Abstract
MicroRNAs (miRNAs) are regulatory molecules that negatively control gene expression by binding to complementary sequences on target mRNAs. The most thoroughly characterized miRNAs, lin-4 and let-7, direct cell fate determination during the larval transitions in C. elegans and act as key regulators of temporal gene expression. lin-4 and let-7 are founding members of two distinct families of miRNA genes sharing strong sequence homology primarily in the 5′ end of the mature miRNAs. In this report, we characterize the temporal and spatial expression patterns of lin-4 and let-7 family members using Northern blot analysis and mir∷gfp fusion studies. Our results show that lin-4 and let-7 homologues possess distinct temporal and spatial expression patterns during nematode development and that known heterochronic genes regulate their expression. We find that certain lin-4 and let-7 family members display overlapping expression patterns in the hypodermis and the reproductive system, suggesting that combinations of miRNAs from across families may control common developmental events.
Keywords: microRNAs, C. elegans, development, lin-4, let-7, expression analysis
Introduction
MicroRNAs (miRNAs), small noncoding RNAs of ∼22 nucleotides, belong to a novel class of gene regulatory molecules found in plants and animals that negatively control gene expression by binding to complementary sequences on target messenger RNAs (mRNAs) (Bartel, 2004). Hundreds of miRNAs have recently been identified in worm, fly, and mammalian genomes through cloning and bioinformatic approaches (Ambros and Horvitz, 1984; Pasquinelli et al., 2000; Reinhart et al., 2000; Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001; Lagos-Quintana et al., 2002; Lee et al., 2002; Aravin et al., 2003; Brennecke et al., 2003; Johnston and Hobert, 2003; Lagos-Quintana et al., 2003; Lai et al., 2003; Lim et al., 2003a; Lim et al., 2003b; Sempere et al., 2003; Xu et al., 2003; Chang et al., 2004). Only a small subset of miRNAs identified have been characterized in animals and these control important developmental events involved in cellular differentiation, proliferation, apoptosis, and fat metabolism (Ambros and Horvitz, 1984; Reinhart et al., 2000; Brennecke et al., 2003; Johnston and Hobert, 2003; Xu et al., 2003; Chang et al., 2004; Chen et al., 2004; Johnson et al., 2005). The remaining uncharacterized miRNAs may also act as gene regulators that direct important biological processes during development and in the adult.
The first miRNAs to be discovered, lin-4 and let-7, were identified through genetic analysis to act as developmental switches that control the timing of cell fate determination during the larval transitions in C. elegans (Chalfie et al., 1981; Ambros and Horvitz, 1984; Reinhart et al., 2000). lin-4 and let-7 loss-of-function mutations result in reiterations of the first larval stage (L1) and the fourth larval stage (L4) fates respectively, and these defects lead to disruptions in cell cycle exit and terminal differentiation. These early studies revealed that the lin-4 and let-7 miRNAs were key regulators in an emerging heterochronic pathway, which governs how tissues and organs are specified at the correct time and synchronized during development (Slack and Ruvkun, 1997; Banerjee and Slack, 2002). lin-4 RNA accumulates during the late L1 stage and is responsible for the L1/L2 transition in nematodes by binding to imperfect complementary sites within the 3′UTRs of its gene targets, lin-14 and lin-28 (repressors of post-L1 fates), and down-regulating their expression at the level of translation (Lee et al., 1993; Wightman et al., 1993; Moss et al., 1997; Olsen and Ambros, 1999; Ambros, 2000). Likewise, let-7 RNA accumulates during the L4 stage and is responsible for the L4/adult transition by inhibiting the expression of its target genes, lin-41 and hbl-1 (Reinhart et al., 2000; Slack et al., 2000; Pasquinelli and Ruvkun, 2002; Abrahante et al., 2003; Lin et al., 2003). The lin-4 and let-7 miRNAs are not exclusively used by nematodes to control developmental timing but are rather found to be evolutionarily conserved and temporally expressed in higher animals, implying a more universal role for these miRNAs during development (Feinbaum and Ambros, 1999; Pasquinelli et al., 2000; Reinhart et al., 2000; Lagos-Quintana et al., 2002; Pasquinelli and Ruvkun, 2002; Lim et al., 2003b; Sempere et al., 2003).
lin-4 and let-7 are members of two distinct miRNA families in C. elegans that include lin-4 and mir-237 in the lin-4 family, and let-7, mir-84, mir-241, and mir-48 in the let-7 family based on sequence homology shared primarily at the 5′ end of the mature miRNAs (Lim et al., 2003b). In addition to lin-4 and let-7, only one other miRNA from this group, mir-84, has been characterized. Recent work from our laboratory has shown that mir-84, the closest let-7 homologue in nematodes, directs proper hypodermal seam cell and vulva morphogenesis, in part through the negative regulation of the let-60/RAS gene in these tissues (Grosshans et al., 2005; Johnson et al., 2005). Members of the lin-4 and let-7 families are found to be temporally regulated during nematode development (Lau et al., 2001; Lim et al., 2003b), however, little is known regarding how the temporal and spatial expression patterns of the lin-4 and let-7 family members overlap or if these genes direct similar biological functions as the known heterochronic miRNAs, lin-4 and let-7.
In this report, we have begun to characterize the expression patterns of the miRNAs belonging to the lin-4 and let-7 families. We show that miRNAs, which are closely related on a sequence level, are not initially expressed at identical stages during C. elegans development based on Northern blot analysis, indicating that miRNA homologues may be functionally distinct. In support of this hypothesis, we observe unique temporal and tissue expression patterns in nematodes among the lin-4 and let-7 family members when miRNA promoter regulatory sequences are fused to the green fluorescence protein (gfp) reporter gene (mir∷gfp). We note that certain members across the lin-4 and let-7 families are expressed similarly in the developing hypodermal seam cells, the gonad, and the vulva, suggesting that unrelated miRNAs may control common developmental processes in these tissues.
