Abstract
A male infant was diagnosed prenatally with a partial ornithine transcarbamylase (OTC) gene deletion and managed from birth. However, he displayed neurological abnormalities and developed pleural effusions, ascites and anasarca not solely explained by OTC deficiency (OTCD). Further evaluation of the gene locus using exon-specific PCR and high density SNP array copy number analysis revealed a 3.9Mb deletion from Xp11.4 to Xp21.1 including five additional gene deletions, three causing the known genetic diseases: Retinitis pigmentosa (RP3), X-linked chronic granulomatous disease (CGD) and McLeod syndrome. The case illustrates (1) the complexities of managing a patient withneonatal onset OTCD, CGD, RP3 and McLeod syndrome, (2) the need for detailed evaluation in seemingly “isolated” gene deletions and (3) the clinical utility of high density copy number analysis for rapidly characterizing chromosomal lesions.
Keywords: ornithine transcarbamylase; OTC; Xp11.4 to Xp21.1 deletion; granulomatous disease, chronic; CGD; CGH; copy number analysis; retinitis pigmentosa; RP3; McLeod syndrome
Introduction
Ornithine transcarbamylase deficiency (OTCD; OMIM 311250), an X-linked disorder, is the most common inherited defect of ureagenesis, affecting 1:14,000 births [1]. Most mutations are ‘private’ single nucleotide substitutions or small insertions or deletions [2–4]. OTCD perturbs urea cycle function and results in variable degrees of hyperammonemia depending on the extent of enzyme deficiency. Complete OTCD in a male infant is usually lethal or results in severe brain damage in survivors due to delay in diagnosis and treatment. When an OTCD diagnosis is known prior to the birth of the child, prospective treatment has been successful [5]. Recent studies have suggested a role for liver transplantation in the management of severe OTCD [6–10] although long-term data are not yet available.
When engaging in the treatment of severe neonatal OTCD understanding the extent of the genomic lesion is critical, as demonstrated by the cases of two brothers with neonatal OTCD resulting from an Xp11.4-p21.1 contiguous gene deletion also including chronic granulomatous disease (CGD; OMIM 306400), Retinitis pigmentosa (RP3; OMIM 312610), and McLeod syndrome (OMIM 314850).
Methods
Case reports
Patient 1 was a term male born via normal vaginal delivery. At birth he demonstrated lethargy and a weak cry. Subsequently, he developed poor feeding, emesis, and progressive tachypnea and cyanosis. He was intubated. and shortly afterward went into coma. Blood ammonia was 1154 µmol/ml (normal 9–33 µmol/ml). Plasma amino acid and urine orotic acid analysis suggested OTCD. The patient died on the 5th day of life. Molecular analysis of the OTC gene revealed a partial gene deletion involving exons 1–8 (of 10 total). The mother was a vegetarian by choice and reported intolerance of dairy. She had a history of occasional headaches. She also reported frequent respiratory tract infections and an “eye problem” not treatable with corrective lenses. Similar symptoms were not present in any other family members. The mother deferred carrier status testing after the death of patient 1. Four years later she presented in the first weeks of pregnancy. Chorionic villus sampling (CVS) confirmed a male fetus, however, OTC gene evaluation was delayed by maternal cell contamination. Final prenatal diagnosis confirmed the presence of the OTC deletion in the fetus and the mother as a carrier. Her ammonia levels remained normal during pregnancy, but rose to 230 µmol/L postpartum.
