Abstract
Pyruvate carboxylase (PC) deficiency (OMIM, 266150) is a rare autosomal recessive disease. The revised PC gene structure described in this report consists of 20 coding exons and four non-coding exons at the 5’-untranslated region (5’-UTR). The gene codes for three transcripts due to alternative splicing: variant 1 (NM_000920.3), variant 2 (NM_022172.2) and variant 3 (BC011617.2). PC deficiency is manifested by three clinical phenotypes - an infantile form (Type A), a neonatal form (Type B), and a benign form (Type C). We report the molecular basis for eight cases (one Type A, five Type B and two Type C) of PC deficiency. Eight novel complex mutations were identified representing different combinations of missense mutations, deletions, a splice site substitution and a nonsense mutation. The classical phenotypes (A, B and C) correlated poorly with clinical outcomes. Mosaicism was found in five cases (one Type A, three Type B and one Type C) and four of these cases had prolonged survival. Death in the fifth case resulted from unrelated medical complications. The discrepancy between the current findings and the existing classification system should be addressed to accommodate these new observations.
Keywords: PC deficiency, lactic acidosis, brain metabolism, developmental encephalopathy
Introduction
Pyruvate carboxylase deficiency (PC deficiency) (OMIM, 266150), is a rare autosomal recessive disease. Three distinct clinical presentations of PC deficiency are known. An infantile form (Type A, North American Form) mostly seen in North American Indians, is characterized by infantile-onset, mild to moderate lactic acidemia, delayed mental and motor development, failure to thrive, apathy, hypotonia, pyramidal tract signs, ataxia, nystagmus, convulsions, and death in infancy or early childhood(1–5). The lactate-to-pyruvate ratio is normal despite acidemia. Episodes of acute vomiting, tachypnea and acidosis are characteristically precipitated by metabolic stress or infections.
The neonatal form (Type B, French Form) (6), presents very early in life with severe lactic acidosis, anorexia, hepatomegaly, convulsions, stupor, hypotonia, rigidity, hypokinesia, bizarre ocular behavior, pyramidal tract signs and severely delayed psychomotor development(5–10). Elevated blood levels of ammonia, citrulline, proline and lysine, an increased lactate-to-pyruvate ratio and decreased 3-hydroxybutyrate to acetoacetate ratio are characteristic. Death usually occurs in the first months of life. In these patients, depletion of intracellular aspartate and oxaloacetate compromises both the synthetic and anaplerotic PC functions.
The benign form (Type C, Benign Form) is represented by four cases, (11–14). The clinical phenotype is characterized by normal or mildly delayed neurological development and episodic metabolic acidosis.
Pyruvate carboxylase (EC 6.4.1.1, PC) plays a pivotal role in intermediary metabolism. It serves a critical anaplerotic function replenishing the Krebs cycle intermediates by catalyzing the conversion of pyruvate to oxaloacetate, a reaction that is activated by elevated acetyl-coenzyme A levels (15). In addition, PC controls the first step of hepatic gluconeogenesis, and is involved in lipogenesis (16, 17). The enzyme is localized within the mitochondrial matrix and is expressed in a tissue-specific manner, with highest activity in liver, kidney, adipose tissue, lactating mammary gland and pancreatic islets, moderate activity in brain, heart and adrenal gland, and low activity in white blood cells and skin fibroblasts(18).
The PC cDNA was first cloned from a human liver cDNA library (19). The mRNA transcript is 4.2 kb and the protein is 125 kDa (20, 21). Southern blotting of human genomic DNA confirmed the PC gene as a single copy with no detectable pseudogenes. Using fluorescence in situ hybridization (FISH) to normal chromosomes and chromosomes carrying the FRA11A (OMIM,136560) fragile site, Walker et al. mapped PC to 11q13, distal to FRA11A, and further localized it to 11q13.4-q13.5(22). Mammalian PC is a homotetramer, each peptide covalently bound to a biotin molecule and capable of both catalytic and regulatory functions(17).
Our study objectives were to validate the PC gene structure and to study the molecular genetics of PC deficiency in eight patients. Several novel complex mutations and somatic mosaicism were confirmed. The amount of PC mRNA and protein expressed in fibroblasts varied from normal to significantly decreased. Long-term survival correlated better with somatic mosaicism than with residual tissue enzyme protein or clinical phenotype.
Material and methods
1. Characterization of the PC gene structure
Human BAC clones (89O10 and 5P19, Catalog #96012) containing the PC gene were obtained from Research Genetics (Huntsville, AL). BAC plasmids were prepared using a Maxi-prep kit from Invitrogen (Carlsbad, CA) for direct sequencing with primers based on the PC cDNA sequence. The intron-exon boundaries were predicted by comparing the genomic DNA sequence from the BAC plasmid with the cDNA sequence.
2. Human subjects
The investigatory studies of skin fibroblasts and mutation screening were approved by the Columbia University Institutional Review Board. Informed consent was obtained from the parents and patients who were participants in this study.
3. PC assay
The PC activity in cultured skin fibroblasts was determined as previously described(5)
4. Clinical features of PC deficiency patients
The clinical information from the eight patients, as summarized in Table 1, was obtained by review of available medical records. All patients had normal birth weights, and there were no complications of pregnancy except patient 5 who had intrauterine growth retardation. Apgar scores were 9 and 10 at one and five minutes after birth in all cases.
Table 1.
Clinical features of PC deficient patients
| 1* | 2 | 3 | 4 | 5 | 6† | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| Phenotype | C | B | B | B | B | A | B | C |
| PC protein | 86% | 8% | 3% | 2% | 44% | 49% | 17% | N/A |
| Pyruvate carboxylase activity** | 0.03 | 0.00 | 0.00 | 0.08 | 0.03 | 0.09 | 0.27 | 0.002 |
| Sex | Female | Male | Male | Female | Male | Male | Male | Male |
| Clinical onset | 7 m | 1 m | 1 h | 24h | 2 d | 14m | 3 h | 12m |
| First symptoms | Vomiting, dehydration | Respiratory distress, Lactic acidosis | Kussmaul respirations, seizures | Tachypnea, dyspnea | Respiratory distress, metabolic acidosis | Vomiting, Gastroenteropathy, Acidosis | Tachypnea, acidosis | Unsteadiness, falls |
| Outcome | Alive at 12 y | Alive at 20y | Died at 8 d | Died at1 m | Alive at 9 y | Alive at 23 y | Died at 6m | Alive at 3 y |
| Major clinical findings | Episodes of metabolic acidosis | Lactic acidosis seizures | Lactic acidosis, hyperammonemia | Lactic acidosis | Metabolic acidosis | Hypotonia, Mental retardation | Hypotonia, rolling eye movements, severe encephalopathy | Episodes of metabolic acidosis with intercurrent illness |
| Blood lactate† | 5.5–7.3 | 13 | 22–27.8 | increased | 19 | 11.5 | 10.4 | 10.2 |
| Blood ammonia† | Normal | Normal | 222 | n/a | 175 | Normal | 77 | 92 |
| Blood alanine† | 852 | Increased | n/a | n/a | 799, 669, 933 | 1116 | 1184, 165 | 465, 350 |
| Blood citrulline† | Normal | n/a | n/a | n/a | 60, 57, 195 | 33 | 43, 132 | 14,16 |
| Blood lysine† | 160 | n/a | n/a | n/a | 288, 298, 945 | 234 | 552 | 108,286 |
| Blood proline† | 281 | Increased | n/a | n/a | 381, 358, 800 | 192 | 730 | 316,274 |
| Urinalysis | Ketones (+) | Lactate Pyruvate | Ketones (++) | Ketones (+++) | Lactate, pyruvate, | Lactate, alanine | Lactate, p-OH-phenyllactate | Lactate, Ketones |
| Neurological imaging | N/A | Normal | N/A | N/A | N/A | Normal | Periventricular cysts | Normal |
| EEG | N/A | Normal | N/A | N/A | Normal | Slowing | Nonspecific abnormalities | N/A |
5. Mutation screening in PC deficiency patients
The coding exons and intron-exon boundaries of the PC gene were amplified (Supplementary Table S1) from genomic DNA extracted from fibroblasts or blood using an Expand Long Template PCR kit (Roche, Indianapolis, IN). All the PCR primers and the size of PCR products are summarized in Supplementary Table S1. PCR conditions: denaturation at 95°C for 30 seconds, annealing at 61–63.5°C for 30 seconds, elongation at 68°C for 2–3 minutes for 35 cycles, with a final extension at 68°C for 8 minutes. All of the expected PCR products were confirmed by 1% agarose gel electrophoresis. The expected PCR products were then purified for direct sequencing. All of the mutations were further confirmed by sequencing independent PCR products.
6. Polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) analysis for quantitation of the mutant DNA with 1892G>A and 2540 C>T
PCR fragments NE3-NE4 (401 bp) covering exon 13 (forward primer 5’-ATCTGTGGGGGTTGGAGCAG-3’ and reverse primer 5’-GGCTGACCGTGGAGAACAGA-3’ and CO1-CO2 (398bp) covering exon16 (forward primer 5’-CTGTCACCTGAGTCAGACAT-3’ and reverse primer 5’-AGGCCAGTGGCAGGTGCATG-3’) were amplified respectively using the above PCR conditions except that the annealing temperature was 65°C. All specific PCR products were confirmed by electrophoresis on 1% agarose gel and further purified for the subsequent digestion. Excess Msp I and Fnu4H I were used for overnight digestion at 37°C. The digested products were verified by electrophoresis on 2% agarose gel. Quantification of mutant DNA was performed by an adaptation of the PCR-RFLP analysis. 32 P labeled d-ATP was added before the last cycle of the PCR. The PCR products were digested with the respective restriction enzymes described above and separated on a 15% nondenaturing polyacrylaminde gel. The data then were analyzed in a phosphorimager (Bio-Rad, Hercules, Califonia) to calculate the mutation portion.
7. RNA extraction, reverse-transcription PCR (RT-PCR) and sequencing of subcloned RT-PCR products
Total RNA was isolated from cultured skin fibroblasts of PC deficient patients using RNAqueous™-4PCR kit (Ambion, Catalog #AM1914, Austin, Texas, 78744). The total RNA was used for the RT-PCR using the Tian one tube RT-PCR kit (Roche). RT-PCR conditions were: 50°C for 30 min, 95 °C for 2 min, followed by 35 cycles of amplification with 95°C for 30 s, 56–62°C for 30 s and 68°C for 2–3 min. The primers used are listed in Supplementary Table S2. The specific RT-PCR products were confirmed by electrophoresis on 1% agarose gel. Some of the RT-PCR products were subcloned using TA Cloning Kit (Invitrogen, Carlsbad, CA). The correct clones were confirmed by digestion with EcoR I and directly sequenced with primers M13f and M13r.
8. Western blot analysis
Mitochondria were isolated for Western blot analysis as described previously (23). The protein concentration was determined with a Bio-Rad protein assay kit (Hercules, CA). 50 µg of mitochondrial protein were loaded on to a 5–20% Ready Gel from Bio-Rad and electrophoresed at 80V for 2 hours. The protein was transferred to an Immobilon-P Transfer membrane (Millipore, Bedford, MA) and blocked overnight at 4° C in 5% dried skim milk in TBS-T. The PC, propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC) membrane bands were then probed with streptavidin-HRP conjugate and visualized using the horseradish peroxidase ECL system (Amersham, Piscataway, NJ) following the protocol. The film was scanned and analyzed with Image J to obtain the relative ratio of the PC protein to the PCC and MCC proteins. The molecular masses of MCC and PCC are 75 KDa and 73KDa respectively.
Results
1. Characterization of the PC gene structure
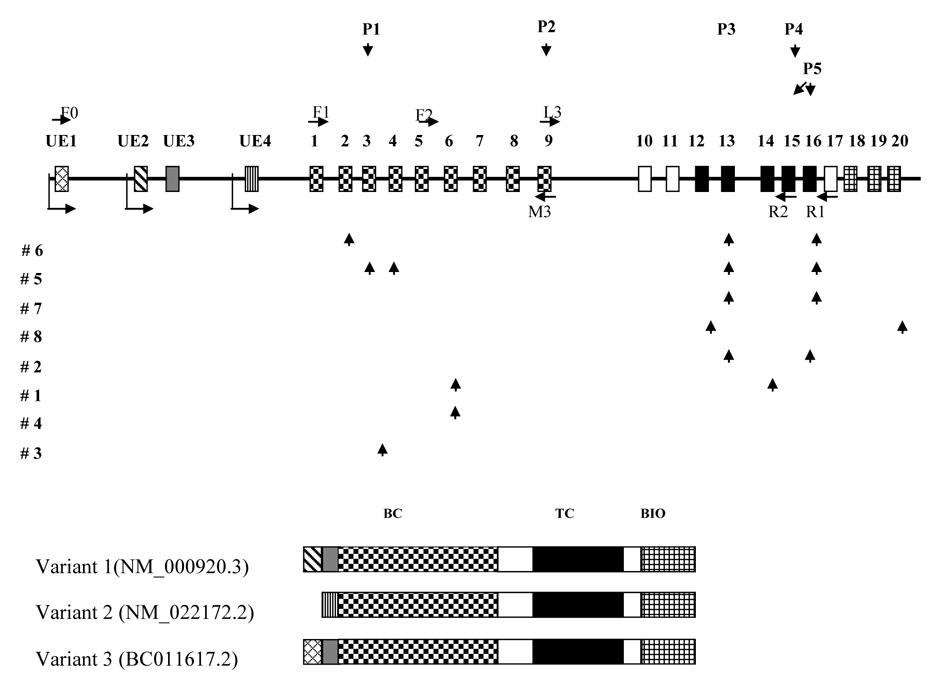
Carbone et al. reported the PC gene structure in 1998 and later revised it (3, 24). Our studies extend the previous findings and refine the size of exons 1–3, 8 and 15, the size of non-coding exon 4(UE4), and the sequences at exon-intron boundaries 5 and 10 based on direct sequencing of the BAC plasmid DNA and the PCR products of patients’ genomic DNA (Supplementary Table S3 and Fig. 1). One additional UE1 has been identified in the human genomic DNA sequence of the chromosome 11 clone (AP003176) by BLAST searching. This refined gene structure is consistent with the structure predicted by http://genome.cse.ucsc.edu/cgi-bin/ and www.ncbi.nih.gov/IEB/Research/Acembly/, except for the UE4 (the non-coding exon of PC variant 2) that was omitted in both websites (Supplementary Table S3 and Fig. 1). There are a total of three tissue-specific PC transcripts: PC variant 1 (NM_000920.3), PC variant 2 (NM_022172.2) and PC variant 3 (BC011617.2). These three transcripts share the same coding region, but differ in the 5’-untranslated region (5’-UTR). The PC variants 1 and 3 are both expressed in brain and liver, confirmed by sequencing the RT-PCR products. The PC variant 2 is expressed in liver and kidney (18, 20, 21)
Fig. 1. PC gene structure, sequence variance distribution and three transcript variants.