Results and Discussion
The lin-4 and let-7 families are differentially expressed during nematode development
miRNAs identified within the C. elegans genome form distinct families based on sequence homology primarily at the 5′ portion of the mature miRNAs (Figs. 1A & 1C; Lim et al., 2003b). In order to explore the possibility that miRNAs within families are regulated in a similar manner, we compared the temporal expression pattern of the lin-4 and let-7 family members using developmental Northern blots. Our results demonstrate that despite the sequence homology shared among family members, their expression patterns were temporally dynamic and differed from one another (Figs. 1 & S1). For example, the lin-4 miRNA is highly expressed during the late L1/eL2 transition through to adulthood with peak expression noted in mid-L3 (Feinbaum and Ambros, 1999; Figs. 1B & S1A). In contrast, we find by Northern blot that the lin-4 homologue, miR-237, was detected at low levels during L2, was upregulated at early L3 and peaked during the L4 stage (Figs. 1B & S1A). We also note that our Northern blot expression data for the lin-4 homologue, miR-237, and the let-7 family members, miR-241, miR-48, and miR-84, differed from those reported by Lim et al., 2003, most likely due to the higher temporal resolution of our Northern blot data and a difference in the sensitivity of our probes. For instance, this previous study reported that the let-7 family members, miR-84, miR-48, and miR-241, were expressed in a similar manner as the let-7 RNA, which was first detected during the L3 stage (Reinhart et al., 2000; Lim et al., 2003b). However, our studies reveal that miR-241 and miR-48 both appeared one stage earlier than let-7 at eL2 with robust expression by L3. miR-84 appeared at low levels in the early L1 stage, was upregulated to an intermediate level in mid-L2 and had a high level of expression in early L4 (Figs. 1D & S1B). It is interesting to note that all let-7 family members showed maximal expression during the L4 stage. In addition, because the miRNA genes, mir-241 and mir-48, are located within 1.7 kb of one another on chromosome V in the C. elegans genome and reside in the same orientation (Fig. S5C), it is assumed that mir-241 and mir-48 are co-transcribed and present within a single pri-miRNA transcript, which is later processed to form two independent 70 nucleotide pre-miRNA precursors. In that case, mir-241 and mir-48 could share common regulatory elements to direct their temporal and spatial expression, analogous to regulatory elements located in the introns of a variety of genes. In support of this hypothesis, we show in our Northern blot analysis that these genes are expressed similarly throughout development (Figs. 1D & S1B). Based on the unique expression patterns of closely related members belonging to the lin-4 and the let-7 families, we predict that these genes may direct distinct biological processes during development.
Figure 1. Temporal expression of the lin-4 and let-7 miRNA families throughout C. elegans development.

A. Sequence alignment of the lin-4 family designated in the 5′ to 3′ orientation. B. Northern blot analysis of lin-4 and miR-237 shows distinct expression patterns at early stages of nematode development. U6 RNA is shown as a loading control. C. Sequence alignment of the let-7 family. mir-84 shares the closest sequence homology with let-7 compared to the other family members. D. Northern blot analysis of let-7, miR-84, miR-48 and miR-241 reveals distinct temporal expression patterns for these closely related miRNAs. U6 is shown as a loading control. Abbreviations: eL1, early larval stage 1; mL1, mid-L1; eL2, early L2; mL2, mid-L2; eL3, early L3; mL3, mid-L3; eL4, early L4, mL4, mid-L4; eAd, early adult stage; mAd, mid-Ad; Ad/egg, late stage adults carrying eggs.
Regulation of the lin-4 and let-7 families by the heterochronic genes, lin-4 and daf-12
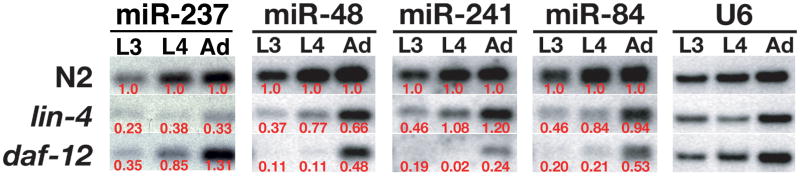
We tested whether miRNAs related to lin-4 and let-7 are regulated by the heterochronic genes during development. Our previous work showed that let-7 miRNA expression is modulated by heterochronic genes that are genetically upstream of let-7 in the heterochronic pathway, i.e. lin-4, lin-14, lin-28 and daf-12 (Johnson et al., 2003). In the present study, we similarly analyzed developmental Northern blots using RNA taken from animals with a loss-of-function mutation in the lin-4 miRNA (lin-4 (e912)), and a ligand-binding domain mutation in the daf-12 nuclear hormone receptor (daf-12 (rh61)) and found that both of these genes were essential for the proper regulation of all the let-7 family members, as well as the regulation of the lin-4 homologue, miR-237 (Figs. 2 & S2). The lin-4 and daf-12 genes are essential components of the heterochronic pathway and are normally required in nematodes to direct the L1/L2 transition (Chalfie et al., 1981) and the L2/L3 transition (Antebi et al., 1998; Grosshans et al., 2005) respectively. As previously reported for let-7 (Johnson et al., 2003), this work shows that mutations in daf-12 also resulted in a decrease of miR-241, miR-48, and miR-84 RNA levels in the late larval and adult stages. However, while the loss of the lin-4 miRNA caused a severe decrease in let-7 RNA (Johnson et al., 2003), the loss of lin-4 resulted in a much less pronounced effect on miR-241, miR-48 and miR-84 in the larval and adult stages. Our results also show that mir-237 is regulated by its earlier expressed homologue, lin-4, and implies that these genes do not function redundantly. We further note that daf-12 is also required for proper expression of mir-237 specifically at the L3 stage, the time when miR-237 levels are upregulated during development in wild type animals (Figs. 1B & S1A). Taken together, these results reveal that the expression of the lin-4 and let-7 family members are regulated by known heterochronic genes and thus may indicate a role for these novel miRNAs in controlling developmental timing.