Patient 2 was delivered at 38 weeks gestation and was noted to be hypotonic. Within 45 minutes of life, he was started on intravenous fluids (dextrose and intralipids) and ammonia scavengers (sodium phenylacetate, sodium benzoate and arginine hydrochloride). His ammonia was 125 µmol/L at 2 hours of life (normal <100). Plasma glutamine was 1087 µmol /L (normal 422–849) with normal ornithine and arginine levels; citrulline was <2.0 µmol /L (normal 0–35) and an initial urine orotic acid level was normal. Albumin was 2.1 g/dL (normal 2.6–3.6) at 2:35 hours of life. Within 12 hours of life, tremors and bilateral ankle cloni were noted. Ammonia levels were easily managed with only mild increases as the dietary protein intake reached 1.7 g/kg/d. Despite relatively well-controlled ammonia levels, the neurologic status was significant for agitation and minimal responsiveness. Several electroencephalograms and a brain MRI were normal. At three weeks of age, he developed pleural effusions and transudative abdominal ascites that progressed to frank anasarca, bowel edema, hepatomegaly and sloughing of skin. Ultrasound of the liver was normal. There was minimal improvement of the anasarca with diuretics and no evidence of unusual enteric or renal protein losses. Initially, the edema was thought to be due to hypoalbuminemia. However, increased protein intake and intravenous albumin normalized the serum albumin level but the anasarca only improved moderately and worsened again with subsequent infections. Postnatal analysis of the OTC gene confirmed the deletion of the first 8 of the 10 OTC exons. Given the presence of neurologic findings despite rather well-controlled ammonia levels and the persistent edema not easily explained by OTCD, we hypothesized that the deletion encompassed more than just OTC and initiated further analysis of the OTC locus in this patient.
Study subjects were enrolled in an IRB-approved protocol of written informed consent at the Children’s Hospital of Philadelphia.
Genomic Analysis
Whole genome SNP genotyping was performed using the Illumina (San Diego, CA) Infinium HumanHap550 Beadchip Array according to the manufacturer’s protocol. Targeted copy number analysis was performed for Xp by analyzing the B-allele frequency and log R ratio tracts of the accompanying Illumina BeadStudio software (ver. 2.3) as described in [11]. For the X chromosome in a male, deletions result in log R ratios below -1 and B allele frequencies < 0.5 for all SNP’s involved.
Amplification of exons of all genes residing in the interval was done by PCR. PCR primers were designed using ExonPrimer (http://ihg.gsf.de/ihg/ExonPrimer.html) and Primer3 [12] accessed via an interface with the UCSC Genome Browser [13] to amplify OTC exons 1, 9, and 10, RPGR exon 1, SYTL5 exon 16, CYBB exon 13, XK exon 2, PRRG1 exon 3, TMEM47 exon 2, and DMD exon 3. Primer sequences are available upon request. Amplifications were performed with Amplitaq Gold™ (Applied Biosystems, Branchburg, NJ) and products visualized via electrophoresis on a 2% agarose gel.