The coding exons of PC gene are represented by rectangles with different symbols and Arabic numbers on the top. The four non-coding exons are labeled with UE1-4 on the top. The arrows before UE1, UE2 and UE4 represent the transcription initiation sites. Primers used for the RT-PCR F0, F1, L3, M3, R1 and R2 are shown with an arrow head to designate the direction. All previously reported mutations in the literature are shown above the gene structure(3, 24, 33). All the novel complex mutations identified in this study are shown under the gene structure with arrow heads to show the locations of alterations in the gene. Three transcript variants are shown with the same coding region and different non-coding exons in the 5’-UTR. BC: Biotin carboxylation domain. TC: Transcarboxylation domain. BIO: Biotin carboxyl carrier domain
2. Clinical features of PC deficiency
The clinical features of all eight patients are summarized in Table 1. Five patients fulfilled clinical criteria for Type B, one for Type A, and two for Type C.
3. Molecular genetic analysis of PC deficiency
We screened the 20 coding exons and intron-exon boundaries in the eight patients’ DNA samples and found an array of novel mutations as shown in Fig. 1. The nomenclature follows the recommendations for the description of sequence variants (www.hgvs.org/mutnomen/recs.html; http://www.hgvs.org/mutnomen/; http://www.hgvs.org/mutnomen/checklist.html; Hum Mutat 22:181–182, 2003; http://dx.doi.org/10.1002/humu.10262; Hum Mut 15: 7–12, 2000). The coding sequence is NM_000920. 3. Nucleotide +1 is the A of the ATG translation initiation codon. All the sequence variants are summarized in Table 2.
Table 2.
Novel sequence variants identified in PC gene and relative PC protein ratio
| ID | Type | DNA1 | Protein2 | Protein amount3 | PC Activity4 |
|---|---|---|---|---|---|
| 1 | C | c. [796T>G; =] + [=; 2114C>A, =] * | p. [S266A; =] + [=; S705X, =] | 86 ± 5.8% | 1.9% |
| 2 | B | c. [1892G>A, =; =] + [=; 2493_2494delGT, =] | p. [R631Q, =; =] + [=; V831VfsX832,=] | 8 ± 0.2% | 0 |
| 3 | B | c. [321+1G>T] + [321+1G>T] | p. [V105_K107del] + [V105_K107del] | 3 ± 1.4% | 0 |
| 4 | B | c.[806G>A] + [806G>A] | p. [R269Q] + [R269Q] | 2 ± 0.1% | 5% |
| 5 | B | c. [467G>A; 496G>A; 1892G>A, =; 2540C>T, =] + [467G>A; 496G>A; =; =] | p. [R156Q; V166I; R631Q, =; A847V, =] + [R156Q; V166I; =; =] | 44 ± 2.8% | 1.9% |
| 6 | A | c. [184C>T; 1892G>A ; 2540C>T] + [=; 1892G> A, =; 2540C> T, =] | p. [R62C; R631Q; 847V] + [=; R631Q, =; A847V, =] | 49 ± 17.6% | 6% |
| 7 | B | c. [1892G>A; 2540C> T]+ [1892G>A, =; 2540C> T, = ]† |
p. [R631Q; A847V] + [R631Q, =; A847V, =] | 17 ± 8.2% | 17% |
| 8 | C | c. [1705A>G; =] + [=; 3499–3500delCT] | p.[T569A; =] + [=; L1137VfsX1170] | N/A | 1% |
| P1 | A | c. [434T>C] + [434T>C] | p. [V145A] + [V145A] | Barely detectable | 7–25% |
| P2 | A | c. [1351C>T] + [1351C>T] | p. [R451C] + [R451C] | ~100% | 7% |
| P3 | A | c. [1828G>A] + [1828G>A] | p. [A610>T] + [A610T] | ~100% | 1–4% |
| P4 | A | c. [2229G>T] + [2229G>T] | p. [M743I] + [M743I] | ~100% | 1–4% |
| P5 | B | c. [2493_2494delGT] + [2473+2-2473+5delTGCA] | p. [V831VfsX832] + [E825GfsX846] | ~0 | 1–4% |
The nomenclature follows the recommendations for the description of sequence variants (www.hgvs.org/mutnomen/recs.html;http://www.hgvs.org/mutnomen/; http://www.hgvs.org/mutnomen/checklist.html; Hum Mutat 22:181–182, 2003; http://dx.doi.org/10.1002/humu.10262; Hum Mut 15: 7–12, 2000). The coding sequence is NM_000920. 3. Nucleotide +1 is the A of the ATG translation initiation codon. c. [2114C>A, =]
indicates the mosaic state of this allele, more ‘2114C>A’ than ‘=’ (wild-type) in the total DNA level. c. [1892G>A, =; 2540C> T, =]
indicates the mosaic state of the two substitutions 1892G>A and 2549C>T in this allele, which are more abundant than the ‘=’ (wild-type).
Note, changes are deduced based on the findings in DNA level. “0”, indicates no protein; “=”, indicates WT protein synthesized.
The PC /MCC +PCC ratios in PC patients are normalized to the ratio in normal control (Mean ± SD) (Fig. 4).
the PC activity is expressed as the ratio of patient/ control.
Patient 1 (Type C) was previously reported as the first benign case (12). This patient carried a heterozygous 796T>G transversion in exon 6, which resulted S266A in the biotin carboxylation domain of the enzyme. Another heterozygous 2114C>A transversion was identified in exon 14 which substituted amino acid 705 from Ser to a premature stop codon (S705X) in the transcarboxylation domain of the enzyme. An RT-PCR fragment covering exons 6 –14 was directly sequenced and showed the presence of only S266A in exon 6 and wild-type sequence (S705) in exon 14, suggesting that the mutant mRNA carrying 2114C>A (S705X) was unstable and rapidly degraded. This finding was confirmed by sequencing subcloned RT-PCR products as described above. Interestingly, we also identified two out of 20 sequenced clones from the same patient containing only wild-type sequence at S266 and S705. These findings, in the aggregate, allow us to postulate that the two substitutions are in trans and c. 2114C>A (S705X) is a somatic mosaic mutation. The mutations are c. [796T>G; =] + [=; 2114C>A, =] and p. [S266A; =] + [=; S705X, =]. We didn’t have the parents’ DNA for study, but they were clinically normal.
Patient 2 (Type B) had an 1892G>A (CGG>CAG) transition in exon 13 resulting in R631Q and a deletion of two nucleotides (GT) at nucleotide 2493_2494 in exon 16 resulting in a frame shift. The frame shift created a premature termination codon at amino acid position 832 (V831Bfs832). These two mutations were confirmed to be in trans by sequencing cloned PCR fragments covering exons 13 and 16. Direct sequencing of the RT-PCR fragment L3-R1 covering exons 9–16 confirmed the presence of r. 1892g>a and the absence of r. 2493_2494delgt, indicating the instability of the mRNA harboring the premature stop codon. The portion of PCR product containing c. 1892 G>A in the total PCR product was 45.3% as determined by quantitative PCR-RFLP, less than 50% as seen in inherited mutation, indicating the c. 1892 G>A was a spontaneous mutation in a mosaic state. The mutations are c. [1892G>A, =; =] + [=; 2493_2494delGT, =]; p. [R631Q, =; =] + [=; V831BfsX832, =]. We didn’t have the parents’ DNA for study, but they were clinically normal.