Figure 2. Regulation of mir-237 and the let-7 family members by the heterochronic genes, lin-4 and daf-12.
Northern blot analysis of miR-237, miR-84, miR-48, and miR-241 expression during the third larval (L3) stage 3, the forth larval (L4) stage, and in the adult (Ad) with total RNA taken from wild-type worms (N2), lin-4 (e912) loss-of-function mutants, and daf-12 (rh61) ligand binding domain mutants. All the miRNAs analyzed showed some degree of regulation by both heterochronic genes. U6 is shown as a loading control. The numbers shown in red are the normalized ratios of miRNA expression in heterochronic mutants vs. wild-type N2 animals.
Using mir∷gfp fusions to analyze temporal and spatial expression patterns of the lin-4 and let-7 families
In order to determine whether the lin-4 and let-7 family members exhibit distinct temporal and spatial expression patterns during nematode development, we examined the expression of these miRNAs in vivo by fusing mir promoter regulatory sequences to the gfp reporter gene followed by the heterologous 3′UTR of the unc-54 gene (mir∷gfp). (Promoter regions chosen for the lin-4 and let-7 families are graphically depicted in Figs. S3, S4, & S5.) In general, we defined the promoter region of a miRNA as follows: (1) If a rescuing construct was known, we used the minimal DNA sequences in the rescuing construct residing upstream of the mature miRNA sequence, e.g. lin-4 and let-7 (Lee et al., 1993; Reinhart et al., 2000). (2) We used approximately 2.0 kb of genomic DNA residing upstream of the mature miRNA sequence or most genomic sequence up to the next gene, whichever was less, e.g. mir-84 (Johnson et al., 2005), or (3) We chose upstream sequences that possessed regions of conservation between C. elegans and C. briggsae. Our previous work has shown that similar mir∷gfp constructs made for let-7 in C. elegans, which relied solely on the let-7 promoter for regulation, were sufficient to drive temporal expression and suggested that miRNA regulation is controlled at the level of transcription and not by RNA processing and/or miRNA stability (Johnson et al., 2003). This technology has also been used to successfully detect the expression of certain C. elegans neural-specific miRNAs (Johnston and Hobert, 2003; Chang et al., 2004). Furthermore, we found that injection of the empty vector, pPD95.75, which contains only the gfp gene and the unc-54 3′UTR, did not result in background GFP expression (data not shown). Taken together, the mir∷gfp constructs made for the novel lin-4 and let-7 families likely indicate at what time and in which areas these miRNAs are normally expressed. Using this approach, our expression analysis showed that the lin-4 and let-7 families exhibited interesting overlapping expression patterns in the developing hypodermal seam cells, the gonad and the vulva.
lin-4 and let-7 family expression in the hypodermal seam cells during development
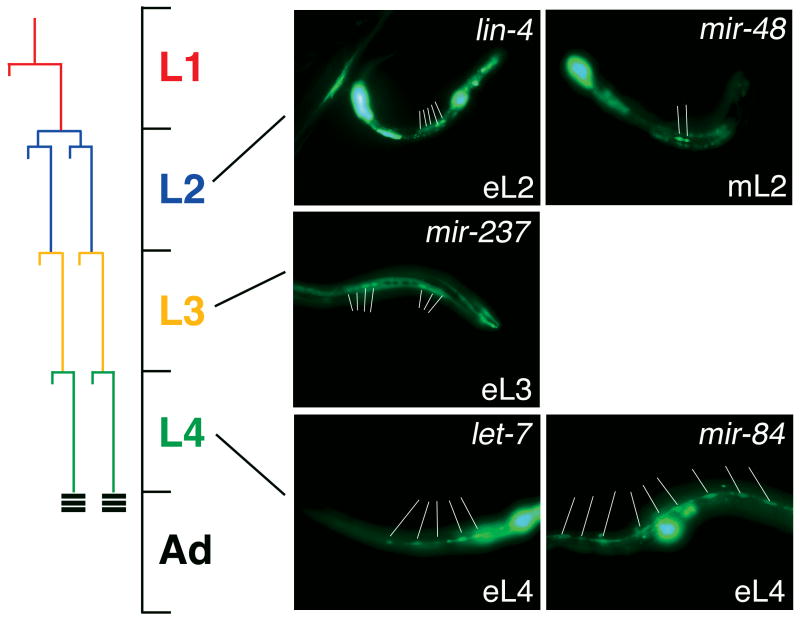
Our mir∷gfp expression studies revealed that the lin-4 and let-7 family members were differentially expressed in the hypodermal seam cells during nematode development. Seam cells, lateral hypodermal cells responsible for secreting the worm cuticle, undergo a characteristic pattern of cell divisions at each larval molt and terminally differentiate by the adult stage (Fig. 3; Rougvie, 2001). The heterochronic genes, including lin-4 and let-7, control the timing of seam cell development and mutations in these genes result in either precocious or retarded seam cell terminal differentiation (Rougvie, 2001). In the present study, we found that lin-4 was temporally expressed in the hypodermal seam cells at the early L2 stage and expression persisted in adulthood (Figs. 3 & S6). These temporal expression patterns closely mirrored those observed for lin-4 by Northern blot analysis (Figs. 1B & S1A; Feinbaum and Ambros, 1999). Furthermore, the spatial patterning of lin-4 in the hypodermal seam cells correlated with the requirement of this gene at the L1/L2 transition to direct normal seam cell development (Ambros and Horvitz, 1984; Feinbaum and Ambros, 1999). However, the lin-4 homologue, mir-237, was not expressed in the seam cells until eL3, which mirrored the up-regulation of this gene at this stage by Northern blot (Figs. 1B & S1A) and implicates a distinct role for mir-237 during seam cell development. We previously reported that the 1.8 kb let-7 promoter directed temporal gfp expression in the seam cells of transgenic animals at the early L4 stage and in the adult (Figs. 3 & S6; Johnson et al., 2003), consistent with the detection of let-7 RNA by Northern blot analysis (Figs. 1D & S1B) and the requirement for let-7 during later stages of seam cell development (Reinhart et al., 2000). We also found that mir-84 temporal expression was also observed in the hypodermal seam cells at the early L4 stage, similar to let-7 seam cell expression (Figs. 3 & S6; Johnson et al., 2005), the time when miR-84 RNA expression begins to peak as shown in our Northern studies (Figs. 1D & S1B). Thus, we find that mir-84 and let-7 display partially overlapping expression patterns and suggest a common role for let-7 and mir-84 in seam cell development. However, animals carrying the mir-48∷gfp construct showed temporal expression in the hypodermal seam cells two stages earlier than its let-7 homologues at the L2 stage (Figs. 3 &S6) correlating with the onset of miR-48 RNA detection by Northern blot analysis (Figs. 1D & S1B). These findings support the notion that closely related miRNA family members may be functionally distinct in select tissues. Our results also suggest that miRNAs may be required at each stage of larval development to control the timing of seam cell division and terminal differentiation.