Results and Discussion
Molecular genetic results
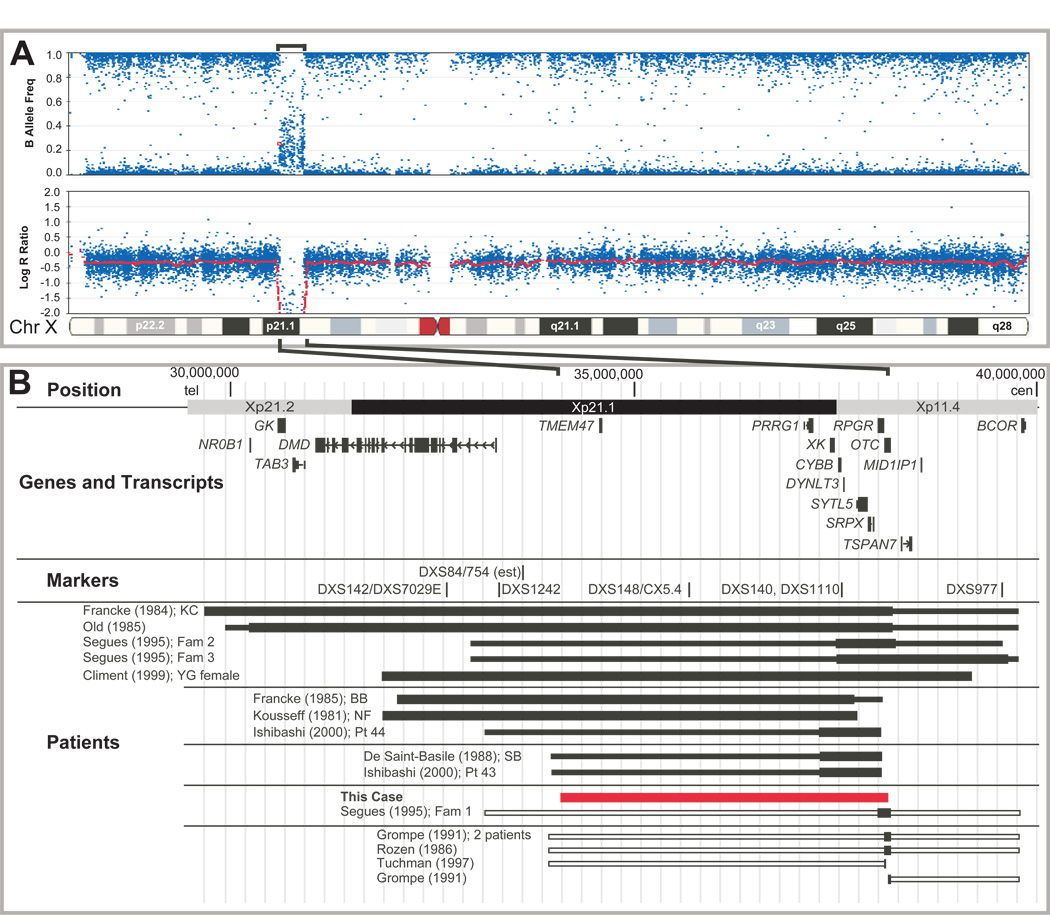
The Illumina Infinium HumanHap550 Beadchip array confirmed and further delineated the OTC/Xp deletion of Patient 2 (Figure 1A). SNPs were identified that defined the maximal and minimal size of the deletion (Supplementary Table 1). The telomeric breakpoint was 975kb centromeric of the Duchenne muscular dystrophy gene (DMD) and the centromeric breakpoint was within intron 8 of OTC (Figure 1) establishing the deletion to be between 3.925 and 3.940Mb in size, spanning from Xp11.4 to Xp21.1. PCR analysis of exons of seven genes between Xp11.4 and p21.1 (cen-OTC, RPGR, SYTL5, CYBB, PRRG1 and TMEM47, DMD-tel) confirmed that exons 9 and 10 of OTC and exon 3 of the DMD gene were present and that OTC exons 1–8, and exons of each of the five other known genes in the interval were absent (Supplementary Figure 1). The deletion thus included RPGR, the retinitis pigmentosa GTPase regulator, mutations of which cause X-linked RP3 with recurrent respiratory infections; SYTL5, synaptotagmin-like 5, which binds several Rab proteins and may serve as a Rab effector protein [14] involved in vesicular traffic; CYBB, cytochrome b-245, Beta polypeptide, mutations of which cause X-linked CGD, due to deficiency of NADPH oxidase resulting in inability to generate superoxide radicals for pathogen killing; XK, the Kell red cell antigen, defects of which cause McLeod syndrome with anemia, acanthocytosis, transfusion complications, late-onset neuromuscular symptoms and chorea; PRRG1, transmembrane gamma-carboxyglutamic acid protein 1, which is highly expressed in the spinal cord, and TMEM47, transmembrane protein 47, highly expressed in the brain (Figure 1 and Table 1A).
Figure 1. Comparison of Xp11.4-21.2 deletions.
A. B-Allele frequencies (top) and Log R ratios (bottom) of SNP copy number analysis for Patient 2. The deleted region is indicated by brackets above and expanded below to show the genomic region. B. Genomic position is indicated at the top with chromosome bands alternately shaded. Genes and selected transcripts are indicated. Markers from previous studies used to compare deletions are indicated. The extent of reported deletions are represented by solid bars to indicate deletions proven by cytogenetic or molecular analysis (wider portion) or by clinical features (narrow portion). Open bars represent additional regions that may be deleted but are undefined molecularly and the patients died too early to define clinically. Authors and specific patients are indicated to the left of each deletion. Patients are clustered as in Table 1B.