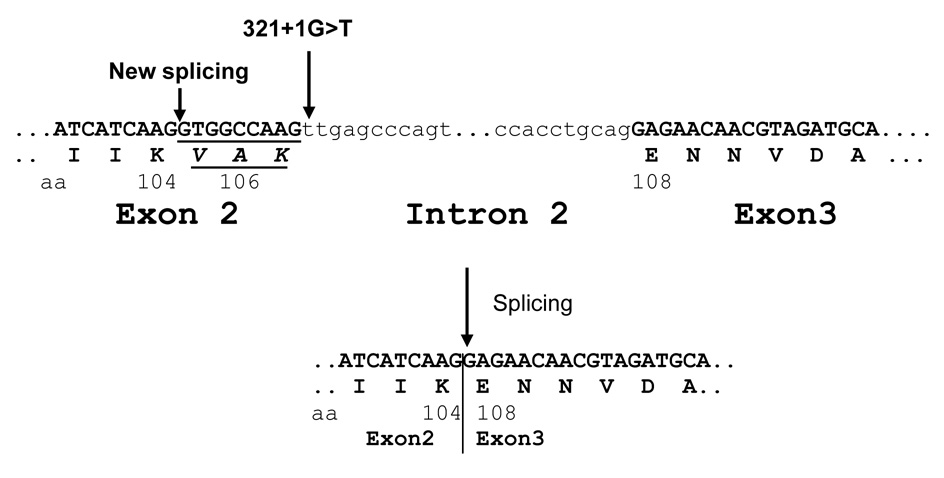
Patient 3 (Type B) was identified to carry a homozygous transversion at the splice donor site c.321+1G>T. Direct sequencing of the RT-PCR fragment F1-M3 showed the deletion of the last 9 nucleotides (GTGGCCAAG) of exon 2, indicating that nucleotides 311–312GT served as an alternative splice donor site. The mutant PC protein would reflect the deletion of the three amino acids (V105, A106 and K107) (Fig. 2). The mutations are c. [321+1G>T] + [321+1G>T]; p. [V105_K107del] + [V105_K107del]. We had no information concerning the parents.
Fig. 2. Splice donor site destroyed in patient 3.
Partial genomic DNA sequence from exons 2 and 3 and intron 2 around the splice donor and acceptor sites are shown. The amino acids (aa) and their locations are shown under the DNA sequence. Part of the mutant mRNA is shown with the deletion of nine nucleotides resulting in the deletion of amino acids V105, A106 and K107.
Patient 4 (Type B) was shown to carry a homozygous transition at nucleotide position 806G>A (CGG>CAG) in exon 4, resulting in R269Q located in the biotin carboxylation domain. This transition was also confirmed in the mRNA by sequencing the RT-PCR fragment F1-M3. Remarkably, amino acid R269 is conserved among many different species. The sequence variants are c. [806G>A] + [806G>A], p. [R269Q] + [R269Q]. We had no information concerning the parents.
Patient 5 (Type B) carried four transitions. The first one was a homozygous transition 467G>A (CGG >CAG), resulting in R156Q in exon 3. The second one also was a homozygous transition 496G>A (GTT>ATT), introducing V166I in exon 4. These two mutations are both located in the biotin carboxylation domain. The third one was a heterozygous transition 1892G>A (CGG>CAG) in exon 13 resulting in R631Q. The fourth one also was a heterozygous transition 2540C>T (GCC>GTC), resulting in A847V. The last two substitutions were located in the transcarboxylation domain. By direct sequencing of the RT-PCR fragments F2-M3 and L3-R1, we only found the presence of “homozygous” r. 467g>a(R156Q) and r. 496g>a(V166I) in the mRNA , suggesting that the mRNA transcripts carrying both r.1892g>a and r. 2540c>u substitutions were not stable. The mutations are c. [467G>A; 496G>A; 1892G>A, =; 2540C>T, =] + [467G>A; 496G>A; =; =], p. [R156Q; V166I; R631Q, =; A847V, =] + [R156Q; V166I; =; =]. Both R156Q and V166I mutations were inherited from the parents who were each heterozygous for both 467G>A (R156Q) and 496G>A (V166I) mutations and were wild-type at nucleotide locations 1892 and 2540. The parents were related as first cousins.
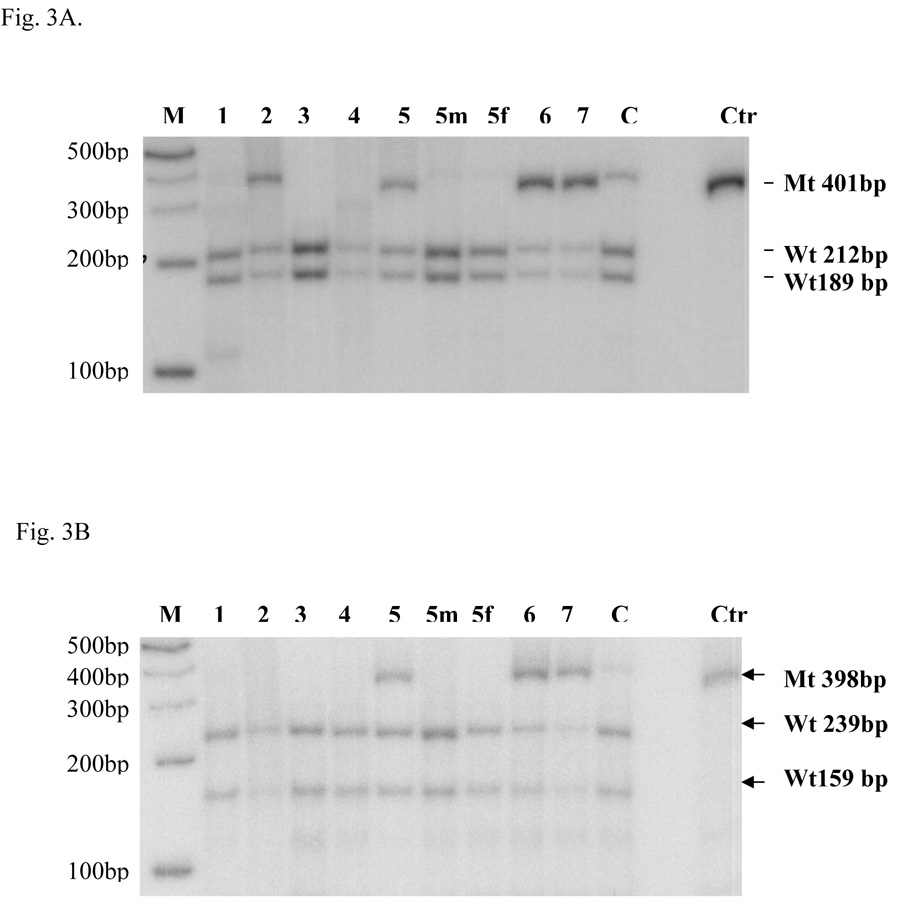
Patient 6 (Type A) was previously reported (25). We found three transitions in the coding region. The first one was a heterozygous c.184C>T (CGC>TGC) transition, causing the R62C in exon 2. The other two transitions were c. 1892G>A (R631Q) and c. 2540C>T(A847V) as in patient 5. Direct sequencing of the PCR fragments showed that these substitutions c. 1892G>A and c. 2540C>T were mixed with wild-type DNA indicating a mosaic state. This observation was confirmed by RFLP analysis (Fig. 3A and 3B). The mutant DNA abundance was 72.9% and 57.9% of the total DNA digested. These percentages are consistent with somatic mosaicism. The possibility of DNA or cultured fibroblast cell contamination was ruled out by quantitative fluorescence PCR using genomic markers on different chromosomes (data not shown). Direct sequencing of RT-PCR fragments only showed the presence of heterozygous r. 184c>u suggesting that the mutant mRNA transcripts containing r. 1892g>a and r. 2540c>u were unstable and rapidly degraded, as was seen in patient 5. As a result, only a fraction of the mRNA with r. 184c>u (R62C) and wild-type mRNA remained detectable. The patient’s mother was wild-type at these locations and the father’s DNA was not available for study. We deduced that this patient’s mutations were c. [184C>T; 1892G>A; 2540C>T] + [=; 1892G>A, =; 2540C>T, =], p. [R62C; R631Q; A847V] + [=; R631Q, =; A847V, =].