Figure 3. Temporal expression of lin-4 and let-7 family members in the hypodermal seam cells during nematode development.
Transgenic animals carrying mir∷gfp fusion constructs for lin-4, mir-237, let-7, mir-84, and mir-48 were analyzed throughout C. elegans development for seam cell expression. The larval stages shown in each panel denotes the earliest stage at which seam cell expression was first detected for the specified mir∷gfp construct. White lines indicate positive expressing seam cells. The schematic diagram to the left depicts the characteristic division pattern of the hypodermal seam cells during larval development, in which after each seam cell division, one daughter cell fuses with the hypodermis (hyp7) and the other daughter cell divides again, terminally differentiating at the adult stage (indicated by the horizontal black bars) (Rougvie, 2001).
miRNA expression in the reproductive system
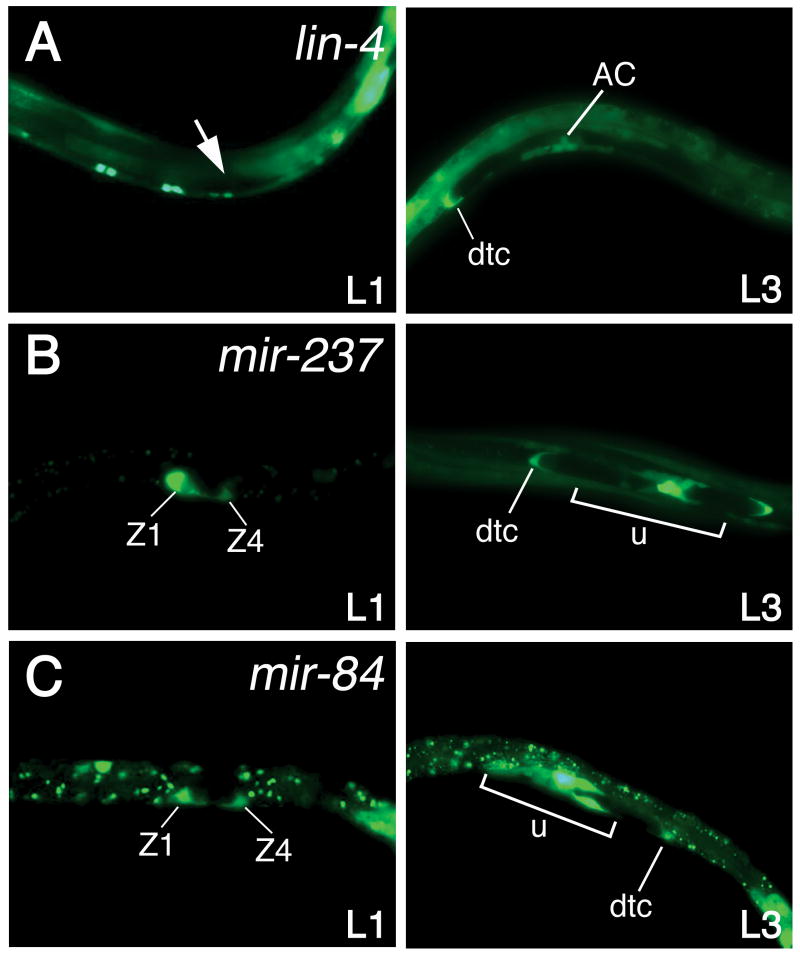
The lin-4 and let-7 miRNA family members also exhibited unique temporal and spatial expression patterns in the reproductive system using mir∷gfp fusion techniques. Our studies revealed that highly related miRNAs were not expressed identically in the gonad during nematode development. For example, lin-4 expression was first observed within the developing gonad at the beginning of L3, specifically in the distal tip cells and the anchor cell of the somatic gonad (Figs. 4A & S7A) and expression in the distal tip cells continued in the adult (data not shown). However, the lin-4 homologue, mir-237, was detected two stages earlier at L1 in the somatic gonad progenitor cells, Z1 and Z4 (Figs. 4B & S7B). This result differs from our Northern blot data, which showed that miR-237 RNA was first detected at L2 (Figs. 1B & S1A). We believe that the mir-237∷gfp expression at the L1 stage is accurate and since mir-237 is only expressed in the Z1 and Z4 cells at this stage, miR-237 RNA is below the levels of detection by Northern blot analysis. Once the Z1 and Z4 somatic gonad derivatives expand at L2, miR-237 RNA can be seen by Northern blot. By the L3 stage, and persisting into the adult (data not shown), mir-237 was expressed in the developing gonad, including the anchor cell, a subset of unidentified uterine cells, and the distal tip cells (Figs. 4B & S7B). The up-regulation of mir-237 expression in animals carrying the mir-237∷gfp construct at the L3 stage in a variety of tissues, such as the gonad and the hypodermis, mirrors the increased miR-237 RNA levels observed at L3 by Northern blot analysis (Fig. 1B & S1A). Our studies reveal that mir-237 is expressed in a distinct temporal and spatial pattern compared to lin-4 and supports our hypothesis that although family members share strong sequence homology, they may direct unique developmental events.
Figure 4. Distinct temporal and spatial patterning of the lin-4 and let-7 family members in the gonad throughout nematode development.
Animals carrying mir∷gfp fusion constructs for lin-4 (A), the lin-4 homologue, mir-237 (B), and the let-7 homologue, mir-84 (C), were analyzed for expression in the developing gonad at the first larval (L1) stage and the third larval (L3) stage 3. Note that lin-4 was not detected in the gonad at the L1 stage (white arrow). Abbreviations: AC, anchor cell; dtc, distal tip cell; u, uterine cells including the anchor cell; Z1 and Z4, somatic gonad progenitor cells.
We have also shown that miRNAs across families display similar spatial expression patterns in the developing gonad, suggesting that unrelated miRNAs could direct common biological processes in this tissue. The lin-4 family member, mir-237, and the let-7 family member, mir-84, were both expressed at the L1 stage in the Z1 and Z4 cells of the gonad, and during the L3 stage in the distal tip cells and uterine cells of the somatic gonad (Fig. 4B, 4C, S7B & S7C). These results suggest that mir-237 and mir-84 may have overlapping functions or work in a similar pathway to control proper gonad formation at early larval stages. However, by the adult stage, mir-84 and mir-237 exhibited distinct expression patterns in the somatic gonad, and mir-84 expression was additionally observed in the sheath cells, the spermatheca, the uterine cells surrounding the eggs, as well as the distal tip cells, where-as mir-237 expression was only detected in the distal tip cells at this stage (data not shown), implying that these two genes may have distinct roles in the gonad in the adult. Strikingly, we found that the other let-7 family members were expressed quite differently to mir-84 in the gonad, and let-7 was observed in the anchor cell at L3 (Figs. 5 & S8) and in the distal tip cells at the adult stage (data not shown), and mir-48 was not detected in the gonad at any stage (data not shown). Again, these results imply that the let-7 homologues may not be functionally redundant.
Figure 5. Differential expression patterns of the lin-4 and let-7 family members in the developing vulva and anchor cell.
Animals carrying mir∷gfp fusion constructs for lin-4, the lin-4 homologue, mir-237, let-7, and the let-7 family members, mir-84 and mir-48 were analyzed for expression in the vulval precursor cells (VPCs), P3.p-P8.p, and in the anchor cell of the somatic gonad (white arrows) at third larval (L3) stage. A schematic diagram of the spatial expression patterns for the lin-4 and let-7 family members during vulva development at the L3 stages is shown to the right. The morphogenic signal, LIN-3, secreted by the anchor cell (AC) induces the closest vulva precursor cell, P6.p, to adopt the 1° cell fate and the two adjacent VPCs, P5.p and P7.p, to adopt the 2° cell fates. 1° and 2° cells will later differentiate into the mature vulva (V). VPCs that are not induced by the anchor cell (P3.p, P4.p, and P8.p) will take on non-vulval 3° cell fates, fuse with the hyp 7 cell, and become part of the outlaying epidermis (E). Our mir∷gfp fusion studies for lin-4 (blue), let-7 (blue), mir-237 (green), mir-84 (green), and mir-48 (red) showed unique and partially overlapping expression patterns in both the anchor cell of the somatic gonad and the VPCs, indicating that these miRNAs may play an important role during cell fate specification and vulva formation. The (*) indicates that mir-84 shows dynamic expression in the VPCs and by the very late L3 stage, mir-84 expression was noted exclusively in the P5.p, P6.p, and P7.p cells (Johnson et al., 2005). Note that Pn.p cells depicted in panels for lin-4, let-7, and mir-48 have undergone one cell division.
Overlapping miRNA expression patterns in vulval cells
During our analysis of the lin-4 and let-7 miRNA families using mir promoter∷gfp fusion constructs, we were struck by the unique expression patterns these miRNAs exhibited in the vulval precursor cells (Figs. 4 & S8; Johnson et al., 2005). Our results suggest that a complex circuitry of miRNAs specifies vulval cell fates. It is well established that at the L3 stage, signaling from the anchor cell of the somatic gonad (via LIN-3) is responsible for the specification of six hypodermal precursor cells (P3.p-P8.p) to adopt either 1°, 2° or 3°cell fates. The 1° and 2° cells will later differentiate into the vulva, a specialized hypodermal structure that allows the passage of eggs from the gonad into the external environment. The 3° non-vulval cells will fuse with the multinucleated hypodermal cell, hyp7, and become part of the epidermis (Fig. 4; Sulston and Horvitz, 1977; Sulston and White, 1980; Kornfeld, 1997). Past work has revealed that the miRNAs, lin-4, let-7 and mir-84, are important for normal vulval morphogenesis (Chalfie et al., 1981; Ambros and Horvitz, 1984; Feinbaum and Ambros, 1999; Reinhart et al., 2000; Slack et al., 2000; Johnson et al., 2005). We observed partially overlapping expression patterns of the lin-4 and let-7 family members at L3 in the anchor cell and the vulval precursor cells (VPCs) (Figs. 4 & S8; Johnson et al., 2005). Intriguingly, both lin-4 and let-7 were weakly expressed in the anchor cell, which is required for proper vulval patterning, and in the P5.p, P6.p, and P7.p cells that will later differentiate into the mature vulva. Moreover, specific expression of mir-48 was observed in the P5.p and P7.p cells that will assume 2° vulval cell fates. In addition, the lin-4 family member, mir-237, and the let-7 family member, mir-84, were both expressed in the anchor cell and in the VPCs except P6.p at the mL3 stage. However, expression of mir-84 in the vulval precursor cells appeared dynamic in nature and was detected in the P5.p, P6.p, and P7.p descendants but down regulated in the P3.p, P4.p, and P8.p descendents by the end of the L3 stage (Johnson et al., 2005). It is interesting to note that expression of mir-237, mir-84, and mir-48 was absent in the P6.p cell during the mid- to late L3 stage, and expression of mir-237 and mir-84 but not mir-48 was present in the anchor cell. The partially overlapping expression patterns of the lin-4 and let-7 family members seen in the vulva precursor cells and the anchor cell may reveal a combinatorial regulating code, a “miRNA code”, reminiscent of the LIM homeobox code that specifies neuronal differentiation in the vertebral spinal cord (Tsuchida et al., 1994; Thor et al., 1999; Hobert, 2004). We propose that these miRNAs and their homologues act to fine-tune vulval patterning by controlling gene targets in the vulval precursor cells.
Conclusion
This report reveals that miRNAs belonging to the lin-4 and let-7 families are regulated in specific temporal and spatial patterns during C. elegans development. The dynamic expression of the lin-4 and let-7 family members, detected by Northern blot analysis and mir∷gfp technology, suggests that these miRNA homologues may not act redundantly but rather perform distinct biological functions. Moreover, our studies imply that miRNAs, which are unrelated and belong to different families, are similarly expressed and may work together to direct common developmental processes such as seam cell, gonad and vulva formation. We also present evidence that the lin-4 paralogue, mir-237, and the let-7 family members are regulated by a subset of heterochronic genes that control developmental timing in nematodes. Interestingly, we found that lin-4 is important for the proper expression of mir-237, further supporting the notion that these miRNAs possess distinct developmental functions. These studies are an important step in understanding which developmental processes miRNAs control and identifying candidate gene targets.
Our studies show that miRNA homologues exhibit distinct temporal and tissue specific expression patterns during development and that unrelated miRNAs belonging to different families exhibit overlapping expression patterns. If miRNAs are expressed with such varying spatial and temporal expression patterns, then even a small number of miRNAs could achieve a diverse level of regulation. We predict that combinations of miRNAs may be required to regulate a single gene target and initiate a given biological response, such as vulva formation (see above). Many examples exist in which the 3′ UTRs of bona fide lin-4 and let-7 targets contain multiple miRNA complimentary sites belonging to different families. For instance, the 3′ UTRs of lin-14 and lin-28, both known targets of lin-4, have conserved let-7 complementary sites that are thought to be functionally important. The converse is true with the 3′ UTRs of lin-41 and hbl-1, targets of let-7, which also contain lin-4 complementary elements. Moreover, additional members of the lin-4 and let-7 families may bind to the lin-4 and let-7 complementary sites in these 3′ UTRs, or possibly complementary sites for unrelated miRNAs may also be present. We propose that distinct family members may bind with different affinities to the same or multiple complementary sites in a given 3′ UTR, which could achieve varying degrees of translational regulation in the tissues where the miRNAs are expressed. Both lin-4 and let-7 are evolutionarily conserved in mammals, with humans possessing three lin-4 homologues, mir-125a, mir-125b1, and mir-125b2, and over ten let-7 homologues (Lagos-Quintana et al., 2002; Lim et al., 2003b). Due to the apparent complex regulation observed for the lin-4 and let-7 family members in C. elegans, there is a potential for an even higher level of miRNA-mediated gene regulation in mammals. Taken together, our results suggest an intricate system of negative regulation directed by miRNAs to specify the proper patterning, differentiation, and morphogenesis of a variety of structures such as the seam cells, gonad and vulva in the nematode. The next challenge will be to determine exactly which genes the lin-4 and let-7 miRNA families are controlling during these developmental processes.
Experimental Procedures
Northern blot analysis
Approximately 20.0 μg of total RNA was obtained from N2 wild type worms, lin-4 loss-of-function mutants (lin-4(e912)), and daf-12 ligand binding domain mutants (daf-12(rh61)) animals for Northern blot analysis using methods described previously by Reinhart et al., 2000. Probes used to detect RNA levels of lin-4 (5′-TCACACTTGAGGTCTCAGGGA-3′), mir-237 (5′-AAGCTGTTCGAGAATTCTCAGGGA-3′), mir-84 (5′-TACAATATTACATACTACCTCA-3′), mir-241 (5′-TCATTTCTCGCACCTACCTCA-3′), and mir-48 (5′- TCGCATCTACTGAGCCTACCTCA-3′) were made using the StarFire Oligonucleotide Labeling System (IDT). Probes p249N (5′-AACTATACAACCTACTACCTCACCGGATCC-3′) and pU6 (5′-GCAGGGGCCATGCTAATCTTCTCTGTATTG-3′), used to detect let-7 and U6 RNAs respectively (Reinhart et al., 2000), were 5′-end labeled with γ-32P ATP using the KinaseMax Kit (Ambion). Northern blots analyzing miRNA expression compared lin-4 and daf-12 mutant samples to wild type (N2) samples at the identical developmental stage in order to derive the relative intensity of the labeled probe bound to the miRNA band. All Northern blots probed with a given miRNA were subsequently stripped and reprobed with U6 to normalize lanes for loading.
Expression analysis in animals carrying mir∷gfp constructs
mir∷gfp constructs for the miRNA genes lin-4, mir-237, let-7, mir-84, mir-241, and mir-48 were constructed to include the miRNA promoter upstream of the gfp gene followed by the heterologous 3′ UTR from the unc-54 gene. Lin4GFPAS (lin-4∷gfp) was made by amplifying 507 bp of genomic sequence (base pairs −513 to −7) upstream of the mature lin-4 sequence from N2 genomic DNA and adding a SmaI site and an AgeI site to the 5′ and 3′ ends, respectively, using the polymerase chain reaction (PCR) with primers LIN4LB3 and LIN4LB1 (Table 1). This product was digested with SmaI and AgeI and then cloned into the pPD95.70 vector (gift from A. Fire), which was also digested with SmaI and AgeI. Digestion of pPD95.70 with SmaI and AgeI removed the nuclear localization signal (NLS) from this vector. PSJ840 (mir-84∷gfp) was made using a similar cloning strategy as described and consisted of 2.2 kb of genomic sequence (base pairs -2201 to −9) upstream of the mir-84 mature sequence and was amplified using the PCR primers MIR84UP and MIR84DN (Table 1). PSJ11 (let-7∷gfp) was made by amplifying 1.8 kb of genomic sequence (base pairs -1762 to −1) upstream of the mature let-7 sequence as previously described (Johnson et al., 2003) using the PCR primers LET7SMJ2 and LET7SMJ3 (Table 1). MIR241-2kb (mir-241∷gfp) consisted of 2.0 kb of genomic sequence (-2036 to −1) upstream of the mature mir-241 sequence and was amplified from N2 genomic DNA using the PCR primers JRP7 and JRP2 (Table 1), which added a BamH1 site to the 5′ and 3′ fragment ends. The resulting PCR product was then digested with BamH1 and cloned into the pPD95.70 vector. A similar cloning strategy as described for mir-241∷gfp was used to create MIR48-1KB (mir-48∷gfp), which consisted of 1.1 kb of genomic sequence (-1147 to −1) upstream of the mature mir-48 sequence amplified using the PCR primers JRP6 and JRP3 (Table 1). Animals carrying the mir-241∷gfp showed non-temporal expression patterns, which varied between lines and were not further characterized in this study. Due to the close proximity of mir-241 and mir-48 in the genome, mir-241 and mir-48 may share regulatory elements located between these two genes in order to direct their proper expression. Our mir-241∷gfp construct would have lacked these shared regulatory sequences. The mir-237∷gfp construct was generated using the PCR-Fusion-Based Protocol as previously described (Hobert, 2002), which consisted of 1.7 kb of genomic sequence (base pairs -1749 to −1) upstream of the mature mir-237 sequence. In short, a PCR fragment of 1.9 kb consisting of the gfp gene and the 3′ UTR from the unc-54 gene was amplified from the pPD95.75 vector (gift from A. Fire) using the PCR primers GFPAK9 (5′-AGCTTGCATGCCTGCAGGTCGACT-3′) and GFP2C (5′-GGAAACAGTTATGTTTGGTATATTGGG-3′). In parallel, a PCR fragment of 1.7 kb was amplified using the PCR primers MIR237AK5 and MIR237AK6 (Table 1) from N2 genomic DNA (base pairs −1749 to −1) that consisted of the mir-237 promoter region. The 3′ PCR primer (MIR237AK6) for the mir-237 promoter contained a 24-nucleotide overhang for the gfp gene (underlined in Table 1). In the second PCR reaction, equal amounts of the above PCR products were added for primer extension using the PCR primers MIR237AK5 and GFP2C, resulting in a 3.6 kb mir237∷gfp fusion construct. Recent annotation for the C. elegans genome in WormBase revealed two tRNA genes approximately 1kb upstream of the mature mir-237 sequence that were included in the original mir-237 promoter construct (Fig. S4B). Since the tRNA genes reside in the opposite orientation to the mir-237 gene, their presence is not believed to drive GFP expression.
Table 1. Oligonucleotide sequences of PCR primers used for the mir∷gfp constructs.
| PCR primer | Name | Sequence (5′ - 3′) |
|---|---|---|
| lin-4 forward | LIN4LB3 | CAACAACCCGGGGTCGACGAGACGCCGAGTCTCCC |
| lin-4 reverse | LIN4LB1 | CAACAAACCGGTAGGCCGGAAGCATAAACTCATAAACC |
| mir-237 forward | MIR237AK5 | CTGAATCGACTTCTCTAGGAATCC |
| mir-237 reverse + gfp overhang | MIR237AK6 | AGTCGACCTGCAGGCATGCAAGCTCCACGCAATGTAGAAGTTTTGAAC |
| let-7 forward | LET7SMJ2 | GGTACCCTCCCTCTTTTAAGCCTG |
| let-7 reverse | LET7SMJ3 | CAACAAACCGGTCCGGATCCACAGTGTAGACCGTCC |
| mir-84 forward | MIR84UP | CAACAACCCGGGGTGCAACGAGCTCTGGAGCATAAG |
| mir-84 reverse | MIR84DN | CAACAAACCGGTAGGCAGACGTATGATGAATAGTAG |
| mir-241 forward | JRP7 | CGGGATCCCGGTGTCGTCGTGTTCTAAATGTTCC |
| mir-241 reverse | JRP2 | CGGGATCCACTTTGACACCCCCGCGGTTTG |
| mir-48 forward | JRP6 | CGGGATCCCGCCATATTTTCGATAGCACAGGAAGG |
| mir-48 reverse | JRP3 | CGGGATCCAGTTCCCGGGAGTTTCAATTGG |
All experimental constructs were sequenced and injected at 50 ng/μl together with the co-transformation marker, pRP4 [rol-6(su1006)] (100 ng/μl) into the gonads of early Adult (eAd) stage N2 worms and transgenic lines were obtained. At least three independent lines for each mir∷gfp were examined at every stage of nematode development (L1-Adult) and the temporal and spatial expression patterns were compared.
Visualization of Worms
Worms were mounted on agarose pads for viewing as described (Schnabel, 1999). Worms were viewed on an Axioplan2 imaging microscope (Zeiss) using either Normarski optics and Kohler illumination or florescent imaging for GFP. All pictures were taken with an AxioCam using AxioVision version 2.0.5 (Zeiss).
Supplementary Material
Quantification of the expression profiles shown in Figures 1B & 1D for the lin-4 (A) and the let-7 (B) families respectively throughout development normalized to the U6 loading control. Note that all members of the let-7 family show peak expression levels at the mid-L4 stage of larval development.