Table 1.
Summary of Xp11.4-p21.1 genes and deletions.
| A. Genes involved in patients Xp11.4-p21.1 deletion (this report) | ||
|---|---|---|
| Involved Gene | Functional Implications | |
| TMEM47 | Transmembrane protein 47 | Expressed in brain at high levels |
| PRRG1 | Transmembrane gamma-carboxyglutamic acid protein 1 | Expressed in the spinal cord |
| LANCL3 | LanC lantibiotic synthetase component C-like 3 | Unknown |
| *XK | Membrane transport protein XK | Kell blood group 'precursor substance' (Kx). Mutations associated with McLeod syndrome, characterized by neuromuscular and hematopoietic abnormalities |
| *CYBB | Cytochrome b-245, beta polypeptide | CYBB deficiency causes chronic granulomatous disease (CGD) with decreased phagocyte (NADPH oxidase-dependent) killing of intracellular bacteria |
| DYNLT3 | Dynein light chain Tctex-type 3 (T-complex-associated testis-expressed 1-like) | Unknown |
| SYTL5 | Synaptotagmin-like 5 | May play a role in vesicle trafficking[14–16]. Expressed in placenta and liver. |
| SRPX | Sushi-repeat-containing protein, X-linked | May be involved in phagocytosis |
| *RPGR | Retinitis pigmentosa GTPase regulator isoform C | Mutations cause ‘X-linked retinitis pigmentosa’ (XLRP) |
| *OTC | Ornithine carbamoyltransferase | Catalyzes formation of Citrulline from carbamyl phosphate and ornithine Deficiency causes urea cycle disorder with hyperammonemia |
| B. Summary of Xp11.4-p21.1 deletions. | ||||
|---|---|---|---|---|
| Category | Authors/Case | Involved Genes | Clinical Features | Comments |
| B1. Deletions approx 10Mb including more than DMD through OTC | ||||
| [17–19] (KC) | NR0B1 through OTC (complex rearrangement) | Female heterozygote | ||
| [20] | NR0B1through OTC | Male neonatal lethal with AHC, GKD and OTCD | ||
| [21] (Families 2 and 3) | DMD through OTC | Male neonatal lethal OTCD | ||
| [22] (YG) | DMD through OTC | Female with only OTCD symptoms | ||
| B2. Deletions >5Mb from DMD telomeric to OTC | ||||
| [18, 19, 23–25] (BB) | DMD through RPGR | Male with DMD, CGD, retinal pigment atrophy, anemia, acanthocytosis, mild MR. Also had intestinal pseudoobstruction, chronic abdominal distension and diarrhea. Mild MR. Died at 15y due to MVA. | Case facilitated identification of dystrophin [26] and X-linked CGD [27] | |
| [23, 28, 29] (NF) | DMD through CYBB | Male with CGD, McLeod, choroidoretinal atrophy | Used in identification of CYBB | |
| [30] (Patient 44) | ?DMD through RPGR | Male with McLeod, CGD, retitinis pigmentosa and muscle weakness | ||
| B3. Deletions <5Mb centromeric of DMD including OTC | ||||
| → | This report | TMEM47 through OTC | Severe neonatal OTCD, CGD during lifetime, died at 3 mo. | |
| [21]/ (Family 1) | RPGR (?SRPX) through OTC | Female with OTCD, died at 22 mo. | ||
| B4. Deletions <5Mb from centromeric of DMD to telomeric of OTC | ||||
| [31] (SB) | Likely TMEM47 through RPGR | CGD, McLeod and choroidoretinal atrophy | Case allowed identification of RPGR as causing RP3 and XK as causing McLeod [23, 32, 33] | |
| [30] (Patient 43) | XK through RPGR | Male with McLeod, CGD and retitinis pigmentosa | ||
| B5. Uncharacterized Deletions reported involving terminal exons of OTC | ||||
| [34] (2 patients) | OTC exons 1-10 | Severe neonatal OTCD | ||
| [3] (1 patient) | ||||
| [4] (1 patient) | ||||
| [35] (1 patient) | OTC ~ exons 1-10 | Severe neonatal OTCD | Excluded markers L1.28 and 754 | |
| [2] | OTC exons 1-3 | Severe neonatal OTCD | ||
| [4] | OTC exons 1-8 | Severe neonatal OTCD | ||
| [4] | OTC exons 1-9 | Severe neonatal OTCD | ||
| [34] | OTC exons 9-10 | Severe neonatal OTCD | ||