Fig. 3. Quantitative PCR-RFLP.
Quantitative PCR-RFLP was done as described in the Materials and Methods section. Molecular weight markers and patients’ codes are indicated on the top part of the figure. The marker ladder is spaced 100bp apart. 5m and 5f are the mother and father of patient 5. C is a carrier control and Ctr is the undigested control. A. The PCR mutant fragment (Mt) NE3-NE4 (401bp) with substitution c. [1892G>A] could not be digested by Msp I, whereas the wild-type DNA (Wt) NE3-NE4 fragment could be cut into two fragments (189bp and 212bp) as shown by an arrow head on the right side. B. The CO1-CO2 (398bp) mutant fragment (Mt) containing c. [2540C>T] could not be cut by Fnu4H I, whereas the wild-type fragment(Wt) could be cut into two fragments (239 bp and 159bp) as shown by the arrow head on the right side.
Patient 7 (Type B) carried two transitions c.1892G>A (R631Q) and c.2540C > T (A847V) as seen in patients 5 and 6. The mutant DNA represented 79.1% and 71.3% respectively of the total DNA digested during RFLP analysis, again suggesting a somatic mosaic state (Fig.3A and 3B). Based on the direct sequencing of the RT-PCR fragment L3-R1, we arrived at the same conclusion that mutant mRNA with r. 1892g>a and r. 2540c > u was rapidly degraded as was true with patients 5 and 6. The mutations are c. [1892G>A; 2540C>T] + [1892G>A, =; 2540C> T, =], p. [R631Q; A847V] + [R631Q, =; A847V, =]. No parental DNA was available for study, but both parents were normal clinically. DNA or fibroblast cell culture contamination of samples from patients 5, 6 and 7 was ruled out by quantitative fluorescence PCR using genomic markers on different chromosomes
Patient 8 (Type C) carried two heterozygous mutations, c.1705A>G in exon 12 resulting in T569A, and c. 3499-3500delCT in exon 20 resulting in L1137VfsX1170. The T569A was inherited from his mother and the 3499–3500delCT was inherited from his father.
4. Quantitative PCR-RFLP
We used quantitative PCR-RFLP to measure the percentages of mutated DNA in the NE3-NE4 PCR products that contained the 1892G>A(R631Q) and the CO1-CO2 fragments that contained the 2540C>T( A847V). If R631Q and A847V were inherited, we would expect 50% mutated DNA in the quantitative PCR-RFLP. The mutated DNA ratios for 1892G>A(R631Q) were 45.3% in patient 2, 45.4% in patient 5, 72.9% in patient 6, and 79.1% in patient 7. The mutated DNA ratios for 2540C>T(A847V) were 33.4% in patient 5, 57.9% in patient 6, and 71.4% in patient 7. These percentages suggest the possibility of somatic mosaicism.
5. Western blot analysis
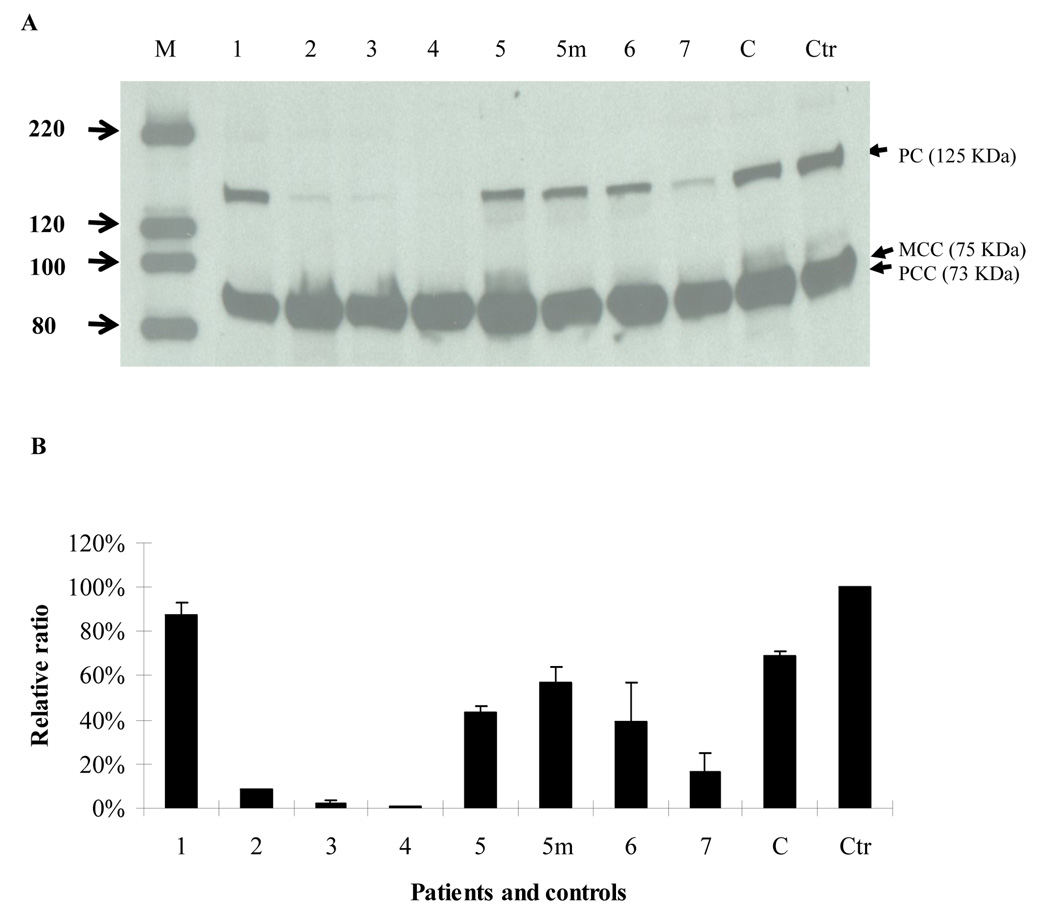
The amount of PC protein in patients 2, 3, 4 and 7 was significantly decreased to 8%, 3%, 2% and 17% of control values, respectively, while patients 5, 5m( mother of Patient 5), and 6 displayed 44%, 57%, and 49% of control values, respectively( Fig. 4A and B). Patient 1 exhibited 86% of control values.
Fig. 4. Western blot analysis.
Western blot was done as described in the Materials and Methods section. The membrane was then probed with streptavidin-HRP conjugate. PC, MCC and PCC were visualized using the horseradish peroxidase ECL system (Amersham, Piscataway, NJ) following the protocol. C is a carrier control and Ctr is normal control. A. molecular weight markers and patient codes are shown on the top. 5m is the mother of patient 5. On the left side of the figure the molecular weight of each band in the marker are labeled with arrow heads (in KDa). The location of PC, MCC, and PCC are shown by arrow heads. B. the western blot was analyzed using Image J. The density of PC protein was normalized by the total density of MCC and PCC, further normalized to the control (100%). The data are shown as mean ± SD, n =3.
Discussion
1. PC gene structure
Carbone et al. reported the PC gene structure in 1998 and later revised it (3, 24). Our studies extend the Carbone findings and refine the size of exons. There are a total of three tissue-specific PC transcripts: PC variant 1 (NM_000920.3), PC variant 2 (NM_022172.2) and PC variant 3 (BC011617.2) (Supplementary Table S3, Fig.1). The PC variants 1 and 3 are both expressed in brain and liver, confirmed by sequencing the RT-PCR products. The PC variant 2 is expressed in liver and kidney (18, 20, 21). These variants, resulting from alternative splicing by use of three promoters in the PC gene, differ only in the 5’UTR and share the same coding region. Alternative splicing is one of the most important molecular mechanisms for generating a large number of mRNA transcripts and proteins in the human genome(26). In the rat, the PC gene consists of 19 coding exons and four non-coding exons in the 5’-UTR. Alternative transcription from two tissue-specific promoters is responsible for the production of five different transcripts containing the same coding region (18). In domestic cow, six transcripts with the same coding region have been reported(27). These findings likely account for the observed tissue specific PC expression in mammals (18). In rat, there are two distinct promoters which control the alternative transcripts. The proximal promoter plays a role in gluconeogenesis and lipogenesis while the distal promoter is important for anaplerosis in pancreatic β-cells and other tissues (18). Distinct translation regulation by alternative 5’ UTRs has been reported for a stress-responsive protein –dPrxI (28), for human neuronal nitric-oxide synthase (29), and for human proinsulin gene (30). The three promoters in the human PC gene giving rise to three different PC isoforms may allow for tissue specificity, for differential functional capacity, or for developmental stage-specificity as is the rule with other human and mammalian genes (18, 26, 31).
2. Molecular genetics of PC deficiency
Type A genotype
Previous studies with skin fibroblasts from Type A cases indicate the presence of a mature, biotin-containing protein of correct molecular weight and mRNA(32). These mutant enzymes possess enough residual catalytic activity to sustain the anaplerotic role of PC. Four homozygous mutations p. [V145A] + [V145A]; p. [R451C] + [R451C]; p. [A610>T] + [A610T]; p. [M743I] + [M743I] have been identified in this group of patients (3, 33). Our Type A case (Patient 6) (Table 1) carried novel mutations p. [R62C; R631Q; A847V] + [=; R631Q, =; A847V, =]. The c.1982G>A( CGG>CAG) and c. 2540C>T( GCC>GTC) substitutions were found in two other patients (5 and 7), suggesting a common substitution caused by methylation-mediated deamination of 5-methylcytocine in the CpG dinucleotide with a mutation rate 8.5 times higher than other dinucleotides(34). Residual wild-type DNA in cultured fibroblasts was 27.1% based on quantitative PCR-RFLP analysis suggesting somatic mosaicism. The amount of wild-type DNA will vary from tissue to tissue based on this finding, offering a possible explanation as to why patient 6 exhibits a relatively mild clinical phenotype. Unfortunately, we could not obtain more tissues from this patient to confirm our speculation. We ruled out the possibility of DNA or fibroblast cell contamination in samples from patients 2, 5, 6 and 7 by comparing the quantitative fluorescence PCR results using genomic markers on different chromosomes between patient 5’s parents and the other patients.
Type B genotype
Absent or low levels of PC protein and mRNA transcript correlate with the Type B phenotype (32) . To date, only one case (two brothers) has been reported with compound heterozygosity c. [2493_2494delGT] + [2473+2_2473+5delTGCA] (24). We report another five Type B cases (Patients 2, 3, 4, 5 and 7 ) (Table 1 ) with novel complex mutations including missense mutations, deletions and splice donor site mutations in the form of homozygosity, compound heterozygosity and mosaicism. These findings expand the previous statements that insertions or deletions result in premature termination codons leading to mRNA degradation (24).
Three Type B patients (Patients 3, 4, and 7) died between age 8 days and 6 months whereas two Type B patients (Patient 2 and 5) remain alive at ages 20 and 9 years. Patient 3 carried c. [321+1G>T] + [321+1G>T], resulting in the deletion of the last 3 amino acids (V105_K107) of exon 2. This substitution disrupted the protein stability, severely decreasing the amount of residual protein (3%) in skin fibroblasts (Fig.4A and 4B) Patient 4 carried a homozygous transition 806G>A in both genomic DNA and mRNA, and the protein abundance was 1% of control suggesting a profound instability of the R269Q mutant protein. Patient 7 carried two transitions c.1892G>A and c.2540C > T existing in a somatic mosaic state (Fig.3A and 3B). The mutant DNA represented 79.1% and 71.3% respectively of the total DNA digested. Direct sequencing of the RT-PCR products of L3-R1 indicated that the mutant mRNA with r. 1892g>a and r. 2540c >u was rapidly degraded leaving only a small residual amount of wild-type protein (17% of control) (Fig.4 A and 4B). The patient died at age six months of bronchopneumonia complicated by progressive cardiac failure and renal insufficiency. These molecular and biochemical findings would explain the more severe clinical phenotype exhibited by patients 3 and 4. It is uncertain clinically whether patient 7 had a milder PC phenotype and died with an unrelated medical complication.
Patient 2 carried compound heterozygous mutations p. [R631Q, =] + [=, V831BfsX832]. The portion of 1892 G>A (R631Q) in the PCR product was 45.3%, indicating the1892G>A was a somatic mutation. Again, the RT-PCR sequencing results suggested the mutant mRNA carrying 2493_2494delGT (V831BfsX832) was not stable, which is consistent with previous findings in a patient with 2493_2494delGT (24). The mutant PC protein with R631Q was not stable, a finding that was similar to the other patients (5 and 7) in this group. The residual protein amount (8%) correlated with the amount of wild-type PC genomic DNA (4.7%). This finding and the mosaic state probably explains the prolonged survival in this patient.
Patient 5 carried complex substitutions p. [R156Q; V166I; R631Q, =; A847V, =] + [R156Q; V166I; =; =]. The c.1892G>A (R631Q) and c. 2540C>T (A847I) mutations caused a rapid degradation of mutant mRNA. DNA with the 1982G>A (R631Q) mutation was 45.3% of the total PC genomic DNA. DNA with the 2540C>T (A847V) mutation was 33.4% of the total DNA. We predicted that mutant PC protein with R156Q and V166I could be synthesized from mRNA with R156Q and V166I that was transcribed from 54.7% (100%-45.3%) of total PC genomic DNA. mRNA with R156Q, V166I and R631Q, transcribed from 11.9% (45.3%-33.4%) of total PC genomic DNA, would be hard to detect by direct sequencing of RT-PCR products. mRNA transcripts carrying R156Q, V166I, R631Q and A847V derived from 33.4% PC genomic DNA were quickly degraded according to the sequencing result of RT-PCR products. The mutant mRNA transcripts with R156Q and V166I ( derived from 54.7% PC gene ) produced 44% mutant PC protein as compared to the control, but the fibroblast PC activity was very low (0.03nmol/min/mg protein). As the R631Q and A847V were somatic mosaic mutations, we expect that the mosaic R631Q and A847V mutant PC gene percentage could vary from tissue to tissue. However, we can not explain why the R156Q and V166I and the low fibroblast PC enzyme activity is associated with a less severe clinical phenotype and prolonged survival. Further studies will be necessary to resolve this apparent paradox.
Type C genotype
Only four cases of PC deficiency have been reported with the benign phenotype (12, 13). We studied the molecular basis in patient 1 (12) and identified p.[S266A; =] + [=; S705X, =]. Only S266A and wild-type sequences S705 were identified in an RT-PCR fragment sequencing analysis suggesting that the mRNA carrying S705X was unstable and rapidly degraded. We postulate that the two substitutions are in trans and S705A is a somatic mosaic transversion. If true, the mosaic S705X mutant PC gene percentage could vary from tissue to tissue. This patient enjoyed normal development possibly attributable to the uneven distribution of the somatic mosaic mutation c. 2114C>G in tissues. However, based on Western blot analysis, we believe that the half-life of the mutant PC protein (S266A) is longer than the wild-type protein as the mixture still shows 86% PC protein (Fig. 4).
Patient 8 was identified to carry two heterozygous mutations in trans, T569A and L1137VfsX1170. These two mutations were inherited from his carrier parents who were clinically normal. The L1137VfsX1170 mutation resulted in the replacement of the last 42 amino acids by 33 different amino acids. The fibroblast PC enzyme activity was very low and we could not explain the benign phenotype in this patient. Further studies will be needed to address these discordant findings.
Genotype-phenotype correlation
There was no correlation between the clinical phenotype and residual fibroblast enzyme activity in our eight patients or in other patients reported previously(3, 9, 13, 32, 33, 35). It is known that the endogenous PC activity in fibroblasts is low compared to other tissues (18). It is not known which PC transcripts are expressed in human fibroblasts, or how genetic and environmental factors influence PC gene expression in different tissues. It is generally accepted that the amount of PC protein and residual tissue enzyme activity influence the severity of the clinical phenotype (13, 33, 36). Mosaicism may, in fact, exert an equivalent influence on phenotypic expression as suggested in this series of PC deficient patients and explain the discordance between protein amount and clinical phenotype in some cases
Seven mutations R62C, R631Q, A847V, V145A, R451C, A610T and M743I were responsible for the Type A phenotype in five patients (one from this report and four other patients previously reported) (Table 1). The type B phenotype correlates with complex missense mutations, deletions and splice donor site mutations in the form of homozygosity, compound heterozygosity and mosaicism (Table 2). An heterozygous mutation S266A and mosaic state S705X was the molecular basis for the first Type C patient (12), and compound heterozygosity T569A and L1137VfsX1170 was responsible for the second Type C patient‥
Mosaicism was found in five cases (Type A: 6; Type B: 2, 5, and 7; Type C: 1) and four had prolonged survival. Death in the sixth case (Type B 7) resulted from unrelated medical complications. The deaths of the more severely affected Type B patients correlated with homozygous mutations and with very low amounts of fibroblast PC protein of 2% and 3% (Patient 3 and 4)( Table 2). Age of onset is an important criterion in the clinical classification (neonatal-Type B, infantile-Type A and intermittent /benign -Type C); but prolonged survival appears to be determined by mosaicism, presence of mRNA and residual enzyme activity. We suggest that the discrepancy between prognostic expectation and clinical classification imply the presence of somatic mosaicism and appropriate molecular studies should be conducted to confirm this suggestion. Gene dosage will be determined by the post-implantational timing of the somatic mutation, and clinical expression will be influenced by the distribution of the mutation in the developing organism. We believe that this genetic mechanism is more common than currently realized and may obtain in all genetically-determined diseases where initial long-term expectancies do not match observed clinical outcomes.
We suggest that the discrepancy between the current findings and the existing classification system should be addressed by revising the classification system to accommodate these newer findings.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by NS37949 and 1 UL1 RR024156-01(DCD), the Will and the Colleen Giblin Foundations (DCD), K12-NS01698 (DW), and RPB, Inc. We thank Drs. W. Krause, R. Blackston, G. Herman, S. Simonetti, A. Palomeque, M. Pineda and J. Campistol for their clinical assessments of the PC patients. We thank Dr. Den Dunnen for his kind assistance in the nomenclatures of the complex sequence variants.
Abbreviations
- PC
Pyruvate carboxylase
- FISH
fluorescence in situ hybridization
- PCR
Polymerase chain reaction
- RFLP
Restriction fragment length polymorphism
- RT-PCR
Reverse-transcription PCR
- PCC
Propionyl-CoA carboxylase
- MCC
3-methylcrotonyl-CoA carboxylase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yoshida T, Tada K, Konno T, Arakawa T. Hyperalaninemia with pyruvicemia due to pyruvate carboxylase deficiency of the liver. Tohoku J Exp Med. 1969;99:121–128. doi: 10.1620/tjem.99.121. [DOI] [PubMed] [Google Scholar]
- 2.Atkin BM, Buist NR, Utter MF, Leiter AB, Banker BQ. Pyruvate carboxylase deficiency and lactic acidosis in a retarded child without Leigh's disease. Pediatr Res. 1979;13:109–116. doi: 10.1203/00006450-197902000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Carbone MA, MacKay N, Ling M, Cole DE, Douglas C, Rigat B, Feigenbaum A, Clarke JT, Haworth JC, Greenberg CR, Seargeant L, Robinson BH. Amerindian pyruvate carboxylase deficiency is associated with two distinct missense mutations. Am J Hum Genet. 1998;62:1312–1319. doi: 10.1086/301884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert EF, Arya S, Chun R. Leigh's necrotizing encephalopathy with pyruvate carboxylase deficiency. Arch Pathol Lab Med. 1983;107:162–166. [PubMed] [Google Scholar]
- 5.De Vivo DC, Haymond MW, Leckie MP, Bussmann YL, McDougal DB, Pagliara AS. The clinical and biochemical implications of pyruvate carboxylase deficiency. Journal of Clinical Endocrinology&Mrtabolism. 1977;45:1281–1296. doi: 10.1210/jcem-45-6-1281. [DOI] [PubMed] [Google Scholar]
- 6.Saudubray JM, Marsac C, Cathelineau CL, Besson Leaud M, Leroux JP. Neonatal congenital lactic acidosis with pyruvate carboxylase deficiency in two siblings. Acta Paediatr Scand. 1976;65:717–724. doi: 10.1111/j.1651-2227.1976.tb18009.x. [DOI] [PubMed] [Google Scholar]
- 7.Coude FX, Ogier H, Marsac C, Munnich A, Charpentier C, Saudubray JM. Secondary citrullinemia with hyperammonemia in four neonatal cases of pyruvate carboxylase deficiency. Pediatrics. 1981;68:914. [PubMed] [Google Scholar]
