated goat anti-rabbit IgG (Sigma) as the secondary antibody. Chemiluminescence was accomplished using the ECL Plus Western Blotting Detection System (Amersham, GE Healthcare) according to the manufacturer's directions.
DNA sequencing and analysis
DNA sequencing was performed by the University of Michigan DNA sequencing core. Sequences were analyzed using the DNASTAR Lasergene suite software, version 7.0. Translated mrpJ paralogs were aligned using the CLUSTALW algorithm in Lasergene.
Transmission electron microscopy
Ten μl of an overnight culture was added to the surface of 300 mesh copper grids (Electron Microscopy Sciences) and allowed to bind for 3 min. Liquid was wicked off with filter paper, and 10 μl of fixative (2.5% glutaraldehyde, 2% paraformaldehyde in PBS) was added to each grid for 5 min. Grids were dipped in distilled water 10 times, and a 1% phosphotungstic acid stain was applied for 30 sec. Remaining liquid was wicked away with filter paper, and the grids were allowed to dry for 10 min. Grids were viewed using a Philips CM-100 transmission electron microscope.
Mutagenesis of mrpJ
Site-directed mutagenesis of mrpJ was accomplished using the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's directions (mutagenic primers in Supplementary Table II). Random 15-nt insertions in mrpJ were constructed using the GPS-LS linker scanning system (NEB) according to the manufacturer's protocol. In both cases, the mrpJ overexpression plasmid pLX3805 was used as the template for mutagenesis. Mutation of pLX3805 was confirmed using nucleotide sequence analysis. Mutated mrpJ plasmids were introduced to the mrpJ null mutant HI4320ΔmrpJ by electroporation. Site-directed mutant plasmids are listed in Table I; the numerous linker insertion mutant plasmids are omitted.
Electrophoretic mobility shift assays
Purification of His-tagged MrpJ protein from E. coli M15 containing pLX2501 has been described (Li et al., 2001). His-tagged UcaJ protein was obtained by cloning ucaJ into the NcoI and HindIII sites of pBAD/Myc-His A (Invitrogen) and purifying using the same method of Li et al. (Li et al., 2001). Purified protein was desalted using 3500 MWCO dialysis cassettes (Pierce). Gel shifts using the PCR-amplified gene promoter region of flhDC or the flhD coding sequence (primers in Supplementary Table III) and MrpJ-His6 or UcaJ-His6 were accomplished using the DIG gel shift kit (Roche) according to the manufacturer's recommendations.
Quantitative real-time PCR (qRT-PCR)
HI4320 containing pLX3607 or pLX3805 (+mrpJ) was cultured overnight in LB broth supplemented with ampicillin. These cultures were diluted 1:100 into fresh medium and grown to logarithmic phase (OD600 = 1.0). RNA protect (2 ml) (Qiagen) was added to 1 ml of culture, and RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer's instructions. DNA was digested using Turbo-DNAfree DNase (Ambion). RNA was used as the template for cDNA synthesis using the Superscript First-Strand Synthesis System (Invitrogen) according to the manufacturer's protocol. PCR with primers specific to the rpoA gene (RNA polymerase A) was performed on cDNA samples prepared with and without reverse transcriptase to confirm no genomic DNA contamination of the RNA preparations. Each qRT-PCR reaction was set up in duplicate and consisted of 30 ng cDNA template, 150 nM of each primer, and 12.5 2X SYBR Green PCR master mix (Stratagene). Target genes were amplified using an Mx3000P thermal cycler (Stratagene). Melting curve analysis was used to confirm a lack of primer dimers or genomic DNA contamination of reagents. Data were normalized to rpoA, and analyzed by the 2-ΔΔCT method (Livak and Schmittgen, 2001). Primers used for qRT-PCR are listed in Supplementary Table III.
Supplementary Material
Acknowledgements
The authors thank Dotty Sorenson of the Microscopy and Image Analysis Laboratory at the University of Michigan for expert assistance with transmission electron microscopy. This work was funded by Public Health Service grant AI059722 from the National Institutes of Health (H.L.T.M.). M.M.P. was supported in part by the Molecular Mechanisms of Microbial Pathogenesis training grant (T32 AI7528) and National Research Service Award F32 AI068324.
References
- Adegbola RA, Old DC, Senior BW. The adhesins and fimbriae of Proteus mirabilis strains associated with high and low affinity for the urinary tract. Journal of Medical Microbiology. 1983;16:427–431. doi: 10.1099/00222615-16-4-427. [DOI] [PubMed] [Google Scholar]
- Akerley BJ, Cotter PA, Miller JF. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- Bahrani FK, Johnson DE, Robbins D, Mobley HL. Proteus mirabilis flagella and MR/P fimbriae: isolation, purification, N-terminal analysis, and serum antibody response following experimental urinary tract infection. Infect Immun. 1991;59:3574–3580. doi: 10.1128/iai.59.10.3574-3580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani FK, Cook S, Hull RA, Massad G, Mobley HL. Proteus mirabilis fimbriae: N-terminal amino acid sequence of a major fimbrial subunit and nucleotide sequences of the genes from two strains. Infection and Immunity. 1993;61:884–891. doi: 10.1128/iai.61.3.884-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani FK, Mobley HL. Proteus mirabilis MR/P fimbriae: molecular cloning, expression, and nucleotide sequence of the major fimbrial subunit gene. J Bacteriol. 1993;175:457–464. doi: 10.1128/jb.175.2.457-464.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani FK, Massad G, Lockatell CV, Johnson DE, Russell RG, Warren JW, Mobley HL. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infection and Immunity. 1994;62:3363–3371. doi: 10.1128/iai.62.8.3363-3371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma IG, van Dijk L, Kusters JG, Gaastra W. Nucleotide sequences of two fimbrial major subunit genes, pmpA and ucaA, from canine-uropathogenic Proteus mirabilis strains. Microbiology. 1995;141(Pt 6):1349–1357. doi: 10.1099/13500872-141-6-1349. [DOI] [PubMed] [Google Scholar]
- Bryan A, Roesch P, Davis L, Moritz R, Pellett S, Welch RA. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect Immun. 2006;74:1072–1083. doi: 10.1128/IAI.74.2.1072-1083.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg S, Hughes KT. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:1209–1213. doi: 10.1128/jb.184.4.1209-1213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker C, Poore CA, Li X, Mobley HL. Pathogenesis of Proteus mirabilis urinary tract infection. Microbes.Infect. 2000;2:1497–1505. doi: 10.1016/s1286-4579(00)01304-6. [DOI] [PubMed] [Google Scholar]
- Cook SW, Mody N, Valle J, Hull R. Molecular cloning of Proteus mirabilis uroepithelial cell adherence (uca) genes. Infect Immun. 1995;63:2082–2086. doi: 10.1128/iai.63.5.2082-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel CL, Mekalanos JJ. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Snyder HA, Forst S. Unique organization and regulation of the mrx fimbrial operon in Xenorhabdus nematophila. Microbiology. 2004;150:1439–1446. doi: 10.1099/mic.0.26853-0. [DOI] [PubMed] [Google Scholar]
- Heimer SR, Mobley HL. Interaction of Proteus mirabilis urease apoenzyme and accessory proteins identified with yeast two-hybrid technology. J Bacteriol. 2001;183:1423–1433. doi: 10.1128/JB.183.4.1423-1433.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AM, Lockatell CV, Johnson DE, Mobley HL. Visualization of Proteus mirabilis morphotypes in the urinary tract: the elongated swarmer cell is rarely observed in ascending urinary tract infection. Infect Immun. 2003;71:3607–3613. doi: 10.1128/IAI.71.6.3607-3613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MC, Simms AN, Mobley HL. Complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J Bacteriol. 2007;189:5523–5533. doi: 10.1128/JB.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta RK, Grondin A, Jarrell HC, Nicholls GR, Bérubé LR. Differential expression of nonagglutinating fimbriae and MR/P pili in swarming colonies of Proteus mirabilis. J Bacteriol. 1999;181:3220–3225. doi: 10.1128/jb.181.10.3220-3225.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rasko DA, Lockatell CV, Johnson DE, Mobley HL. Repression of bacterial motility by a novel fimbrial gene product. EMBO Journal. 2001;20:4854–4862. doi: 10.1093/emboj/20.17.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lockatell CV, Johnson DE, Mobley HL. Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. Mol.Microbiol. 2002;45:865–874. doi: 10.1046/j.1365-2958.2002.03067.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Macnab RM. Flagella and motility. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D.C.: 1996. pp. 123–145. [Google Scholar]
- Massad G, Bahrani FK, Mobley HL. Proteus mirabilis fimbriae: identification, isolation, and characterization of a new ambient-temperature fimbria. Infection and Immunity. 1994a;62:1989–1994. doi: 10.1128/iai.62.5.1989-1994.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massad G, Lockatell CV, Johnson DE, Mobley HL. Proteus mirabilis fimbriae: construction of an isogenic pmfA mutant and analysis of virulence in a CBA mouse model of ascending urinary tract infection. Infection and Immunity. 1994b;62:536–542. doi: 10.1128/iai.62.2.536-542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslet-Cladiere LM, Pimenta A, Duchaud E, Holland IB, Blight MA. In vivo expression of the mannose-resistant fimbriae of Photorhabdus temperata K122 during insect infection. J Bacteriol. 2004;186:611–622. doi: 10.1128/JB.186.3.611-622.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley HL, Warren JW. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J Clin Microbiol. 1987;25:2216–2217. doi: 10.1128/jcm.25.11.2216-2217.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley HL, Belas R, Lockatell V, Chippendale G, Trifillis AL, Johnson DE, Warren JW. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infection and Immunity. 1996;64:5332–5340. doi: 10.1128/iai.64.12.5332-5340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather PN, Parkhill J, Mobley HL. The complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol. 2008 doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather PN. Swarmer cell differentiation in Proteus mirabilis. Environ Microbiol. 2005;7:1065–1073. doi: 10.1111/j.1462-2920.2005.00806.x. [DOI] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Römling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Tolson DL, Barrigar DL, McLean RJ, Altman E. Expression of a nonagglutinating fimbria by Proteus mirabilis. Infect Immun. 1995;63:1127–1129. doi: 10.1128/iai.63.3.1127-1129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray SK, Hull SI, Cook RG, Barrish J, Hull RA. Identification and characterization of a uroepithelial cell adhesin from a uropathogenic isolate of Proteus mirabilis. Infect Immun. 1986;54:43–49. doi: 10.1128/iai.54.1.43-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Li X, Johnson DE, Blomfield I, Mobley HL. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Molecular Microbiology. 1997;23:1009–1019. doi: 10.1046/j.1365-2958.1997.2791645.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.