Quantification of normalized RNA expression of lin-4 and let-7 family members in heterochronic mutant animals shown in Figure 2.
miRNA promoter regions (red) were fused to the green fluorescent protein (gfp) reporter gene (green) followed by the unc-54 3′UTR (white). The size of the genomic sequence encoding the mir promoter region is shown below each construct.
The promoter sequences for lin-4 (A) and mir-237 (B) used for the mir∷gfp constructs are shown in the context of the C. elegans genome, in which the location of the miRNA, gene models, operons, and conservation between C. elegans and C. Briggsae are indicated. In section A, the entire lin-4 rescuing construct (693 bp) is depicted as the lin-4 gene (Lee et al., 1993). Note that the upstream sequences used for the mir-237∷gfp construct includes two tRNA genes, which are transcribed in the opposite orientation to mir-237 and are not believed to effect GFP expression (B). The genome map was kindly provided by the WormBase web site, http://www.wormbase.org, release WS144.
The promoter sequences for let-7 (A; Johnson et al., 2003), mir-84 (B; Johnson, et al., 2005), mir-241 and mir-48 (C) are shown in relation to their gene location. Note that mir-241 and mir-48 reside within 1.7 kb of one another and are predicted to be co-transcribed and therefore may share regulatory elements located between these genes in order to direct their proper expression. The genome map was kindly provided by the WormBase web site, http://www.wormbase.org, release WS144.
Identical to Figure 3 with the corresponding DIC images.
Identical to Figure 4 with the corresponding DIC images.
Identical to Figure 5 with the corresponding DIC images.
Acknowledgments
We thank Andrew Fire (Stanford University School of Medicine) for providing the pPD95.70 and pPD95.75 vectors used to make the mir∷gfp constructs. The daf-12 mutant (daf-12(rh61)) was kindly obtained from Adam Antebi (Max-Planck Institute for Molecular Genetics). We also thank Helge Grosshans, Katherine Carter, Diya Banerjee and Monica Vella for critical reading of the manuscript. This work was supported by NSF Grant IBN0344429 and NIH Grant 1RO1GM64701 (to F.J.S.) and A.E.-K. was supported by NIH/NSRA Grant F32GM071157.
Grant Information: NSF: IBN0344429 (to F.J.S.); NIH:1RO1GM64701 (to F.J.S.); NIH/NSRA: F32GM071157 (to A. E.-K.).
References
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ambros V. Control of developmental timing in Caenorhabditis elegans. Curr Opin Genet Dev. 2000;10:428–433. doi: 10.1016/s0959-437x(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Antebi A, Culotti JG, Hedgecock EM. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119–129. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Feinbaum R, Ambros V. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev Biol. 1999;210:87–95. doi: 10.1006/dbio.1999.9272. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Kornfeld K. Vulval development in Caenorhabditis elegans. Trends Genet. 1997;13:55–61. doi: 10.1016/s0168-9525(97)01005-6. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003a;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003b;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The Cold Shock Domain Protein LIN-28 Controls Developmental Timing in C. elegans and is Reglated by th lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 Regulatory RNA Controls Developmental timing in Caenorhabditis elegans by Blocking LIN-14 Protein Synthesis after the Initiation of Translation. Developmental Biology. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A, Reinhart B, Slack F, Maller B, Ruvkun G. Conservation across animal phylogeny of the sequence and temporal regulation of the 21 nucleotide C. elegans let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz R, Ruvkun G. The 21 nucleotide let-7 RNA regulates C. elegans developmental timing. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rougvie AE. Control of developmental timing in animals. Nat Rev Genet. 2001;2:690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- Schnabel R. Microscopy. In: Hope IA, editor. C elegans: A Practical Approach. New York: Oxford University Press; 1999. pp. 119–141. [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Slack F, Ruvkun G. Temporal pattern formation by heterochronic genes. Annu Rev Genet. 1997;31:611–634. doi: 10.1146/annurev.genet.31.1.611. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the lin-29 transcription factor. Molec Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, White JG. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev Biol. 1980;78:577–597. doi: 10.1016/0012-1606(80)90353-x. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin- 4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila MicroRNA Mir-14 Suppresses Cell Death and Is Required for Normal Fat Metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of the expression profiles shown in Figures 1B & 1D for the lin-4 (A) and the let-7 (B) families respectively throughout development normalized to the U6 loading control. Note that all members of the let-7 family show peak expression levels at the mid-L4 stage of larval development.
Quantification of normalized RNA expression of lin-4 and let-7 family members in heterochronic mutant animals shown in Figure 2.
miRNA promoter regions (red) were fused to the green fluorescent protein (gfp) reporter gene (green) followed by the unc-54 3′UTR (white). The size of the genomic sequence encoding the mir promoter region is shown below each construct.
The promoter sequences for lin-4 (A) and mir-237 (B) used for the mir∷gfp constructs are shown in the context of the C. elegans genome, in which the location of the miRNA, gene models, operons, and conservation between C. elegans and C. Briggsae are indicated. In section A, the entire lin-4 rescuing construct (693 bp) is depicted as the lin-4 gene (Lee et al., 1993). Note that the upstream sequences used for the mir-237∷gfp construct includes two tRNA genes, which are transcribed in the opposite orientation to mir-237 and are not believed to effect GFP expression (B). The genome map was kindly provided by the WormBase web site, http://www.wormbase.org, release WS144.
The promoter sequences for let-7 (A; Johnson et al., 2003), mir-84 (B; Johnson, et al., 2005), mir-241 and mir-48 (C) are shown in relation to their gene location. Note that mir-241 and mir-48 reside within 1.7 kb of one another and are predicted to be co-transcribed and therefore may share regulatory elements located between these genes in order to direct their proper expression. The genome map was kindly provided by the WormBase web site, http://www.wormbase.org, release WS144.
Identical to Figure 3 with the corresponding DIC images.
Identical to Figure 4 with the corresponding DIC images.
Identical to Figure 5 with the corresponding DIC images.