- 8.Bartlett K, Ghneim HK, Stirk JH, Dale G, Alberti KG. Pyruvate carboxylase deficiency. J Inherit Metab Dis. 1984;7 Suppl 1:74–78. doi: 10.1007/BF03047379. [DOI] [PubMed] [Google Scholar]
- 9.Robinson BH, Oei J, Sherwood WG, Applegarth D, Wong L, Haworth J, Goodyer P, Casey R, Zaleski LA. The molecular basis for the two different clinical presentations of classical pyruvate carboxylase deficiency. Am J Hum Genet. 1984;36:283–294. [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Cazorla A, Rabier D, Touati G, Chadefaux-Vekemans B, Marsac C, de Lonlay P, Saudubray JM. Pyruvate carboxylase deficiency: Metabolic characteristics and new neurological aspects. Ann Neurol. 2006;59:121–127. doi: 10.1002/ana.20709. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton J, Rae MD, Logan RW, Robinson PH. A case of benign pyruvate carboxylase deficiency with normal development. J Inherit Metab Dis. 1997;20:401–403. doi: 10.1023/a:1005350600278. [DOI] [PubMed] [Google Scholar]
- 12.Van Coster RN, Fernhoff PM, De Vivo DC. Pyruvate carboxylase deficiency: a benign variant with normal development. Pediatr Res. 1991;30:1–4. doi: 10.1203/00006450-199107000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Stern HJ, Nayar R, Depalma L, Rifai N. Prolonged survival in pyruvate carboxylase deficiency: lack of correlation with enzyme activity in cultured fibroblasts. Clin Biochem. 1995;28:85–89. doi: 10.1016/0009-9120(94)00059-5. [DOI] [PubMed] [Google Scholar]
- 14.Arnold GL, Griebel ML, Porterfield M, Brewster M. Pyruvate carboxylase deficiency. Report of a case and additional evidence for the "mild" phenotype. Clin Pediatr (Phila) 2001;40:519–521. doi: 10.1177/000992280104000909. [DOI] [PubMed] [Google Scholar]
- 15.Attwood PV. The structure and the mechanism of action of pyruvate carboxylase. Int J Biochem Cell Biol. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-r. [DOI] [PubMed] [Google Scholar]
- 16.Mackall JC, Lane MD. Role of pyruvate carboxylase in fatty acid synthesis: alterations during preadipocyte differentiation. Biochem Biophys Res Commun. 1977;79:720–725. doi: 10.1016/0006-291x(77)91171-8. [DOI] [PubMed] [Google Scholar]
- 17.Barden RE, Taylor BL, Isoashi F, Frey WH, Zander G, Lee JC, Utter MF. Structural properties of pyruvate carboxylases from chicken liver and other sources. Proc Natl Acad Sci U S A. 1975;72:4308–4312. doi: 10.1073/pnas.72.11.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jitrapakdee S, Walker ME, Wallace JC. Identification of novel alternatively spliced pyruvate carboxylase mRNAs with divergent 5'-untranslated regions which are expressed in a tissue-specific manner. Biochem Biophys Res Commun. 1996;223:695–700. doi: 10.1006/bbrc.1996.0958. [DOI] [PubMed] [Google Scholar]
- 19.Freytag SO, Collier KJ. Molecular cloning of a cDNA for human pyruvate carboxylase. Structural relationship to other biotin-containing carboxylases and regulation of mRNA content in differentiating preadipocytes. J Biol Chem. 1984;259:12831–12837. [PubMed] [Google Scholar]
- 20.Wexler ID, Du Y, Lisgaris MV, Mandal SK, Freytag SO, Yang BS, Liu TC, Kwon M, Patel MS, Kerr DS. Primary amino acid sequence and structure of human pyruvate carboxylase. Biochim Biophys Acta. 1994;1227:46–52. doi: 10.1016/0925-4439(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 21.MacKay N, Rigat B, Douglas C, Chen HS, Robinson BH. cDNA cloning of human kidney pyruvate carboxylase. Biochem Biophys Res Commun. 1994;202:1009–1014. doi: 10.1006/bbrc.1994.2029. [DOI] [PubMed] [Google Scholar]
- 22.Cole JA, Walker RE, Yordy MR. Hyperglycemia-induced changes in Na+/myo-inositol transport, Na(+)-K(+)-ATPase, and protein kinase C activity in proximal tubule cells. Diabetes. 1995;44:446–452. doi: 10.2337/diab.44.4.446. [DOI] [PubMed] [Google Scholar]
- 23.Old SE, De Vivo DC. Pyruvate dehydrogenase complex deficiency: biochemical and immunoblot analysis of cultured skin fibroblasts. Ann Neurol. 1989;26:746–751. doi: 10.1002/ana.410260610. [DOI] [PubMed] [Google Scholar]
- 24.Carbone MA, Applegarth DA, Robinson BH. Intron retention and frameshift mutations result in severe pyruvate carboxylase deficiency in two male siblings. Hum Mutat. 2002;20:48–56. doi: 10.1002/humu.10093. [DOI] [PubMed] [Google Scholar]
- 25.Pineda M, Campistol J, Vilaseca MA, Briones P, Ribes A, Temudo T, Pons M, Cusi V, Rolland MO. An atypical French form of pyruvate carboxylase deficiency. Brain Dev. 1995;17:276–279. doi: 10.1016/0387-7604(95)00057-i. [DOI] [PubMed] [Google Scholar]
- 26.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Agca C, Bidwell CA, Donkin SS. Cloning of bovine pyruvate carboxylase and 5' untranslated region variants. Anim Biotechnol. 2004;15:47–66. doi: 10.1081/ABIO-120037897. [DOI] [PubMed] [Google Scholar]
- 28.Chiu LL, Tsai YL, Lee WC, Cho YM, Ho HY, Chen SM, Chen MT, Kuo CH. Acute effect of exercise-hypoxia challenge on GLUT4 protein expression in rat cardiac muscle. High Alt Med Biol. 2005;6:256–262. doi: 10.1089/ham.2005.6.256. [DOI] [PubMed] [Google Scholar]
- 29.Newton DC, Bevan SC, Choi S, Robb GB, Millar A, Wang Y, Marsden PA. Translational regulation of human neuronal nitric-oxide synthase by an alternatively spliced 5'-untranslated region leader exon. J Biol Chem. 2003;278:636–644. doi: 10.1074/jbc.M209988200. [DOI] [PubMed] [Google Scholar]
- 30.Shalev A, Blair PJ, Hoffmann SC, Hirshberg B, Peculis BA, Harlan DM. A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology. 2002;143:2541–2547. doi: 10.1210/endo.143.7.8920. [DOI] [PubMed] [Google Scholar]
- 31.Ayoubi TA, Van De Ven WJ. Regulation of gene expression by alternative promoters. Faseb J. 1996;10:453–460. [PubMed] [Google Scholar]
- 32.Robinson BH, Oei J, Saudubray JM, Marsac C, Bartlett K, Quan F, Gravel R. The French and North American phenotypes of pyruvate carboxylase deficiency, correlation with biotin containing protein by 3H-biotin incorporation, 35S-streptavidin labeling, and Northern blotting with a cloned cDNA probe. Am J Hum Genet. 1987;40:50–59. [PMC free article] [PubMed] [Google Scholar]
- 33.Wexler ID, Kerr DS, Du Y, Kaung MM, Stephenson W, Lusk MM, Wappner RS, Higgins JJ. Molecular characterization of pyruvate carboxylase deficiency in two consanguineous families. Pediatr Res. 1998;43:579–584. doi: 10.1203/00006450-199805000-00004. [DOI] [PubMed] [Google Scholar]
- 34.Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 35.Robinson BH, Oei J, Saunders M, Gravel R. [3H]biotin-labeled proteins in cultured human skin fibroblasts from patients with pyruvate carboxylase deficiency. J Biol Chem. 1983;258:6660–6664. [PubMed] [Google Scholar]
- 36.Robinson BH. Lactic acidemia: disorders of pyruvate carboxylase and pyruvate dehydrogenase. New York: McGraw-Hill; 2000. pp. 2275–2295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.