Abstract
Cofilin is essential for cell viability and for actin-based motility. Cofilin severs actin filaments to enhance the dynamics of filament assembly. We investigated the mechanism of filament severing by cofilin with direct fluorescence microscopy observation of single actin filaments in real time. In cells, actin filaments are likely to be attached at multiple points along their length, and, we found that attaching filaments in such a manner greatly increased the efficiency of filament severing by cofilin. Cofilin severing increased and then decreased with increasing cofilin concentration. Together, these results indicate that cofilin severs the actin filament by a mechanism of allosteric and cooperative destabilization. Severing is more efficient when relaxation of this cofilin-induced instability of the actin filament is inhibited by restricting the flexibility of the filament. These conclusions have particular relevance to cofilin function during actin-based motility in cells and in synthetic systems.
Keywords: actin, cofilin, gelsolin, severing, flexibility
INTRODUCTION
Many cellular processes involve rapid remodeling of the actin cytoskeleton and changes in the pools of assembled and monomeric actin1. Not surprisingly, rapid cycles of actin polymerization/depolymerization2 that occur at the leading edge of motile cells depend on factors accelerating both the polymerization and depolymerization of actin filaments. The cofilin/ADF (Actin Depolymerizing Factor) family of proteins appears to be a major factor contributing to actin depolymerization in cells, which is essential for recycling actin subunits to support new filament growth. Cofilin can sever actin filaments, creating free barbed and pointed ends available for polymerization or depolymerization, depending on the local actin monomer concentration3–5. Cofilin also appears to increase the rate of loss of actin subunits from the pointed end of the actin filament6. In vivo, actin filaments are likely to be capped at one or both ends over time, suggesting that severing may be crucial for disassembly of actin filaments and turnover of the actin filament network5. Indeed, cofilin is essential for viability in a number of cell systems, based on its function in actin filament dynamics and actin-based motility7,8.
When actin-based motility was reconstituted from pure proteins, cofilin was a necessary component, along with Arp2/3 complex and capping protein. Increasing concentrations of cofilin first increased, and then decreased the rate of actin-based motility9, showing that the balance of filament assembly and disassembly is important. Correspondingly, in cells, the motility of glioblastoma tumor cells was recently reported to increase and then decrease with increasing overexpression of cofilin10. These observations also correlate with the bell-shaped dependence of the rate of actin filament treadmilling on cofilin concentration6,11.
Cofilin binding to the side of an actin filament results in conformational changes that have been proposed to account for increased severing and pointed-end dissociation, the two mechanisms of cofilin-induced actin depolymerization. These conformational changes in the actin filament include a shift in the mean twist of actin filaments12,13 and substantial changes in longitudinal and lateral interprotomer contacts13, which are predicted to lead to filament destabilization. Accordlngly, filament flexibility has been suggested as an important variable for the frequency of severing events along the filament, be those spontaneous14 or mediated by cofilin6. On the other hand, recent differential scanning calorimetry (DSC) studies suggest a dual role for cofilin in actin turnover, revealing stabilization or destabilization of the filament, depending on the molar ratio of cofilin to actin15,16. Such F-actin stabilization should occur locally, at the filament sites directly in contact with cofilin17.
Cofilin binds cooperatively to the actin filament, and cooperative effects of cofilin on the structure of the filament have been revealed in measurements of the twist and torsional flexibility of the filament13,18,19, and through the presence of “tilted” filament structure in segments free of cofilin20. Gelsolin also affects the actin filament conformation cooperatively21,22, causing severing of the filament based on breaks in longitudinal actin-actin contacts combined with distortions of lateral, across-strands bonds23.
In this work, we asked how filaments are severed by cofilin and gelsolin. We performed direct observations of individual actin filaments with real-time fluorescence microscopy, which allows one to visualize severing events. More important, this approach allowed us to vary the flexibility of the filaments by changing their attachment to the substrate. Our results show that filament flexibility and degree of saturation with cofilin and gelsolin have important and different impacts on how the two proteins sever actin filaments.
RESULTS
The goal of this study was to investigate the relationship between cofilin interactions with actin filaments and the severing of these filaments. We have focused on the effects of filament flexibility and filament saturation by cofilin on the severing process, using gelsolin for comparison. We used TRC-labeled actin for fluorescence imaging of actin filaments in lieu of the commonly employed rhodamine phalloidin, because phalloidin competes with cofilin for binding to F-actin24. Conveniently, the addition of TRC label at Gln-41 of actin (as well as other probes at this site25,26 lowers the critical concentration for actin polymerization, stabilizes filaments, and reports on the cofilin binding without impairing that process27. Thus, TRC~actin filaments could be easily viewed in a fluorescence microscope at nanomolar concentration (5.0 – 20 nM) without significant depolymerization28. For the observation of severing, TRC~actin filaments were attached to a glass coverslip surface through binding to HMM, α-actinin, or CapZ adsorbed to that surface. Addition of yeast cofilin in assay buffer (pH 6.0 – 8.0) led to appearance of filament fragments due to break points introduced along the entire filament within the first 60 to 120 seconds (Fig. 1). Over time, these fragments underwent additional severing and shortening until almost complete disappearance (the detached filament debris diffused away from the focal plane). Severing activity was measured only on those filaments that could be identified in fluorescence images taken before and after cofilin addition. Although the progress of severing with time was qualitatively visible in all early time points, full statistical analysis was carried out for filaments exposed to cofilin for 2 minutes, at which point their fragmentation could be measured best.
Figure 1. TRC~F-actin severing by cofilin.
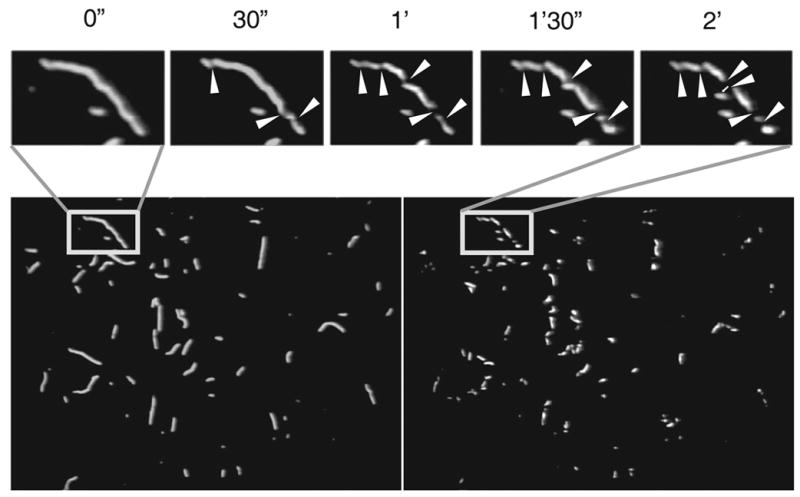
Snapshot images of TRC~F-actin filaments bound to the HMM-covered surface were taken before addition of cofilin and during the incubation for 2 minutes with yeast cofilin. The top panel shows magnified images of a single filament degradation taken every 30 seconds after addition of cofilin. The assay buffer contained 2.0 mM K+-EGTA, 20 mM KCl, 2.0 mM MgCl2, 10 mM DTT, 25 mM MOPS (total ionic strength - 50 mM), 14 mM glucose, 9×10003 units of catalase/mL, and 240 units of glucose oxidase/mL at pH 8.0. The glass surface was coated with HMM at a concentration of 5 μg/mL. White arrowheads indicate sites of severing. Identical data was obtained at pH 6.0.
Importantly, TRC-labeling of actin did not impede or change significantly the severing of filaments by cofilin. This conclusion is supported by measuring the severing of 25% TRC-labeled filaments (co-polymers of fully labeled and unlabeled actin), which resulted in similar, albeit somewhat smaller changes in filament length distribution as those observed for fully labeled actin (Fig. 2).
Figure 2. Severing of actin filaments tethered (via CapZ) and attached (via HMM) to the glass surface.
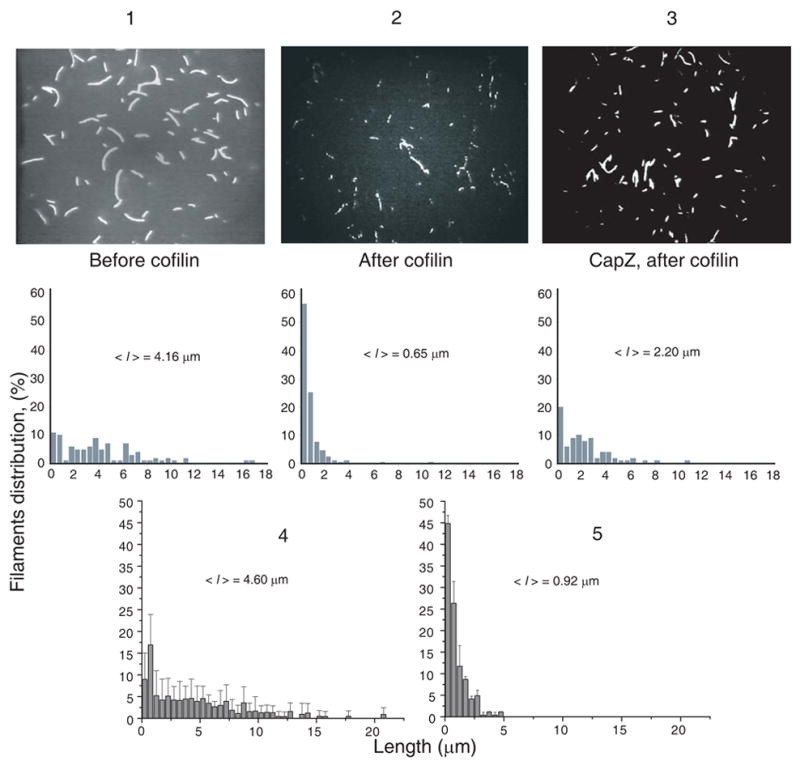
Several snapshots of TRC~F-actin being held by HMM (2) and CapZ (3) were taken before the addition of 50 nM cofilin (1) and after 2 minutes of incubation with cofilin at 25 °C and pH 8.0 (for filaments held to the surface by HMM (2) and CapZ (3)). Snapshot images of TRC~F-actin filaments were enhanced and processed using the Sigma Scan Pro 5 image analysis program. The length of at least 150 filaments was measured in each case after the screen was calibrated using a grid-containing slide. Length distributions were plotted in lower panels 1–3. The mean filament length was 2–3 times smaller when the severed filaments were attached to the surface by HMM (2) rather than tethered via their barbed ends to CapZ (3). Length distributions are shown also for TRC-F-actin filaments co-polymerized from 25% fully labeled TRC-G-actin and 75% unlabeled G-actin before the addition of 20 nM cofilin (4) and after their incubation with 20 nM cofilin for 2 minutes at 25°C and pH 8.0 (5).
The ability to detect the severed filament fragments in our system rests on the diffusional motion of fragment ends, which generates observable gaps between them. As stated above, and documented in a prior study showing pH-independence of filament severing by yeast cofilin28, depolymerization was unlikely to shorten significantly the TRC-actin filaments in our short-time-scale experiments (up to 2 minutes). Nevertheless, we did not measure the kinetic rates of actin severing by cofilin since that would require more accurate accounting of all factors that may affect the data. Instead, we chose a single optimal time-point for a data-based, qualitative analysis of the effects of actin flexibility and cofilin concentration on actin severing.
F-actin flexibility and the severing by cofilin and gelsolin
To investigate the role of filament flexibility in the severing process, we varied the concentration of HMM or α -actinin adsorbed to the coverslip as the tethering protein. In most of our experiments, we used the lowest amount of HMM (~5μg/ml) that could hold filaments at several points on the working surface. Using the results of previous studies29, we estimated that the approximate density of HMM molecules attached to the working surface in our experiments was: ~111±33 μm−2.
Given that actin filaments are 6 nm in diameter30, and assuming that the only crossbridges that interact with actin filament are those that are within 10 nm from it, (i.e., within a 26 nm wide “band” could reach the filament29). Using this assumption, we calculated the number of myosin heads that were able to interact with 1 μm length of actin filament to be ~ 6 ± 2 μm−1. High HMM concentrations led to tethering at multiple sites along the filament, thus decreasing filament flexibility and preventing reliable observation of separate filament fragments during its severing by cofilin. At a constant cofilin concentration, increasing concentrations of HMM (up to 300 μg/mL) led to easily observed increases in the rate of actin filament severing. However, because at higher HMM concentrations the diffusional motion of filament fragments (which create the gaps between them) is reduced, these observations probably underestimate the actual severing increase, and thus, are not analyzed in quantitative terms. In addition, we noted that those free actin filaments in the solution that had not been attached or washed away, were severed at a much slower rate than were the surface-attached filaments. This suggests that the mode of filament attachment and the resulting variation in filament flexibility may play an important role in severing by cofilin.
To provide maximum flexibility of actin filaments while tethering them to the surface, we employed CapZ, which was able to hold actin filaments onto the surface through their barbed end. To reduce the Brownian motion of the filaments and thereby keep them in focus, methylcellulose was added (up to 0.2 %) to the assay solution (methylcellulose had no effect on the severing of HMM-tethered F-actin). Upon addition of yeast cofilin, we observed shortening of the tethered filaments (Fig. 2). Some severed filament fragments diffused away, so we could not image all severing events. Indeed, the debris of short filament fragments became very noticeable above the surface plane, as a result of filament severing. Severing of CapZ-tethered filaments was decreased compared to HMM-bound filaments, as revealed by analysis of filament length distributions after 2 minutes incubation with 50 nM cofilin (Fig. 2). Qualitatively and quantitatively, the filaments tethered by one end, with CapZ, were severed less efficiently than those tethered by multiple side attachments, by HMM.
Cofilins can also promote depolymerization by increasing the off rate of actin protomers from the pointed end of the filament. In our experimental approach, this potential complication was minimized by the use of TRC-labeled actin, which is depolymerized by cofilin much less than is unlabeled F-actin28. In previous work with TRC-actin, we found that the decrease in filament length caused by cofilin corresponded closely with the increase in the number of filaments, implying little, if any, effect of filament depolymerization28. In addition, in that study, TRC-labeling of actin did not appear to change its severing by cofilin, as actin labeled at a different amino acid residue with a different fluorophore behaved in a similar manner. This conclusion is supported by showing similar fragmentation of fully and partially labeled filaments by cofilin (Fig. 2). Also, to exclude the possibility that depolymerization contributed to the filament shortening observed above, we repeated the experiments in the presence of the pointed-end capper tropomodulin. With both CapZ and tropomodulin present, filaments on the coverslip surface shortened, and short fragments of filament were observed above the surface, revealing severing activity.
We also tested the effect of filament flexibility on severing with capped filaments. Actin filaments were prepared in solution in the presence of CapZ and tropomodulin, to cap both ends, and then applied to an HMM-covered surface, as above, to restrict flexibility. Again, we observed that severing induced by cofilin was much faster than when filaments were tethered to the surface only by one end via CapZ (same as in Fig. 2). This result confirms that restriction of filament flexibility promotes the severing process by cofilin.
The preceding experiments were performed with budding yeast cofilin. We repeated the key experiments with cofilin from nematodes (C. elegans UNC-60B) and from mice. Again, the severing was greater when the actin filaments were bound to the surface via multiple side attachments with HMM, as opposed to single-end attachments with CapZ (data not shown). Thus, the effect of filament flexibility on severing appears to be a common property of cofilins in general.
To determine whether severing of actin filaments increases with decreased filament flexibility in general, we tested the action of gelsolin in similar experiments. Unlike with cofilin, increasing concentrations of HMM applied to the glass surface decreased severing by gelsolin (Fig. 3). In contrast, when CapZ was used for tethering actin filaments to the glass surface (equivalent to 0 μg/mL of HMM in Fig. 3), the gelsolin-caused severing was fast and complete. These results are the opposite of what we observed with cofilin, pointing to the different mechanisms of severing by cofilin and gelsolin.
Figure 3. Changes in the mean actin filaments length vs. HMM concentration (applied to the glass surface) after 2 minutes incubation with 100 pM gelsolin.
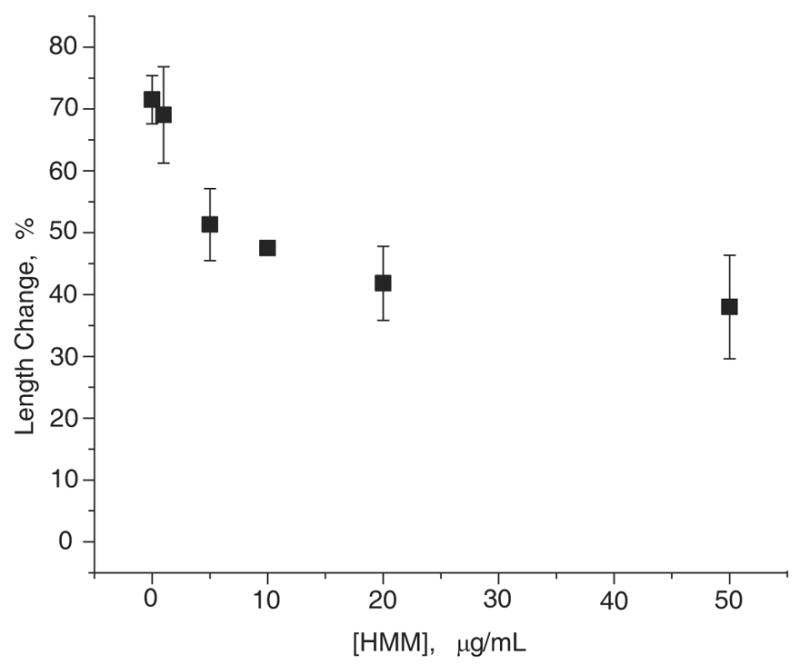
Several snapshots of TRC~F-actin filaments tethered by CapZ (0 μg/mL of HMM) and attached to HMM were taken before the addition of 100 pM gelsolin, and after 2 minutes incubation with gelsolin at 25 °C. Images were enhanced and processed as in Fig. 2. The length of at least 150 filaments was measured in each case. The change in the filament mean length was plotted vs. the concentration of HMM applied to the glass surface prior to actin addition. Maximum filament length change was observed when the severed filaments were tethered via their barbed ends to CapZ.
Complex dependence of F-actin severing on cofilin concentration
To gain further insight into the mechanism of cofilin’s severing action on F-actin, we used fluorescence microscopy to test the functional implications of previous calorimetry results showing actin filament destabilization and stabilization, depending on filament saturation by cofilin15,16. To this end, we varied the concentration of various cofilins (from 20 nM to 1 μM) added to the actin filaments immobilized at a constant density of HMM. Initial increases in cofilin concentration, from zero, caused a sharp rise in the severing activity (Fig. 4), but further increases caused a gradual decrease in severing activity. We repeated these experiments using CapZ instead of HMM to tether actin filaments via their barbed ends to the glass surface, in order to avoid any competition between cofilin and HMM for binding sites on the actin filament and to retain maximal torsional flexibility of actin filaments. The severing activity of cofilin was lower with CapZ tethering, at any given concentration, compared to that with HMM tethering (Fig. 4). However, the shape of the profile of severing activity vs. cofilin concentration was similar in the two cases. Mouse (data not shown) and nematode (Fig. 4) cofilins had different levels of severing activities than did yeast cofilin, but they all showed a similar concentration dependence with severing activity, reaching a maximum at some optimal concentration and decreasing at higher cofilin concentrations (Fig. 4). Thus, high cofilin concentrations stabilized actin filaments15,16.
Figure 4. Change of actin filaments mean length after 2 minutes incubation with cofilin from different sources.
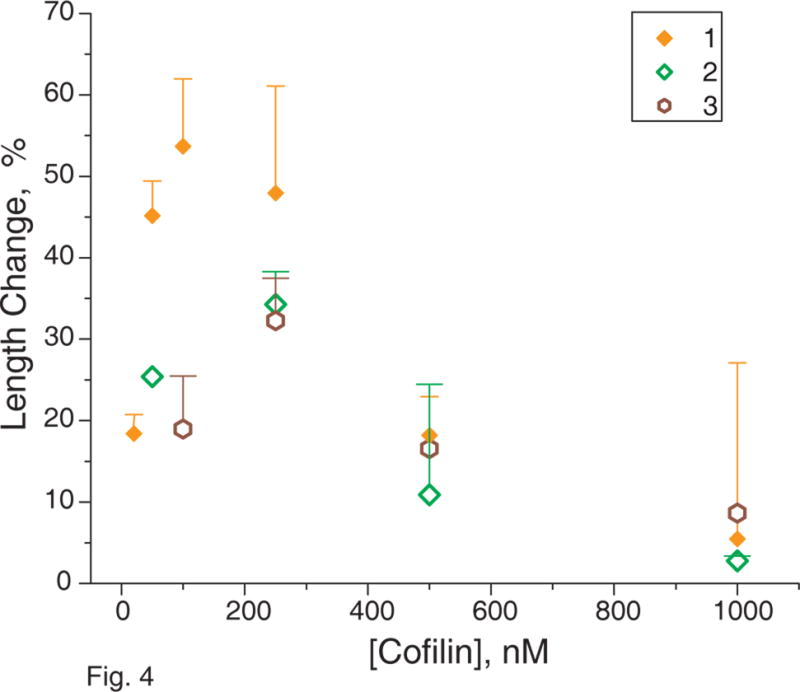
Several snapshots of TRC~F-actin filaments being held by HMM or CapZ were taken before the addition of cofilin from different sources, and after 2 minutes incubation with cofilin at 25 °C. Images were enhanced and processed as in Fig. 2. The length of at least 150 filaments was measured in each case. The change in the filament mean length distributions was plotted vs. concentration of cofilin added to the immobilized actin filaments. 1 - yeast cofilin was added to actin filaments attached to the glass surface by HMM; 2 - yeast cofilin was added to actin filaments attached to the glass surface by CapZ; 3 - nematode cofilin was added to actin filaments attached to the glass surface by CapZ. The data obtained for S3C mouse cofilin is not shown.
We also tested filament severing in solution, where the actin : cofilin stoichiometry is known accurately. In these experiments, TRC~F-actin (2 μM) was gently mixed with cofilin (0.25–5.0 μM), and after 30 and 180 seconds aliquots of the mixture were diluted (200 fold) with buffer containing 1 μM phalloidin, to stop the severing process. The filaments were then applied to HMM-coated coverslips in order to observe the filament lengths. Increasing cofilin concentrations increased the filament severing rate, reaching a maximum at approximately 1 : 2 molar ration of cofilin : actin, and then decreasing gradually with further increases in the cofilin : actin ratio, consistent with previous reports on cofilin effect on actin treadmilling and depolymerization6,11. The magnitude of the decrease in the filament length was less than in the case of “surface” experiments, perhaps because of incomplete stopping of the severing reaction by phalloidin, but the overall effect was similar. The results of these functional assays support the conclusions of the calorimetric studies in which actin filaments were stabilized when saturated by cofilin and destabilized at lower cofilin concentration15,16.
We next tested the effect of gelsolin concentration on filament severing activity in solution. In contrast to cofilin, the gelsolin severing rate increased with increasing gelsolin concentration (50 pM – 100 nM).
DISCUSSION
Filament flexibility and severing
The most important result of this study is the experimental confirmation of a prior suggestion6 that actin filament flexibility affects the ability of cofilin to sever the filaments. It appears that when the actin filament is tethered to a glass surface only via its barbed ends, the filament can dissipate the structural strain caused by the binding of cofilin. We found that these filaments, tethered at one end, were severed much slower than filaments attached at multiple points along their sides, which did not allow dissipation of the induced strain. Two different filament side-binding proteins, HMM and α-actinin, showed this effect, with the severing rate increasing when the density of attachments was increased. Similar results were obtained for cofilins from different sources. For the myosin complex with F-actin, several solution studies indeed reported a decreased flexibility of filaments31–33, albeit another study34 did not detect such myosin-induced changes in actin. Clearly, in our case, with filaments tethered to the glass slide at multiple points the resulting decrease in filament flexibility would be greater than in the solution.
Our interpretation of these results, in line with the speculation of Carlier et al6 is that multiple attachments of actin filaments restrict the relaxation of the cofilin-induced torsional strain, accounting for more efficient severing. This mechanism may be important in cells, where cofilin is essential for actin filament dynamics and cytoskeleton reorganization. Tethering or cross-linking of actin filaments is a common feature at the leading edge of cells35, where actin assembly is prominent. Nevertheless, as demonstrated here, filament severing by cofilin also occurs in filaments that are unconstrained by attachments.
In contrast to cofilin, gelsolin severed unconstrained filaments much faster and more efficiently than those held by multipoint attachments to the glass surface. This basic mechanistic difference between filament severing by gelsolin and cofilin may be related to their different tasks, perhaps with one of the proteins participating in actin recycling in static cells, and the other at the leading edge of motile cells36.
Filament saturation by cofilin and severing
Another important experimental finding of this study is that cofilin severs actin filaments most efficiently at subsaturating concentrations. Similar optimal effects of substoichiometric ratios of cofilin to actin on filament treadmilling rate and the in vitro motility of a reconstituted system have been reported before6,9,11, but these effects were not connected directly to the severing by cofilin or the mechanism of its action. Preferential filament severing at substoichiometric cofilin to actin ratios was proposed recently16,37, but without a direct observation provided here. Our observation is consistent with the recent discovery of dual effects of cofilin on actin filaments in calorimetry studies15,16. Those studies revealed allosteric and cooperative destabilization of interprotomer contacts in segments of actin filament free of cofilin, coupled with steric stabilization at the sites of cofilin binding via an interprotomer actin-cofilin-actin bridge between subdomain 2 and subdomain 1 on adjacent protomers. The calorimetry results showed that actin filaments saturated with cofilin were stabilized against thermal denaturation. On the other hand, subsaturating cofilin concentrations yielded two populations of actin filaments: one in which the filaments were stabilized, as in the case of filaments saturated with cofilin, and another one in which the filaments were cooperatively destabilized. A critical prediction of these calorimetry results is that filament severing should achieve a maximum rate at subsaturating cofilin concentrations and decline with further increases in cofilin concentration. Our results here, with direct observation of filament severing, confirm this prediction (Fig. 4). The same conclusion was also reached from examining the severing of actin filaments in solution, which was maximal at approximately 1 : 2 cofilin to actin mole ratio, suggesting that the severing occurs preferentially at a site neighboring to the cofilin-bound actin. This result is important because the binding ratios of cofilin to actin are well defined in solution, but potentially less certain for filaments on glass surfaces. Again, cofilin from different sources yielded similar results in all cases. In addition, gelsolin did not show this dependence of filament severing on concentration, confirming that cofilin and gelsolin have different mechanisms for severing.
A model for the mechanism of actin filament severing
Several distinctive features of the interaction of cofilin with actin filaments must be included when one considers models for the mechanism of filament severing:
The high stability of severed and short cofilin-actin filaments11
The cooperative disruption of interprotomer contacts and the cooperative increase in torsional motion of filaments upon cofilin binding13,16,18,38
The facilitation of filament assembly from polymerization-impaired actin by cofilin, based on the bridging of adjacent actin protomers17,19
The altered filament conformations in the presence of cofilin (tilted state, torsional strain)18,19
The allosteric destabilization of unoccupied filament segments at low cofilin densities15,16,18,19.
To account for these observations, we have confirmed that the actin filaments break preferentially at sites unoccupied by cofilin, but neighboring or close to the cofilin-bound actin16,24,37. At these sites, the structural perturbation (or strain) propagated from the cofilin-bound protomers is maximal, not yet dissipated over the length of the filaments. At these weak spots, energy from Brownian motion may suffice to sever the filaments. The results of the present study are consistent with this strain-based severing model, since flexible filaments, tethered only by one end, should dissipate the cofilin-induced structural strain far more effectively than do less flexible, filaments attached at multiple points along their length.
More importantly, the severing activity of several cofilins showed a biphasic concentration dependence, first increasing, and then decreasing with cofilin concentration. This result provides support for the model of preferential filament severing at the unoccupied sites, assuming that the cumulative torsional strain increases up to a certain level of cofilin saturation18, while the probability of conformational fluctuations that can sever filaments at the sites occupied by cofilin is reduced. We note that these results and our model may account for the recently reported increase and then decrease of glioblastoma tumour cells motility with increased overexpression of cofilin10. The strong severing inhibition seen at high cofilin concentration is most likely a “by-product” of filament stabilization by TRC attached to Gln-41. Clearly, cofilin severs also fully saturated filaments, as documented before11 and in our solution tests. This severing may arise from similar, strained twist states of F-actin, especially when the stabilizing cofilin bridge between two actin protomers is transiently broken, leaving cofilin bound to only one protomer.
In striking contrast to cofilin, but not surprisingly, gelsolin does not show such a complex severing behavior. Filament flexibility may have little effect on gelsolin action, and the apparent inhibition of severing with increasing density of F-actin attachments to HMM (Fig. 3) may result from decreased diffusional motion of filament fragments, thereby impeding detection of severing events. Our results are consistent with the prevailing model in which the initial binding of gelsolin segment 2 (GS2) to F-actin, and the subsequent structural changes in gelsolin, culminate in the intercalation of gelsolin segment 1 (GS1) between two actin protomers and thus, filament severing23.
Addendum
While this manuscript was under review, a study reporting similar observations was published by Andrianantroandro and Pollard39. Their results are consistent with our data on a complex mechanism of actin filaments severing by cofilin, showing more efficient filament severing at low cofilin concentrations and less efficient at high concentrations. This study showed also increased probability of filament severing with their increased length, but did not consider the effects of filament flexibility on the severing.
MATERIALS AND METHODS
Proteins
Skeletal myosin, actin, and heavy meromyosin (HMM) were prepared as reported previously40. α-actinin was purchased from Cytoskeleton Inc. (Denver, CO). Gelsolin was purchased from Sigma Chemical Co. (St. Louis, MO) or purified from bovine plasma41.
The labeling of Gln-41 on skeletal actin with TRC was performed using Ca2+-independent bacterial transglutaminase (TGase) (a generous gift from Dr K. Seguro; Ajinomoto Co, Kawasaki, Japan) as previously described27. The extent of labeling (~100%) was determined using TRC extinction coefficient of = 78 mM 1 cm 1. Gelsolin-capped filaments were obtained by polymerizing actin in the E544 nm presence of gelsolin (1:3000 – 1:200 mole ratios of gelsolin to actin) with 1.0 mM CaCl2, 50 mM KCl and 2.0 mM MgCl2.
WT yeast cofilin was expressed and purified as previously described42. Nematode cofilin from Caenorhabditis elegans (UNC-60B) was a generous gift from Dr. Shoichiro Ono (Emory University). S3C mouse (Mus Musculus) cofilin was a generous gift from Dr. Gerard Marriott (University of Wisconsin). The concentrations of cofilin and unlabeled α-actin were determined spectrophotometrically, using extinction coefficients E2801%=9.2 cm−1 and E2901%=11.5 cm−1, respectively.
Tropomodulin was a generous gift from Dr. Velia M. Fowler (The Scripps Research Institute). Mouse actin capping protein (CapZ), (α1 and β 2 subunits), was expressed and purified as described by Palmgren43.
In Vitro Severing Assays
Fluorescence microscopy and actin covalently labeled at Gln41 with tetramethylrhodamine cadaverine (TRC) were used for direct observation of filament severing. In vitro severing assays were performed similarly to the previously described in vitro motility experiments40. A glass slide with spacers of double-side Scotch tape and a nitrocellulose-coated coverslip formed a ~70 μl chamber open on two sides. HMM (1 – 300 μg/mL), α-actinin (1.0 μM), CapZ (0.6 – 5.0 μM), or tropomodulin (0.6 μM) were applied to the chamber; then BSA (2.0 mg/mL) was added to block the glass surface. TRC-labeled F-actin (TRC-FA) (20 nM) was then applied in an assay buffer (2.0 mM K+-EGTA, 20 mM KCl, 2.0 mM MgCl2, 10 mM DTT, 25 mM MOPS, pH 7.4; total ionic strength 50 mM) containing an oxygen scavenging system44 at 25 °C. The unbound filaments were washed off with the assay buffer. The slide was placed on a temperature-controlled stage (25 °C) of a Leica fluorescence microscope and fixed with a Scotch tape. The TRC-labeled actin filaments were visualized using epifluorescence illumination (100-watt mercury arc lamp and a 100× , numeric aperture = 1.3 objective), were imaged on a VE 1000 SIT camera (DAGE-MTI, Michigan City, IN), and were recorded without enhancement on a Panasonic VHS VCR. Severing was initiated by adding cofilin or gelsolin in an assay buffer at the desired pH (6.0 – 8.0), at concentrations ranging from 20 nM to 1.0 μM for cofilin and from 50 pM to 100 nM for gelsolin. Assay buffer containing cofilin or gelsolin was carefully applied to an open side of the working chamber, and the excess liquid was removed from the other side, while the field of view remained unchanged and the filaments on the surface remained in focus. During image recording by VCR, the filaments were exposed to light for short periods of time (10–15 sec, every 30–60 seconds) to avoid photobleaching. Several snapshots were taken before cofilin addition, and over the first 2–5 minutes after its addition, using ATI TV-tuner. The recorded images were averaged, enhanced, and analyzed using the SigmaScan Pro 5 program (SPSS Inc.).
Experimental approach limitations
The goal of real-time observation of the effects of cofilin concentration and actin filament flexibility on its severing was achieved by using TRC-labeled actin in fluorescence microscopy experiments. The severed filament fragments were detected and measured because of the gaps generated between them through filament severing. As concluded before, for TRC-F-actin these gaps stem mainly from diffusional motion of the ends of tethered fragments, rather than from the depolymerization of these fragments (within our short observation times)28. Although the evaluation of cofilin concentration effect on filament severing was carried out at an empirically determined optimal (and constant) tethering HMM concentration – to allow for diffusional motion of filament fragment ends – our data are interpreted mainly in terms of qualitative trends and the overall behavior of the actin-cofilin system. This approach is dictated by experimental limitations i.e., the inability to measure very short filament fragments either because of their diffusion from the observation field or the size that is smaller than detection limit (~250 nm). Similarly, in experiments involving changes in HMM (or α–actinin) concentrations, the effect of such changes on the detection accuracy of filament severing measurements is difficult to calibrate, hence the results are presented either in qualitative terms (for cofilin) or are considered in terms of measurement limitations (for gelsolin).
Acknowledgments
We thank Professor Earl Homsher for his help in data acquisition and Mai Phan for excellent technical assistance.
ABBREVIATIONS LIST
- TRC
tetramethylrhodamine cadaverine
- HMM
heavy meromyosin
- ADF
actin depolymerizing factor
Footnotes
This work was supported by grants from the USPHS to E. R. (GM-077190) and J. C. (GM38542) and the NSF to E. R. (MCB 0316269).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pollard TD, Earnshaw WC. Cell Biology. Saunders; Philadelphia, PA: 2004. [Google Scholar]
- 2.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh M, Ichetovkin I, Song X, Condeelis JS, Lawrence DS. A new strategy for caging proteins regulated by kinases. J Am Chem Soc. 2002;124:2440–2441. doi: 10.1021/ja017592l. [DOI] [PubMed] [Google Scholar]
- 4.Ichetovkin I, Han J, Pang KM, Knecht DA, Condeelis JS. Actin filaments are severed by both native and recombinant dictyostelium cofilin but to different extents. Cell Motil Cytoskel. 2000;45:293–306. doi: 10.1002/(SICI)1097-0169(200004)45:4<293::AID-CM5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Maciver SK, Pope BJ, Whytock S, Weeds AG. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur J Biochem. 1998;256:388–397. doi: 10.1046/j.1432-1327.1998.2560388.x. [DOI] [PubMed] [Google Scholar]
- 6.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono K, Parast M, Alberico C, Benian GM, Ono S. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. JCell Sci. 2003;116:2073–2085. doi: 10.1242/jcs.00421. [DOI] [PubMed] [Google Scholar]
- 8.Ono S. Regulation of Actin Filament Dynamics by Actin Depolymerizing Factor/Cofilin and Actin-Interacting Protein 1: New Blades for Twisted Filaments. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- 9.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 10.Yap CT, Simpson TI, Pratt T, Price DJ, Maciver SK. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell Motil Cytoskel. 2005;60:153–165. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- 11.Pope BJ, Gonsior SM, Yeoh S, McGough A, Weeds AG. Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J Mol Biol. 2000;298:649–661. doi: 10.1006/jmbi.2000.3688. [DOI] [PubMed] [Google Scholar]
- 12.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sept D, Xu J, Pollard TD, McCammon JA. Annealing Accounts for the Length of Actin Filaments Formed by Spontaneous Polymerization. Biophys J. 1999;77:2911–2919. doi: 10.1016/s0006-3495(99)77124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dedova IV, Nikolaeva OP, Mikhailova VV, dos Remedios CG, Levitsky DI. Two opposite effects of cofilin on the thermal unfolding of F-actin: a differential scanning calorimetric study. Biophys Chem. 2004;110:119–128. doi: 10.1016/j.bpc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative Effects of Cofilin (ADF) on Actin Structure Suggest Allosteric Mechanism of Cofilin Function. J Mol Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 17.Kudryashov DS, Sawaya MR, Adisetiyo H, Norcross T, Hegyi G, Reisler E, Yeates TO. The crystal structure of a cross-linked actin dimer suggests a detailed molecular interface in F-actin. P Natl Acad Sci. 2005;102:13105–13110. doi: 10.1073/pnas.0506429102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prochniewicz E, Janson N, Thomas DD, De La Cruz EM. Cofilin Increases the Torsional Flexibility and Dynamics of Actin Filaments. J Mol Biol. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Galkin VE, Orlova A, VanLoock MS, Shvetsov A, Reisler E, Egelman EH. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J Cell Biol. 2003;163:1057–1066. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galkin VE, VanLoock MS, Orlova A, Egelman EH. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin's sequence and structure. Curr Biol: Cb. 2002;12:570–5. doi: 10.1016/s0960-9822(02)00742-x. [DOI] [PubMed] [Google Scholar]
- 21.McGough A, Chiu W, Way M. Determination of the Gelsolin Binding Site on F-actin: Implications for Severing and Capping. Biophys J. 1998;74:764–772. doi: 10.1016/S0006-3495(98)74001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orlova A, Prochniewicz E, Egelman EH. Structural dynamics of F-actin: II. Cooperativity in structural transitions. J Mol Biol. 1995;245:598–607. doi: 10.1006/jmbi.1994.0049. [DOI] [PubMed] [Google Scholar]
- 23.McGough AM, Staiger CJ, Min JK, Simonetti KD. The gelsolin family of actin regulatory proteins: modular structures, versatile functions. FEBS Lett. 2003;552:75–81. doi: 10.1016/s0014-5793(03)00932-3. [DOI] [PubMed] [Google Scholar]
- 24.Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9:364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- 25.Takashi R. A novel actin label: a fluorescent probe at glutamine-41 and its consequences. Biochemistry. 1988;27:938–943. doi: 10.1021/bi00403a015. [DOI] [PubMed] [Google Scholar]
- 26.Kim E, Motoki M, Seguro K, Muhlrad A, Reisler E. Conformational changes in subdomain 2 of G-actin: fluorescence probing by dansyl ethylenediamine attached to Gln-41. Biophys J. 1995;69:2024–32. doi: 10.1016/S0006-3495(95)80072-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhlrad A, Kudryashov D, Michael Peyser Y, Bobkov AA, Almo SC, Reisler E. Cofilin Induced Conformational Changes in F-actin Expose Subdomain 2 to Proteolysis. J Mol Biol. 2004;342:1559–1567. doi: 10.1016/j.jmb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Severing of F-actin by yeast cofilin is pH-independent. Cell Motil Cytoskel. 2006;63:533–542. doi: 10.1002/cm.20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uyeda TQP, Kron SJ, Spudich JA. Myosin Step Size: Estimation From Slow Sliding Movement of Actin Over Low Densities of Heavy Meromyosin. J Mol Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 30.Wakabayashi T, Huxley HE, Amos LA, Klug A. Three-dimensional image reconstruction of actin-tropomyosin complex and actin-tropomyosin-troponin T-troponin I complex. J Mol Biol. 1975;93:477–497. doi: 10.1016/0022-2836(75)90241-7. [DOI] [PubMed] [Google Scholar]
- 31.Thomas DD, Seidel JC, Gergely J. Rotational dynamics of spin-labeled F-actin in the sub-millisecond time range. J Mol Biol. 1979;132:257–273. doi: 10.1016/0022-2836(79)90259-6. [DOI] [PubMed] [Google Scholar]
- 32.Hegyi G, Belagyi J. Intermonomer cross-linking of F-actin alters the dynamics of its interaction with H-meromyosin in the weak-binding state. FEBS J. 2006;273:1896–1905. doi: 10.1111/j.1742-4658.2006.05197.x. [DOI] [PubMed] [Google Scholar]
- 33.Prochniewicz E, Walseth TF, Thomas DD. Structural Dynamics of Actin during Active Interaction with Myosin: Different Effects of Weakly and Strongly Bound Myosin Heads. Biochemistry. 2004;43(33):10642–10652. doi: 10.1021/bi049914e. [DOI] [PubMed] [Google Scholar]
- 34.Burlacu S, Borejdo J. Motion of Actin-Filaments in the Presence of Myosin Heads and ATP. Biophys J. 1992;63:1471–1482. doi: 10.1016/S0006-3495(92)81744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of Cofilin in Epidermal Growth Factor-stimulated Actin Polymerization and Lamellipod Protrusion. J Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Southwick FS. Gelsolin and ADF/cofilin enhance the actin dynamics of motile cells. P Natl Acad Sci. 2000;97:6936–6938. doi: 10.1073/pnas.97.13.6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De La Cruz EM. Cofilin Binding to Muscle and Non-muscle Actin Filaments: Isoform-dependent Cooperative Interactions. J Mol Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 38.McGough A, Chiu W. ADF/cofilin weakens lateral contacts in the actin filament. J Mol Biol. 1999;291:513–519. doi: 10.1006/jmbi.1999.2968. [DOI] [PubMed] [Google Scholar]
- 39.Andrianantoandro E, Pollard TD. Mechanism of Actin Filament Turnover by Severing and Nucleation at Different Concentrations of ADF/Cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Miller CJ, Wong WW, Bobkova E, Rubenstein PA, Reisler E. Mutational analysis of the role of the N terminus of actin in actomyosin interactions. Comparison with other mutant actins and implications for the cross-bridge cycle. Biochemistry. 1996;35:16557–65. doi: 10.1021/bi962388+. [DOI] [PubMed] [Google Scholar]
- 41.Kurokawa H, Jujii W, Ohmi K, Sakurai T, Nonomura Y. Simple and rapid purificiation of brevin. Biochem Bioph Res Co. 1990;168:451–457. doi: 10.1016/0006-291x(90)92342-w. [DOI] [PubMed] [Google Scholar]
- 42.Bobkov AA, Muhlrad A, Kokabi K, Vorobiev S, Almo SC, Reisler E. Structural effects of cofilin on longitudinal contacts in F-actin. J Mol Biol. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 43.Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155:251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umemoto S, Sellers JR. Characterization of in vitro motility assays using smooth muscle and cytoplasmic myosins. J Biol Chem. 1990;265:14864–9. [PubMed] [Google Scholar]