Genes with demonstrated clinical relevance to the management of patients in this family‥
Abbreviations: NRB01=DAX1, AHC=adrenal hypoplasia congenita, GKD=glycerol kinase deficiency, OTCD=ornithine transcarbamylase deficiency, CGD=chronic granulomatous disease, MR=mental retardation, MVA=motor vehicle accident
Implications of gene deletions for clinical management
OTCD might have been cured by liver transplant [6–9] but anasarca, respiratory compromise and fungal sepsis made the patient unsuitable for immediate consideration. Patient 2 developed Candida glabrata sepsis at 11 weeks of age that precipitated disseminated intravascular coagulation (DIC), hypotension, renal insufficiency and oliguria. Clinical testing demonstrated lack of an oxidative burst confirming chronic granulomatous disease. Infectious disease complications were assumed to be worsened by the patients CGD. Allogenic bone marrow transplant following myeloablative conditioning was considered, but deferred due to poor clinical status. Furthermore, he was felt to be at high risk of rejection due to the deletion of the XK gene, a red cell antigen, which also results in McLeod syndrome, . To treat his CGD acutely, granulocyte infusions were conducted while Amphotericin B was administered to treat the fungemia. The patient showed signs of a cytotoxic response to this regimen developing marked hyperammonemia despite IV ammonia scavengers and hemodialysis. Interferon gamma administration was also considered but not used due to concerns that it could worsen the cytotoxic response. He subsequently died at 12 weeks of age.
Comparison with other Xp11.4-Xp21.2 deletion cases
The contiguous gene deletion present in these two brothers manifested with a complex but definable clinical picture and their course poignantly illustrates the challenges in managing a neonate with a contiguous gene deletion involving Xp11.4-Xp21.1. These patients manifested OTCD, CGD , RP3, McLeod syndrome and had potentially disease causing deletions of at least 3 additional genes.
Several patients with deletions of Xp11.4-Xp21.2 have been reported. These cases are well-known and were instrumental in identifying the NR0B1 (DAX1), GK, DMD, CYBB, and RPGR genes (Figure 1, Table 1; and references therein). In addition, a number of cases resulted in neonatal lethal OTCD caused by deletions of OTC, but minimal further analysis was performed [2–4, 34, 35] . Upon review of the literature, the two cases reported here appear to be the only males reported with a deletion of Xp11.4-Xp21.2 centromeric of DMD and including OTC. Interestingly, deletions that arose telomeric to OTC through DMD resulted in only mild, if any, mental retardation (i.e. loss of CYBB, RP3, and XK, Table 1, Section B). Patient 2 showed neurological abnormalities from birth despite of only mildly increased ammonia levels. Although speculative, it is possible that the altered neurological status in our patients may have been due to the involvement of the TMEM47 or PRRG1 gene as they show high levels of expression in the brain and spinal cord, respectively or an early sign of McLeod syndrome.
With the improving ability to manage OTCD medically and with earlier liver transplantation, it will be important to characterize individuals with phenotypes caused by contiguous gene deletions that include OTC to accurately assess their risk regarding liver transplantation and to facilitate management. As outlined above, the combination of OTC, CGD and McLeod results in a very complicated clinical scenario, with limited therapeutic choices for effective management.
These cases underscore the necessity of detailed molecular analysis to clarify the extent of a genomic lesion for clinical management. The advent of widely available high-density copy number array analysis has immensely facilitated our ability to quickly delineate deletion breakpoints, an analysis previously very time consuming and only possible in research laboratories but not applicable in clinical care. As demonstrated in Patient 2, clinicians can now rapidly and easily determine whether disease-causing genes are deleted in a given patient and appropriately adjust management. Furthermore, it is likely that as this technology becomes engrained in clinical practice, we will learn that microdeletions of individual and contiguous genes are more common than previously appreciated.
Supplementary Material
PCR deletion analysis of OTC and neighboring genes. Photograph of agarose gel electrophoresis of PCR products. Each gene and/or exon is indicated at top of each lane. Control DNA demonstrates presence of each amplimer, while DNA from Patient 2 shows absence of amplimers. ‘No DNA’ indicates negative control containing the PCR cocktail mix but no genomic DNA template.
Acknowledgments
We thank the family who participated in this study. We thank Nancy Spinner, Brian Thiel and the staff of the Center for Applied Genomics for technical assistance. This work was supported by several National Institutes of Health awards: an NHLBI HL074731, a GCRC 2M01RR000240, an NIGMS T-32 Training Award, and an NIH Rare Diseases Clinical Research Center grant (5U54RR019453). Institutional funds were also provided by the Children’s Hospital of Philadelphia.
Additional Abbreviations
- BAC
bacterial artificial chromosome
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Batshaw ML, Lichter-Konecki U, Tuchman M. Inborn Errors of Urea Synthesis. In: Swaiman KF, Ashwal S, Ferriero DM, editors. Pediatric neurology: principles & practice. vol. 2. Philadelphia, PA: Mosby Elsevier; 2006. pp. 2407–2429. [Google Scholar]
- 2.Tuchman M, Morizono H, Rajagopal BS, Plante RJ, Allewell NM. Identification of 'private' mutations in patients with ornithine transcarbamylase deficiency. J Inherit Metab Dis. 1997;20:525–527. doi: 10.1023/a:1005301513465. [DOI] [PubMed] [Google Scholar]
- 3.Tuchman M, Jaleel N, Morizono H, Sheehy L, Lynch MG. Mutations and polymorphisms in the human ornithine transcarbamylase gene. Hum Mutat. 2002;19:93–107. doi: 10.1002/humu.10035. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi S, Brailey LL, Morizono H, Bale AE, Tuchman M. Mutations and polymorphisms in the human ornithine transcarbamylase (OTC) gene. Hum Mutat. 2006;27:626–632. doi: 10.1002/humu.20339. [DOI] [PubMed] [Google Scholar]
- 5.Maestri NE, Hauser ER, Bartholomew D, Brusilow SW. Prospective treatment of urea cycle disorders. J Pediatr. 1991;119:923–928. doi: 10.1016/s0022-3476(05)83044-6. [DOI] [PubMed] [Google Scholar]
- 6.Ensenauer R, Tuchman M, El-Youssef M, Kotagal S, Ishitani MB, Matern D, Babovic-Vuksanovic D. Management and outcome of neonatal-onset ornithine transcarbamylase deficiency following liver transplantation at 60 days of life. Mol Genet Metab. 2005;84:363–366. doi: 10.1016/j.ymgme.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Bachmann C. Long-term outcome of urea cycle disorders. Acta Gastroenterol Belg. 2005;68:466–468. [PubMed] [Google Scholar]
- 8.Saudubray JM, Touati G, Delonlay P, Jouvet P, Narcy C, Laurent J, Rabier D, Kamoun P, Jan D, Revillon Y. Liver transplantation in urea cycle disorders. Eur J Pediatr. 1999;158 Suppl 2:S55–S59. doi: 10.1007/pl00014323. [DOI] [PubMed] [Google Scholar]
- 9.Batshaw ML, Robinson MB, Ye X, Pabin C, Daikhin Y, Burton BK, Wilson JM, Yudkoff M. Correction of ureagenesis after gene transfer in an animal model and after liver transplantation in humans with ornithine transcarbamylase deficiency. Pediatr Res. 1999;46:588–593. doi: 10.1203/00006450-199911000-00016. [DOI] [PubMed] [Google Scholar]
- 10.Morioka D, Kasahara M, Takada Y, Shirouzu Y, Taira K, Sakamoto S, Uryuhara K, Egawa H, Shimada H, Tanaka K. Current role of liver transplantation for the treatment of urea cycle disorders: a review of the worldwide English literature and 13 cases at Kyoto University. Liver Transpl. 2005;11:1332–1342. doi: 10.1002/lt.20587. [DOI] [PubMed] [Google Scholar]
- 11.Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, Cheung SW, Shen RM, Barker DL, Gunderson KL. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 13.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroda TS, Fukuda M, Ariga H, Mikoshiba K. Synaptotagmin-like protein 5: a novel Rab27A effector with C-terminal tandem C2 domains. Biochem Biophys Res Commun. 2002;293:899–906. doi: 10.1016/S0006-291X(02)00320-0. [DOI] [PubMed] [Google Scholar]
- 15.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minshall RD, Malik AB. Transport across the endothelium: regulation of endothelial permeability. Handb Exp Pharmacol. 2006:107–144. doi: 10.1007/3-540-32967-6_4. [DOI] [PubMed] [Google Scholar]
- 17.Francke U. Random X inactivation resulting in mosaic nullisomy of region Xp21.1----p21.3 associated with heterozygosity for ornithine transcarbamylase deficiency and for chronic granulomatous disease. Cytogenet Cell Genet. 1984;38:298–307. doi: 10.1159/000132078. [DOI] [PubMed] [Google Scholar]
- 18.Francke U, Harper JF, Darras BT, Cowan JM, McCabe ER, Kohlschutter A, Seltzer WK, Saito F, Goto J, Harpey JP, et al. Congenital adrenal hypoplasia, myopathy, and glycerol kinase deficiency: molecular genetic evidence for deletions. Am J Hum Genet. 1987;40:212–227. [PMC free article] [PubMed] [Google Scholar]
- 19.McCabe ER, Towbin JA, van den Engh G, Trask BJ. Xp21 contiguous gene syndromes: deletion quantitation with bivariate flow karyotyping allows mapping of patient breakpoints. Am J Hum Genet. 1992;51:1277–1285. [PMC free article] [PubMed] [Google Scholar]
- 20.Old JM, Briand PL, Purvis-Smith S, Howard NJ, Wilcken B, Hammond J, Pearson P, Cathelineau L, Williamson R, Davies KE. Prenatal exclusion of ornithine transcarbamylase deficiency by direct gene analysis. Lancet. 1985;1:73–75. doi: 10.1016/s0140-6736(85)91966-x. [DOI] [PubMed] [Google Scholar]
- 21.Segues B, Rozet JM, Gilbert B, Saugier-Veber P, Rabier D, Saudubray JM, Carre M, Rouleau FP, Menget A, Bonardi JM, et al. Apparent segregation of null alleles ascribed to deletions of the ornithine transcarbamylase gene in congenital hyperammonaemia. Prenat Diagn. 1995;15:757–761. doi: 10.1002/pd.1970150812. [DOI] [PubMed] [Google Scholar]
- 22.Climent C, Garcia-Perez MA, Sanjurjo P, Ruiz-Sanz JI, Vilaseca MA, Pineda M, Campistol J, Rubio V. Identification of a cytogenetic deletion and of four novel mutations (Q69X, I172F, G188V, G197R) affecting the gene for ornithine transcarbamylase (OTC) in Spanish patients with OTC deficiency. Hum Mutat. 1999;14:352–353. doi: 10.1002/(SICI)1098-1004(199910)14:4<352::AID-HUMU15>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 23.Brown J, Dry KL, Edgar AJ, Pryde FE, Hardwick LJ, Aldred MA, Lester DH, Boyle S, Kaplan J, Dufier JL, Ho MF, Monaco AM, Musarella MA, Wright AF. Analysis of three deletion breakpoints in Xp21.1 and the further localization of RP3. Genomics. 1996;37:200–210. doi: 10.1006/geno.1996.0543. [DOI] [PubMed] [Google Scholar]
- 24.Worley KC, Towbin JA, Zhu XM, Barker DF, Ballabio A, Chamberlain J, Biesecker LG, Blethen SL, Brosnan P, Fox JE, et al. Identification of new markers in Xp21 between DXS28 (C7) and DMD. Genomics. 1992;13:957–961. doi: 10.1016/0888-7543(92)90007-f. [DOI] [PubMed] [Google Scholar]
- 25.Francke U, Ochs HD, de Martinville B, Giacalone J, Lindgren V, Disteche C, Pagon RA, Hofker MH, van Ommen GJ, Pearson PL, et al. Minor Xp21 chromosome deletion in a male associated with expression of Duchenne muscular dystrophy, chronic granulomatous disease, retinitis pigmentosa, and McLeod syndrome. Am J Hum Genet. 1985;37:250–267. [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel LM, Monaco AP, Middlesworth W, Ochs HD, Latt SA. Specific cloning of DNA fragments absent from the DNA of a male patient with an X chromosome deletion. Proc Natl Acad Sci U S A. 1985;82:4778–4782. doi: 10.1073/pnas.82.14.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Royer-Pokora B, Kunkel LM, Monaco AP, Goff SC, Newburger PE, Baehner RL, Cole FS, Curnutte JT, Orkin SH. Cloning the gene for an inherited human disorder--chronic granulomatous disease--on the basis of its chromosomal location. Nature. 1986;322:32–38. doi: 10.1038/322032a0. [DOI] [PubMed] [Google Scholar]
- 28.Baehner RL, Kunkel LM, Monaco AP, Haines JL, Conneally PM, Palmer C, Heerema N, Orkin SH. DNA linkage analysis of X chromosome-linked chronic granulomatous disease. Proc Natl Acad Sci U S A. 1986;83:3398–3401. doi: 10.1073/pnas.83.10.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kousseff B. Linkage between chronic granulomatous disease and Duchenne's muscular dystrophy? Am J Dis Child. 1981;135:1149. doi: 10.1001/archpedi.1981.02130360053025. [DOI] [PubMed] [Google Scholar]
- 30.Ishibashi F, Nunoi H, Endo F, Matsuda I, Kanegasaki S. Statistical and mutational analysis of chronic granulomatous disease in Japan with special reference to gp91-phox and p22-phox deficiency. Hum Genet. 2000;106:473–481. doi: 10.1007/s004390000288. [DOI] [PubMed] [Google Scholar]
- 31.de Saint-Basile G, Bohler MC, Fischer A, Cartron J, Dufier JL, Griscelli C, Orkin SH. Xp21 DNA microdeletion in a patient with chronic granulomatous disease, retinitis pigmentosa, and McLeod phenotype. Hum Genet. 1988;80:85–89. doi: 10.1007/BF00451463. [DOI] [PubMed] [Google Scholar]
- 32.Meindl A, Dry K, Herrmann K, Manson F, Ciccodicola A, Edgar A, Carvalho MR, Achatz H, Hellebrand H, Lennon A, Migliaccio C, Porter K, Zrenner E, Bird A, Jay M, Lorenz B, Wittwer B, D'Urso M, Meitinger T, Wright A. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3) Nat Genet. 1996;13:35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 33.El Nemer W, Colin Y, Collec E, Gane P, Cartron JP, Kim CL. Analysis of deletions in three McLeod patients: exclusion of the XS locus from the Xp21.1-Xp21.2 region. Eur J Immunogenet. 2000;27:29–33. doi: 10.1046/j.1365-2370.2000.00188.x. [DOI] [PubMed] [Google Scholar]
- 34.Grompe M, Caskey CT, Fenwick RG. Improved molecular diagnostics for ornithine transcarbamylase deficiency. Am J Hum Genet. 1991;48:212–222. [PMC free article] [PubMed] [Google Scholar]
- 35.Rozen R, