Abstract
A fast and sensitive LC-ESI-MS method is described for the comparative quantification of 16 estrogen metabolites based on the derivatization of estrogens with a novel derivatizing reagent, N-methyl-nicotinic acid N-hydroxysuccinimide ester (C1-NA-NHS). The process introduces a quaternary amine to the analytes, making the analytes permanently charged regardless of the pH of the HPLC mobile phase. This quaternization resulted in a highly efficient separation of 16 estrogen metabolites in 7 minutes at a detection level below 1 ng/mL. By using a deuterated derivatizing reagent (C1-d3-NA-NHS), a complete set of deuterated standards was utilized and used as internal standards in a comparative quantification and recovery study, demonstrating acceptable results over a wide concentration range. A pooled breast cancer serum sample was analyzed using the described method, and 15 estrogens were detected in the range of 80 – 530 pg/mL.
1. Introduction
Estrogens are part of a group of steroids that are known to stimulate secondary sexual characteristics, along with some other physiological effects such as growth and maturation of long bones. But estrogens may be involved in carcinogenesis as well. Over a century ago, endogenous estrogens were postulated to have a critical role in the development of breast cancer [1]. Evidence has accumulated from studies involving cell cultures, animal models, and epidemiology showing a strong linkage between exposure to estrogens and breast cancer risk [2,3]. Estrogens have also been found to be involved in colon cancer [4] and ovarian cancer, especially when connected to postmenopausal estrogen treatment [5,6].
Estrogens generally appear in tissues and biological fluids at low concentrations. When combined with the complexity of the matrix within which they are found, analysis of estrogens in biological samples is challenging. The recent review by Giese shows that immunoassays, high performance liquid chromatography (HPLC) with electrochemical or mass spectrometric (MS) detection, and gas chromatography (GC) with electron capture detection (ECD) or MS detection are among the most widely used methods for estrogen analysis [7]. One complication is the structural diversity of estrogens (Figure 1). Another is the possible conjugation with other species, such as DNA, sulphate and glucuronide [7]. The analytical methodology must deal with this complexity. Immunoassays, such as radioimmunoassay [8], are an important high throughput tool for estrogen analysis, but fail to measure all the forms of estrogen in a single assay. Even when processed with a separation step, such as HPLC, some methods can only accommodate on a single estrogen [9–12]. Xu et al developed several LC-MS methods for the simultaneous measurement of endogenous estrogens [13–16], eventually analyzing 15 different estrogens [14,15]. In these separations, multiple estrogens coeluted, and differentiation with mass spectrometry was required to provide resolution of these coeluted analytes.
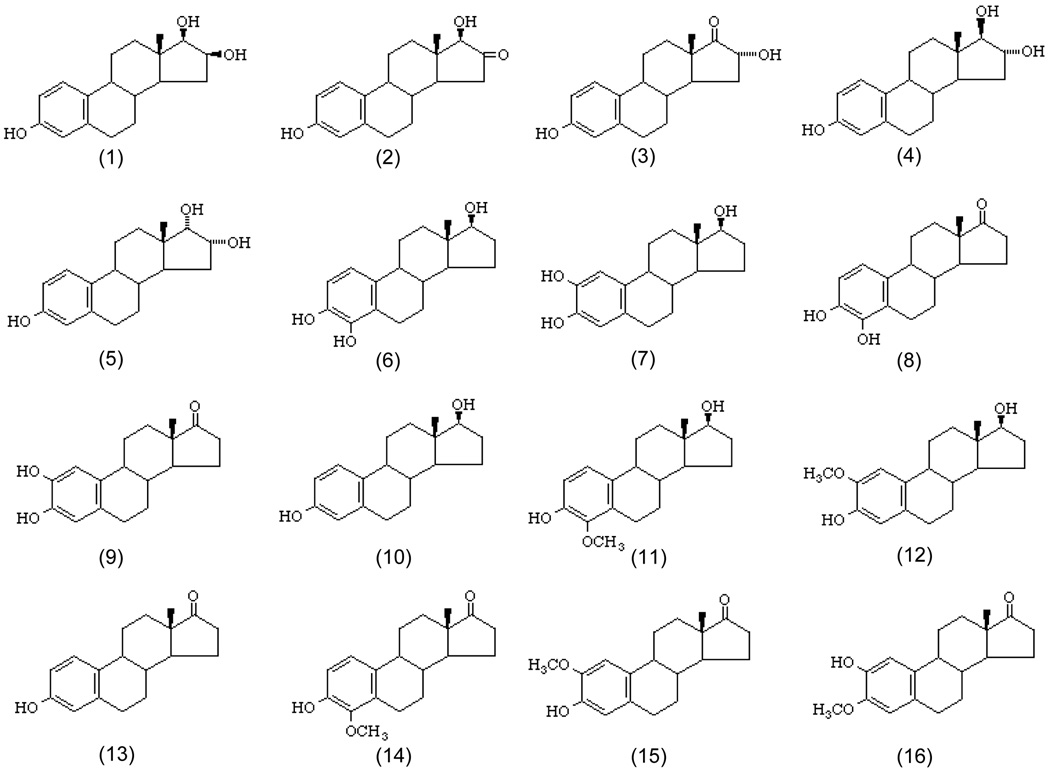
Figure 1.
Structures of the 16 estrogens studied in this work. Compound: 1. 16-epiestriol; 2. 16-keto-17β-estradiol, 3. 16α-hydroxyestrone; 4. estriol; 5. 17-epiestriol; 6. 4-hydroxyestradiol; 7. 4-hydroxyestrone; 8. 2-hydroxyestradiol; 9. 2-hydroxyestrone; 10. 17β-estradiol; 11. 4-methoxyestradiol; 12. 2-methoxyestradiol; 13. estrone; 14. 4-methoxyestrone; 15. 2-methoxyestrone; 16. 2-hydroxyestrone-3-methyl ether.
Another issue that must be addressed is the large number of samples that have to be examined for a meaningful epidemiological analysis. Analytical methods must be relatively fast to meet the throughput needs of such studies. The widely used National Institute of Standard and Technology (NIST) LC-MS/MS method for estradiol requires a 30 minute separation, with additional time for sample derivatization [11]. In comparison, a method for simultaneous determination of 15 estrogens developed by Xu et al requires a 60 minute separation time [15] and can be shortened to roughly 10 min with supercritical fluid chromatography [14]. Furthermore, the concentration of estrogens in physiological samples can be very low, as pointed out earlier, causing analytical difficulties. The circulating estradiol-17β level in postmenopausal women is usually < 73 pmol/L, and concentrations ≤ 18 pmol/L are common [17].
An ultrasensitive chemiluminescent immunoassay for estradiol was reported with detectability of 1.8 pmol/L [17]. Most commercially available estradiol immunoassays have detection limits between 73 and 183 pmol/L [18]. Selected Reaction Monitoring (SRM) in MS based technique is also a very sensitive analytical tool. With this technique, the limit of detection for estradiol after derivatization was decreased to 1 ng/L (3.7 pmol/L) [11].
Still another challenging aspect is the requirement for derivatization as a tool to improve the detection limit. Electrospray ionization (ESI) is a popular ionization source in LC-MS, but has the shortcoming that sensitivity is analyte-dependent. The structure of estrogens suggests they would not readily protonate during ESI. ESI-MS detection is often facilitated by derivatization with easily protonated reagents, such as toluensulfonhydrazide [13,16] or dansyl chloride [11,14,15]. In contrast, derivatization with an electron-capturing moiety such as o-pentafluorobenzyl [19] or 4-nitrobenzenesulfonyl [20] substantially enhances detection in Electron-Capturing Atmospheric Pressure Chemical Ionization (ECAPCI) mass spectrometry. Similarly, electron capture derivatives have been used in gas chromatography with Electron Capture Detection (GC-ECD) [21] and Electron Capture Mass Spectrometry (GC-EC-MS) [22,23]. These methods usually utilize Electron Impact (EI) ionization and silylating derivatization to increase the volatility and stability of estrogens [24–28].
The demand for high throughput analysis, such as in the case of epidemiology studies, as mentioned earlier, has resulted in the use of stable isotope-coded internal standards [16,24,27]. A limitation of stable isotope coding is that appropriate heavy isotope labeled standards may not be commercially available. In this case, an available isotope-coded internal standard is used as a structural analogue for the quantification of other analytes whose isotope-coded versions are unavailable. Quantification can suffer when the separation behavior and relative molar response of the instrumentation for the standard and analytes are very different.
The challenges noted above were addressed in these studies through the development of a comparative HPLC-ESI-MS method centering on a novel stable isotope-coding agent that facilitates ionization and quantification, in addition to accelerating HPLC separation. Stable isotope-based internal standard quantification of 16 unique estrogen metabolites was achieved in less than 7 min at a detection level below 1 ng/mL.
2. Materials and methods
2.1. Chemicals and reagents
Estrogen standards (Figure 1) were purchased from Steraloids Inc (Newport, RI). Iodomethane, dansyl chloride, β-glucuronidase, anhydrous acetonitrile (ACN), dimethylformamide (DMF), dimethyl sulfoxide (DMSO), nicotinic acid dicyclohexylcarbodiimide (DCC), triethylamine (TEA), N-hydroxysuccinimide, and human serum were obtained from Sigma (St. Louis, MO). HPLC grade ACN, acetone and formic acid were obtained from Mallinkrodt Baker (Phillipsburg, NJ). Iodomethane-d3 was a product of Cambridge Isotope laboratories (Andover, MA). Double-deionized water was produced by a Millipore Milli-Q gradient A10 system (Bedford, MA).
Each estrogen standard was prepared in acetonitrile at 0.2 mg/mL as a stock solution. Standard solutions were prepared by mixing equal amounts of corresponding stock solutions and performing serial dilutions with ACN to the desired concentration.
Derivatizing reagents, N-methyl-nicotinic acid N-hydroxysuccinimide ester (C1-NA-NHS) and N-methyl-d3-nicotinic acid N-hydroxysuccinimide ester (C1-d3-NA-NHS), were synthesized as described by Yang et al [29]. Briefly, nicotinic acid was first esterified by N-hydroxysuccinimide through the activation of DCC to form nicotinic acid N-hydroxysuccinimide ester, which was further quaternized by iodomethane or iodomethane-d3.
2.2. Derivatization of estrogens
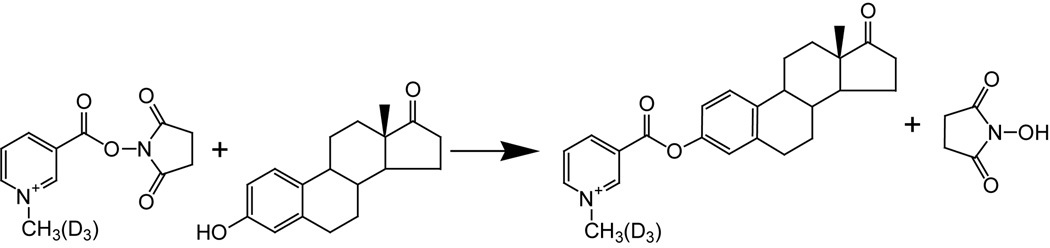
A typical derivatization procedure was as follows: 10 µL of standard solution (12.5 µg/mL each) was added into 37 µL of 50 mg/mL derivatizing reagent, C1-NA-NHS, prepared in dimethyl sulfoxide (DMSO) just before use, followed by the addition of 1 µL TEA. After vortexing the mixture for 30 seconds, 2 µL of formic acid was added to stop the reaction. For comparative quantification, another aliquot of working solution was simultaneously derivatized by C1-d3-NA-NHS. These two derivatized solutions were mixed in specific ratios for the analysis. A representative derivatization reaction is shown in Figure 2.
Figure 2.
Representative derivatization reaction of estrogens.
For comparison, estrogens were also derivatized with dansyl chloride [11]. In this case, 10 µL of standard solution (12.5 µg/mL each) was added into 37 µL of 50 mg/mL dansyl chloride in acetone/0.1 M sodium bicarbonate buffer, pH 10.4 (50:50, v/v), incubated for 5 min at 60°C and kept at 4°C for analysis.
2.3. Recovery studies
Two 0.5 mL-aliquots of human serum labeled as C (control) and S (spiked) were placed into two 15 mL-tubes containing 0.5 mL 0.2 M acetate buffer (pH 4.2), 1 mg of ascorbic acid and 5 µL β-glucuronidase (to release estrogens from glucuronide and sulfate conjugates). Then 8 µL of ACN and 8 µL of standard solution (125 ng/mL each) were added into samples C and S, respectively. After overnight incubation at 37°C, estrogens in samples C and S were extracted by adding 5 mL of dichloromethane and vortexing the samples for 20 min. The lower layers of sample C and S were removed, dried under nitrogen (~ 30 min), and derivatized by adding 10 µL of ACN, 37 µL of freshly prepared C1-d3-NA-NHS (50 mg/mL), 1 µL of TEA, and 2 µL of formic acid after vortexing for 30 seconds. Ten µL of standard solutions (500 ng/mL each) were also derivatized by adding 37 µL of freshly prepared C1-NA-NHS (50 mg/ml), followed by 1 µL of TEA and 2 µL of formic acid after 30 seconds vortexing. A 10 µL of this “light” derivatized standard solution was added to sample S for the recovery studies. Both samples were analyzed by LC-ESI-MS as described below. Recovery was calculated in sample S based on the extracted ion chromatographic peak areas for the pair of deuterated (heavy) and non-deuterated (light) derivatives of estrogens corresponding to their recovered and original spiked amounts, respectively.
2.4. Preparation of breast cancer serum
Two mL of serum from patients with breast cancer was placed into a15 mL-tube containing 0.5 mL 0.2 M acetate buffer (pH 4.2), 1 mg of ascorbic acid and 5 µL β-glucuronidase solution. After overnight incubation at 37°C, the sample was extracted by vortexing for 20 minutes with 5 mL of dichloromethane. The lower layer was removed, dried under nitrogen, and derivatized by adding 10 µL of ACN, 37 µL of freshly prepared C1-NA-NHS in DMSO (50 mg/mL) and 1 µL of TEA followed by 30 seconds vortexing. The reaction was stopped with 2 µL of formic acid. Ten µL of standard solutions (500 ng/mL each) were also derivatized by adding 37 µL of freshly prepared C1-d3-NA-NHS (50 mg/mL), and 1 µL of TEA followed by vortexing for 30 seconds and addition of 2 µL of formic acid to stop reaction. Ten µL of “heavy” labeled standard solution was mixed with 10 µL of "light” labeled human cancer sample solution for the comparative quantification. The mixture was analyzed by the LC-ESI-MS method described below. The concentration factor, 20, was applied for the estimation of estrogen concentration in the original serum. The mixture was analyzed by the LC-ESI-MS method described below. Estrogen concentration (CE) in serum was estimated according to equation:
where Ih and Il represent peak intensities of heavy labeled standard and light labeled estrogen in serum, 500 ng/mL is concentration of a standard and 20 is concentration factor of serum sample.
2.5. LC-MS
HPLC experiments were performed on an Agilent XDB-C18 column (2.1 × 50 mm, 1.8 µm particle size) and an Agilent 1100 Series LC (Agilent Technologies, Wilmington, Delaware). For negative APCI-MS, the mobile phase A consisted of 10 mmol/L ammonium acetate (pH 6.8) containing 5% ACN and mobile phase B was 10 mmol/L ammonium acetate containing 95% ACN. For positive APCI-MS and ESI-MS, the mobile phase A was composed of 5% ACN, 94.95% water and 0.05% formic acid and B was 95% ACN, 4.95% water and 0.05% formic acid. Column elution was achieved in a 30 min linear gradient from 0% to 100% B, at a flow rate of 0.4 mL/min. The injection volume was 5 µL.
MS analysis was carried out on Unique® LC-TOF MS with a high flow ESI and APCI Sources (LECO Corporation, St. Joseph, MI). Operating parameters for positive ESI were as follows: electrospray voltage, 4900 V; desolvation temperature, 370 °C; nebulizer pressure, 400 kPa; interface temperature, 100 °C; nozzle voltage, 230 V; skimmer voltage, 75 V; quad RF voltage, 250 V; quad high voltage, 76 V; quad low voltage, 32 V; quad exit voltage, 32 V; desolvation gas, 6880 mL/min. The main parameters for negative APCI were: desolvation temperature, 300 °C; nebulizer pressure, 350 kPa; interface temperature, 100 °C; corona current, 5 µA; nozzle voltage, −170 V; skimmer voltage, −57 V; quad RF voltage, 180 V; quad high voltage, −54 V; quad low voltage, −45 V; quad exit voltage, −16V. Operating condition for positive APCI were: desolvation temperature, 300 °C; nebulizer pressure, 350 kPa; interface temperature, 100 °C; corona current, 5 µA;nozzle voltage, 170 V; skimmer voltage, 70 V; quad RF voltage, 225 V; quad high voltage, 60 V; quad low voltage, 32 V; quad exit voltage, 29 V. All these parameters were optimized for intensity and resolution using the Agilent tuning solution.
3. Results and discussion
3.1. LC-APCI-MS of underivatized estrogens
Atmospheric pressure chemical ionization (APCI) is being used increasingly for the detection of a wide range of biological substances at low concentrations. In the case of steroids, cholesterols [30], phytoestrol [31], aldosterone [10], and progestogens [32] have been analyzed using APCI-MS or tandem MS in either the positive or negative modes. This suggests that APCI-MS might be useful in the analysis of the estrogens listed in Figure 1.
Negative APCI-MS was used with 10 mM ammonium acetate (pH 6.3) as the elution buffer to assist the ionization of analytes. From the extracted [M-H]− ion chromatograms for individual analyte injections (data not shown),all of the analytes, except 6 and 7 (which have the same retention time and m/z values) could be separated without deliberate optimization of the separation. Elution order clearly reflected analyte hydrophobicity. Table 1 lists the limits of detection (LOD) at a signal to noise ratio of 3 in [M-H]− selected ion monitoring mode. Except for the 4 catechol estrogens, the LODs were not sufficiently low to be useful for analysis of biological samples. [M-H]− ions were the most abundant in mass spectra but many other fragment and adduct ions are formed as well (Table 1). Comparatively, catechol estrogens produce fewer fragments, giving relatively higher detection sensitivity. The negative ion sensitivity problem with APCI-MS has been noted with ethinylestradiol as well ( > 50 ng/mL) [9].
Table 1.
Detection limits of underivatized estrogens with LC-negative APCI-MS.
| Peak number | Estrogens | m/z | Avg LOD (µg/mL, n=5) |
|---|---|---|---|
| 1 | 16-epiestriol | 259.05 (18%), 287.10(100%)*, 288(38%), 317(10%) | 6.60 |
| 2 | 16-keto-17β-estradiol | 285.11(100%), 299.11(60%) | 4.20 |
| 3 | 16α-hydroxyestrone | 279.09(10%), 285.12(100%), 286.08(30%), 298.98(10%), 301.14(15%) | 4.80 |
| 4 | estriol | 259.02(35%), 285.11(40%), 287.13(100%), 301.13(50%) | 15.70 |
| 5 | 17-epiestriol | 259.03(28%), 261.06(38%), 285.12(40%), 287.14(100%), 301.09(32%), 303.00(28%), 305.08(26%) | 11.7 |
| 6 | 4-hydroxyestradiol | 283.06(40%), 287.14(100%) | 0.03 |
| 7 | 4-hydroxyestrone | 285.11(100%) | 0.05 |
| 8 | 2-hydroxyestradiol | 285.07(40%), 287.10(100%), 288.11(50%) | 0.014 |
| 9 | 2-hydroxyestrone | 285.11(100%) | 0.014 |
| 10 | 17β-estradiol | 263(60%), 271(100%) | 0.143 |
| 11 | 4-methoxyestradiol | 259.03(60%), 275.00(100%), 286.10(70%), 301.16(75%) | 19.2 |
| 12 | 2-methoxyestradiol | 246.06(50%), 283.07(90%), 285.06(80%), 287.01(100%), 317.11(90%), 331.13(50%) | 61.5 |
| 13 | estrone | 259.04(18%), 269.10(100%), 285.09(18%) | 37 |
| 14 | 4-methoxyestrone | 248.97(65%), 261.99(85%), 283.03(88%), 299.13(100%) | 8.7 |
| 15 | 2-methoxyestrone | 246.02(65%), 248.07(55%), 274.00(72(%), 284.07(90%), 299.01(100%), 331.17(40%) | 32.4 |
| 16 | 2-hydroxyestrone-3-methyl ether | 246.00(68%), 274.03(60%), 283.05(50%) 284.10(85%), 285.12(66%), 289.05(79%), 299.01(100%), 315.10(52%) | 17.8 |
Underlined ions were used for detection limit determination.
Similar results were observed using 0.05% formic acid in the mobile phase, and positive APCI-MS mode. For example, the spectrum of estradiol chromatographic peaks showed a large number of ions that have not been previously reported: [M+H]+, [M-H2O+H]+, or [M+CH3CN]+ [32]. These discrepancies indicate that formation of ions in an APCI ion source are instrument dependent. The broad distribution of fragment ions reduces detection sensitivity, diminishing the utility of positive ion APCI for the quantification of estrogens in biological samples.
3.2. Derivatization of estrogens
As seen in Figure 1, those endogenous estrogen metabolites [7, 15] possesse the aromatic steroid core structure with phenol and alcohol hydroxyl groups. These hydroxyl groups are very weakly acidic with ionization of the phenol hydroxyl group being greater than the alcohol. At the pH of the HPLC mobile phase, ionization of ethinylestradiol is theoretically less than 0.001% [9]. Because ionization in the gas phase is directly related to ionization in solution [33], the limited ionization of estrogens in solution implies poor ionization efficiency in ESI-MS. One way to enhance ESI sensitivity is to directly introduce a quaternary amine (permanent positive charge) into analytes. This strategy has been applied to amino acids and peptides [29,34,35], alcohols, phenols and thiols [36,37], sugars [38,39], and carboxylic acids [40]. In a previous study from our group, ESI-MS response to tryptophan was enhanced roughly 26 fold after derivatization with the quaternary amine contain reagent C1-NA-NHS compared to the neutral benzoate derivative [29]. In the terms of reactivity, the phenol hydroxyl group in the estrogens is similar to the amine group in amino acids, suggesting that the amino acid derivatization procedure can be used, with few modifications, for estrogen labeling as well. Problems with the solubility of analytes were addressed by changing to a non-aqueous reaction system in which the estrogens are soluble. Small amounts of water (<10% in volume) in the reaction did not adversely affect the reaction (data not shown). The second modification was implemented in order to stabilize the derivatives by neutralizing the reaction with formic acid after the reaction was completed. Although the reaction is initiated in basic medium, the derivatives are not stable at these conditions. Similar problems have been observed in the case of O-acylation of tyrosine with C1-NA-NHS [29]. In neutral solutions, the derivatives are stable for at least one week at 4°C.
Under optimized conditions, C1-NA-NHS only reacted with phenol hydroxyl groups, not with alcohol hydroxyls. This property is significant because it increases the analytical selectivity of estrogens in complex biological matrices. Although alcohol hydroxyl groups are partially derivatized at elevated temperature, this is avoided by performing the reaction at room temperature.
3.3. LC-ESI-MS of estrogen derivatives
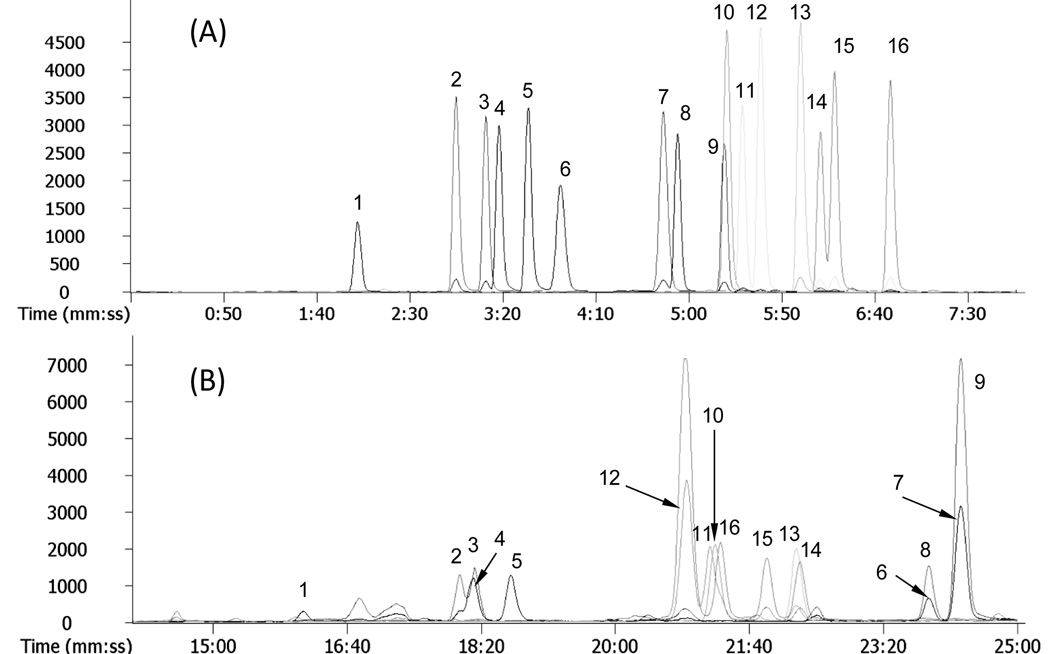
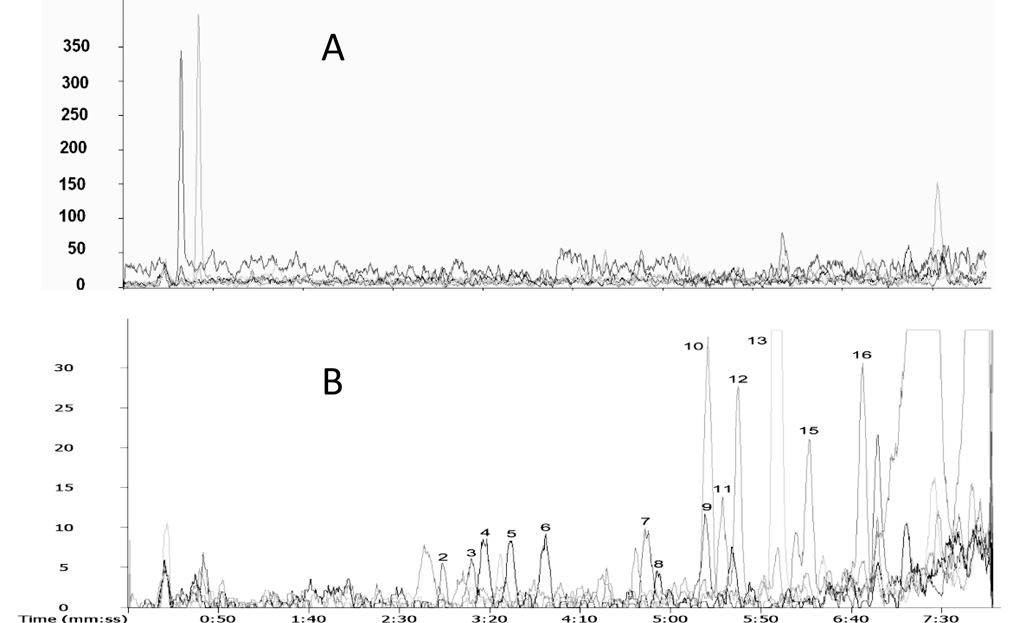
The estrogens listed in Figure 1 are very similar in structure, sharing an aromatic steroid backbone, hydroxyl or methoxyl groups at similar positions, and several pairs of positional isomers or enantiomers. An extracted ion chromatogram (Figure 3A) of the derivatized estrogens shows that except for components 9 and 10, all the other estrogens are well resolved within 7 min. The mass difference between components 9 (2-hydroxyestrone) and 10 (7β-estradiol) is, however, 14 amu and mass spectrometry easily differentiates between the two. Compared to the separation of underivatized estrogens with ammonium acetate mobile phase, these results suggest that derivatization increased the chromatographic selectivity. Presence of the quaternary amine derivatizing tag on the aromatic ring of estrogens diminishes interaction of that portion of the molecule with the C18 stationary phase. Perhaps this increases interaction with asymmetric positions in the B, C, and D rings of estrogens with the column. It is interesting that derivatization did not change the elution order.
Figure 3.
(A) Extracted ion chromatograms of C1-NA-NHS derivatives of estrogens at 6.25 µg/mL, and (B) extracted ion chromatograms of dansyl-chloride derivatives of estrogens at same concentration by positive ESI-MS. The experimental conditions are described in the text. The peak numbers correspond to the structure numbers in Figure 1.
Comparing the reversed phase chromatographic separation of C1-NA-NHS derivatives of estrogens (Figure 3A) with dansyl derivatized estrogens (Figure 3B) under identical conditions, significant differences were observed. First, the C1-NA-estrogens eluted much faster than dansyl-estrogens. The total separation time was shortened from roughly 25 min to 7 min. This is because the dansyl moiety is more hydrophobic than the quaternary amine-containing C1-NA-NHS group [11,15]. Second, C1-NA-NHS derivatization improved peak shape and increased the separation efficiency. Only one pair of C1-NA-estrogens was not resolved (Figure 3A), while multiple dansyl-estrogens coeluted (Figure 3B). This is probably due to the hydrophobic dansyl moiety dominating the interaction of the derivatives with the C18 phase, masking the structural differences of the estrogens.
Differences between the ESI-MS detection sensitivity with C1-NA-estrogens and dansyl-estrogens were small (Figure 3 A and B) except for 2-hydroxyestrone (component 9). A unique characteristic of C1-NA-estrogen spectra was the presence of a single intense quaternary amine molecular ion with no fragmentation or formation of adduct ions (as will be seen in Figure 4B). Consequently, maximum intensity was gained in selected ion monitoring and quantification was enhanced. This will be advantageous when collision-induced dissociation of the parent ion is used to increase detection selectivity. The m/z values of the derivatives summarized in Table 2 were used in constructing extracted ion chromatograms and determining detection limits. Derivatization decreased the detection limit by 1–2 orders of magnitude compared with the published detection limits for underivatized estrogens using ESI-MS (e.g. 10 ng/mL versus 0.44 ng/mL for 17β-estradiol, and 5 ng/mL versus 0.36 ng/ml for estrone – Table 2).
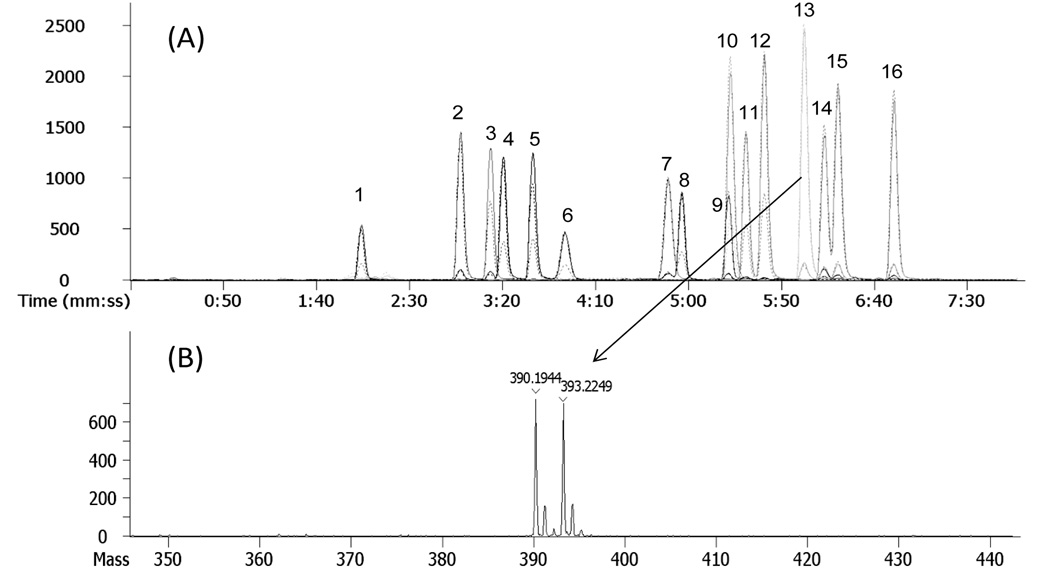
Figure 4.
(A) Extracted ion chromatograms of C1-NA-NHS (solid lines) and C1-NA-NHS-d3 (dashed lines) derivatives of estrogens and (B) the corresponding spectra of C1-NA-NHS and C1-NA-NHS-d3 of estrone. The LC-MS conditions were the same as those in Figure 3.
Table 2.
Linearity and dynamical range for comparative quantification and detection limit.
| m/z | Avg LOD [ng/ml, (pg/injection),n=5] |
||||
|---|---|---|---|---|---|
| d3 labeled | non- d3 labeled | ||||
| Peak number | Derivatized estrogens | Linearity * | |||
| 1 | 16-epiestriol | 411.2911 | 408.2675 | y = 0.9901x + 0.032; r2 = 0.9925 | 2.34 (11.71) |
| 2 | 16-keto-17β-estradiol | 409.2611 | 406.2359 | y = 0.9243x + 0.013; r2 = 0.9953 | 0.39 (1.99) |
| 3 | 16α-hydroxyestrone | 409.2714 | 406.2373 | y = 1.1043x + 0.010; r2 = 0.9989 | 0.62 (3.12) |
| 4 | estriol | 411.2944 | 408.2596 | y = 0.8445x + 0.052; r2 = 0.9969 | 0.69 (3.47) |
| 5 | 17-epiestriol | 411.2924 | 408.2642 | y = 0.8582x + 0.044; r2 = 0.9957 | 0.46 (2.34) |
| 6 | 4-hydroxyestradiol | 411.2786 | 408.255 | y = 0.8376x + 0.047; r2 = 0.9952 | 0.75 (3.75) |
| 7 | 4-hydroxyestrone | 409.2586 | 406.2433 | y = 0.8411x + 0.060; r2 = 0.9885 | 0.56 (2.84) |
| 8 | 2-hydroxyestradiol | 411.2882 | 408.258 | y = 1.1139x + 0.019; r2 = 0.9966 | 0.69 (3.47) |
| 9 | 2-hydroxyestrone | 409.2812 | 406.2363 | y = 0.8512x + 0.054; r2 = 0.9989 | 0.75 (3.75) |
| 10 | 17β-estradiol | 395.2286 | 392.1844 | y = 0.8421x + 0.016; r2 = 0.9988 | 0.44 (2.23) |
| 11 | 4-methoxyestradiol | 425.3775 | 422.3347 | y = 0.8921x + 0.033; r2 = 0.9987 | 0.48 (2.40) |
| 12 | 2-methoxyestradiol | 425.3782 | 422.3412 | y = 0.8656x + 0.041; r2 = 0.9988 | 0.37 (1.87) |
| 13 | estrone | 393.2098 | 390.1731 | y = 0.8797x + 0.039; r2 = 0.9928 | 0.36 (1.80) |
| 14 | 4-methoxyestrone | 423.3415 | 420.3153 | y = 0.8856x + 0.027; r2 = 0.9986 | 0.62 (3.12) |
| 15 | 2-methoxyestrone | 423.3394 | 420.3125 | y = 0.8934x + 0.021; r2 = 0.9980 | 0.46 (2.34) |
| 16 | 2-hydroxyestrone-3-methyl ether | 423.3481 | 420.315 | y = 0.9089x + 0.029; r2 = 0.9989 | 0.62 (3.12) |
x= Concentration ratio of non-d3 labeled to d3-labeled analytes; y= average extracted ion chromatographic peak intensity ratio of these pair of analytes. Linearity range for all is 1:1~1:40.
3.4. Comparative quantification through isotope labeling
Stable isotope coded synthetic standards are frequently used in quantitative analysis. Standards identical in structure to analytes of interest behave in analytical systems like the analytes themselves, except for the mass of the standards. When added in known concentrations to a sample, they serve as an internal standard that compensates for inter-analysis variations in sample manipulation. A limitation of this method is that in the analysis of large numbers of metabolites, isotopically coded standards must be synthesized for each molecule. It has been shown in proteomics that one way to deal with this problem is to label large number of peptides through derivatization of primary amines with stable isotope-coded labeling agents. Peptides in one sample are globally coded with a single isotope of the coding agent (light form) while those of a second sample are globally coded with a second isotope (heavy form) [29,40]. After the two samples are mixed, the relative amount of isotopically coded peptides can be determined in a single analysis [41]. A similar approach was applied to the analysis of estrogens in this work.
A deuterated version of C1-NA-NHS (C1-d3-NA-NHS) was synthesized and used for derivatization of a known amount of estrogen, resulting in a set of heavy-labeled estrogen internal standards. It is desirable for isotope-coded internal standards to coelute with the non-deuterated analyte derivatives to minimize quantification errors. A 1:1 mixture of C1-NA-estrogens and C1-d3-NA-estrogens was examined to determine the degree of analytical similarity. Extracted ion chromatograms of all the derivatives in the mixture and a representative MS spectrum for a single chromatographic peak are shown in Figure 4A and 4B, respectively. The two sets of peaks completely overlapped chromatographically. Doublet clusters of ions separated by 3 atomic mass units were observed in mass spectra of nearly the same peak intensity. When used in comparative quantification, the stable isotope-coded internal standard method was found to be linear over the concentration ratio ranging from 1:1 to 1:40 for all the estrogens (Table 2) by measuring concentration ratio points at 1:1, 1:5, 1:10, 1:15, 1:20, 1:20, 1:25, 1:30, 1:35, 1:40, 1:50, 1:100.
3.5. Analysis of breast cancer serum
Recovery was studied to examine the extraction efficiency of estrogens from blood samples. Aliquot of pooled human serum (Sigma) was extracted, derivatized and analyzed by procedures described in experimental section. No chromatographic peaks corresponding to ion masses of the 16 estrogen standards were found (using extracted ion chromatograms), indicating that concentrations of estrogens in the pooled serum are under detection limits and serum can be used as a blank for this study. To determine recovery, known amounts of estrogen standards were added to the serum, and the new sample carried through the extraction, drying, and C1-d3-NA-NHS (heavy) derivatization process. Another aliquot of the standards was directly derivatized with C1-NA-NHS reagent (light form) and added to “heavy” labeled serum. Recovery was determined by comparing the extracted chromatographic peak area of a deuterated (heavy) and non-deuterated (light) derivatives and showed that except for 16α-hydroxyestrone (42.0 ± 0.53% (mean ± SD, n=3)) and 17-epiestriol (73.0 ± 0.46% (mean ± SD, n=3)), extraction efficiency was > 95%. No substances in the human serum that interfere with the analysis were found. Understanding the lower recovery for 16α-hydroxyestrone and 17-epiestriol is not straightforward. Based on the current recovery study protocol, the extraction and drying steps, rather than derivatization reaction, affect the recovery measurement. The use of the same mixed standard solution for spiking serum after derivatization with C1-d3-NA-NHS, and for direct derivatization with C1-NA-NHS is a possible reason for the additional sample error. We speculate that these two estrogens are less stable or less extractable than the others.
This method was used to determine estrogen levels in serum from breast cancer patients, where estrogen concentrations are known to be increased. To analyze breast cancer serum, 2.0 mL of pooled cancer serum and 15 µL injection volume were used. Figure 5 shows the profile of estrogens in this sample. Except for 16-epiestriol, the other 15 estrogens in the list were identified. Concentrations of estrogens were estimated based on the comparative quantitation protocol. For most of estrogens, the concentrations were in the range of 80 - 530 pg/mL. An exception is estrone, having a concentration of 1.2 ng/mL, which is much higher than the others. This is reasonable, considering it is the major form of estrogen. Accurate quantification was not attempted because the analyte concentrations in this sample are just slightly over the detection limits.
Figure 5.
Extracted ion chromatograms of C1-NA-NHS derivatized (A) pooled healthy human serum and (B) pooled breast cancer serum. The LC-MS conditions were the same as those in Figure 4 except the injection volume was 15 µL.
4. Conclusions
A highly sensitive LC-MS analysis of estrogens at the level needed in life science research highlights a need for chemical derivatization. A novel derivatization reagent introduced in this work, C1-NA-NHS, and the derivatization method benefit LC-ESI-MS analysis of estrogens in several ways: (1) increase detection sensitivity in the positive ion mode of ESI-MS by one to two orders of magnitude, comparing to underivatized estrogens; (2) reduce hydrophobicity, retention time, and band spreading in reverse phase chromatography, and hence provide a higher analysis speed and higher peak resolution than existing methods; (3) make comparative quantitation and recovery studies more efficient, by allowing multiple sample (or standard) analysis to be performed in a single process by combining use of isotopic versions of C1-d3-NA-NHS. While the application of this method to breast cancer serum requires a relative large sample volume (2 ml serum), it could be expected that the combination of this derivatization with a triple quadrupole mass spectrometer will give a more effective approach for detecting cancer related estrogens.
Acknowledgements
The authors would like to thank Dr. Bruce R. Cooper for the discussion of the manuscript. This work was supported by grant number AG13319 from the National Institute of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beatson G. Lancet. 1896;11:104. [Google Scholar]
- 2.Clemons M, Goss P. New England Journal of Medicine. 2001;344:276. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 3.Yager JD, Davidson NE. New England Journal of Medicine. 2006;354:270. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 4.Di Leo A, Messa C, Cavallini A, Linsalata M. Current Drug Targets - Immune Endocrine & Metabolic Disorders. 2001;1:1. doi: 10.2174/1568008013341749. [DOI] [PubMed] [Google Scholar]
- 5.Lacey JV, Jr, Mink PJ, Lubin JH, Sherman ME, Troisi R, Hartge P, Schatzkin A, Schairer C. JAMA. 2002;288:334. doi: 10.1001/jama.288.3.334. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez A, Calle EE, Patel AV, Tatham LM, Jacobs EJ, Thun MJ. American Journal of Epidemiology. 2001;153:145. doi: 10.1093/aje/153.2.145. [DOI] [PubMed] [Google Scholar]
- 7.Giese RW. Journal of Chromatography. A. 2003;1000:401. doi: 10.1016/s0021-9673(03)00306-6. [DOI] [PubMed] [Google Scholar]
- 8.Chard T. Amsterdam: Elsevier Biomedical Press; 1982. [Google Scholar]
- 9.Anari MR, Bakhtiar R, Zhu B, Huskey S, Franklin RB, Evans DC. Analytical Chemistry. 2002;74:4136. doi: 10.1021/ac025712h. [DOI] [PubMed] [Google Scholar]
- 10.Fredline VF, Taylor PJ, Dodds HM, Johnson AG. Analytical Biochemistry. 1997;252:308. doi: 10.1006/abio.1997.2340. [DOI] [PubMed] [Google Scholar]
- 11.Tai SS, Welch MJ. Analytical Chemistry. 2005;77:6359. doi: 10.1021/ac050837i. [DOI] [PubMed] [Google Scholar]
- 12.Yamada H, Yoshizawa K, Hayase T. Journal of Chromatography B: Analytical Technologies in the Biomedical & Life Sciences. 2002;775:209. doi: 10.1016/s1570-0232(02)00292-1. [DOI] [PubMed] [Google Scholar]
- 13.Xu X, Keefer LK, Waterhouse DJ, Saavedra JE, Veenstra TD, Ziegler RG. Analytical Chemistry. 2004;76:5829. doi: 10.1021/ac049405i. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, Roman JM, Veenstra TD, Van Anda J, Ziegler RG, Issaq HJ. Analytical Chemistry. 2006;78:1553. doi: 10.1021/ac051425c. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Veenstra TD, Fox SD, Roman JM, Issaq HJ, Falk R, Saavedra JE, Keefer LK, Ziegler RG. Analytical Chemistry. 2005;77:6646. doi: 10.1021/ac050697c. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Ziegler RG, Waterhouse DJ, Saavedra JE, Keefer LK. Journal of Chromatography B: Analytical Technologies in the Biomedical & Life Sciences. 2002;780:315. doi: 10.1016/s1570-0232(02)00539-1. [DOI] [PubMed] [Google Scholar]
- 17.England BG, Parsons GH, Possley RM, McConnell DS, Midgley AR. Clinical Chemistry. 2002;48:1584. [PubMed] [Google Scholar]
- 18.Lee CS, Smith NM, Kahn SN. Journal of Reproductive Medicine. 1991;36:156. [PubMed] [Google Scholar]
- 19.Singh G, Gutierrez A, Xu K, Blair IA. Analytical Chemistry. 2000;72:3007. doi: 10.1021/ac000374a. [DOI] [PubMed] [Google Scholar]
- 20.Higashi T, Takayama N, Nishio T, Taniguchi E, Shimada K. Analytical & Bioanalytical Chemistry. 2006;386:658. doi: 10.1007/s00216-006-0371-z. [DOI] [PubMed] [Google Scholar]
- 21.Dehennin LA, Scholler R. Steroids. 1969;13:739. doi: 10.1016/0039-128x(69)90070-1. [DOI] [PubMed] [Google Scholar]
- 22.Pinnella KD, Cranmer BK, Tessari JD, Cosma GN, Veeramachaneni DN. Journal of Chromatography. B, Biomedical Sciences & Applications. 2001;758:145. doi: 10.1016/s0378-4347(01)00164-5. [DOI] [PubMed] [Google Scholar]
- 23.Xiao X, McCalley D. Rapid Communications in Mass Spectrometry. 2000;14:1991. doi: 10.1002/1097-0231(20001115)14:21<1991::AID-RCM125>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 24.Adlercreutz H, Gorbach SL, Goldin BR, Woods MN, Dwyer JT, Hamalainen E. Journal of the National Cancer Institute. 1994;86:1076. doi: 10.1093/jnci/86.14.1076. [DOI] [PubMed] [Google Scholar]
- 25.Adlercreutz H, Kiuru P, Rasku S, Wahala K, Fotsis T. Journal of Steroid Biochemistry & Molecular Biology. 2004;92:399. doi: 10.1016/j.jsbmb.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Castagnetta LA, Granata OM, Arcuri FP, Polito LM, Rosati F, Cartoni GP. Steroids. 1992;57:437. doi: 10.1016/0039-128x(92)90097-s. [DOI] [PubMed] [Google Scholar]
- 27.Fotsis T. Journal of Steroid Biochemistry. 1987;28:215. doi: 10.1016/0022-4731(87)90380-3. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Duncan AM, Merz-Demlow BE, Phipps WR, Kurzer MS. Journal of Clinical Endocrinology & Metabolism. 1999;84:3914. doi: 10.1210/jcem.84.11.6134. [DOI] [PubMed] [Google Scholar]
- 29.Yang WC, Mirzaei H, Liu XP, Regnier FE. Analytical Chemistry. 2006;78:4702. doi: 10.1021/ac0600510. [DOI] [PubMed] [Google Scholar]
- 30.Tian Q, Failla ML, Bohn T, Schwartz SJ. Rapid Communications in Mass Spectrometry. 2006;20:3056. doi: 10.1002/rcm.2700. [DOI] [PubMed] [Google Scholar]
- 31.Valentin-Blasini L, Blount BC, Rogers HS, Needham LL. Journal of Exposure Analysis and Environmental Epidemiology. 2000;10:799. doi: 10.1038/sj.jea.7500122. [DOI] [PubMed] [Google Scholar]
- 32.Diaz-Cruz MS, Lopez de Alda MJ, Lopez R, Barcelo D. Journal of Mass Spectrometry. 2003;38:917. doi: 10.1002/jms.529. [DOI] [PubMed] [Google Scholar]
- 33.Kebarle P. Journal of Mass Spectrometry. 2000;35:804. doi: 10.1002/1096-9888(200007)35:7<804::AID-JMS22>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 34.Ren DY, Julka S, Inerowicz HD, Regnier FE. Analytical Chemistry. 2004;76:4522. doi: 10.1021/ac0354645. [DOI] [PubMed] [Google Scholar]
- 35.Stewart NA, Pham VT, Choma CT, Kaplan H. Rapid Communications in Mass Spectrometry. 2002;16:1448. doi: 10.1002/rcm.726. [DOI] [PubMed] [Google Scholar]
- 36.Quirke JME, Adams CL, Vanberkel GJ. Analytical Chemistry. 1994;66:1302. [Google Scholar]
- 37.Quirke JME, Van Berkel GJ. Journal of Mass Spectrometry. 2001;36:1294. doi: 10.1002/jms.233. [DOI] [PubMed] [Google Scholar]
- 38.Broberg S, Broberg A, Duus JO. Rapid Communications in Mass Spectrometry. 2000;14:1801. doi: 10.1002/1097-0231(20001015)14:19<1801::AID-RCM96>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 39.Hsu J, Chang SJ, Franz AH. Journal of the American Society for Mass Spectrometry. 2006;17:194. doi: 10.1016/j.jasms.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Yang WC, Adamec J, Regnier FE. Analytical Chemistry. 2007;79:5150. doi: 10.1021/ac070311t. [DOI] [PubMed] [Google Scholar]
- 41.Ji J, Chakraborty A, Geng M, Zhang X, Amini A, Bina M, Regnier F. Journal of Chromatography. B, Biomedical Sciences & Applications. 2000;745:197. doi: 10.1016/s0378-4347(00)00192-4. [DOI] [PubMed] [Google Scholar]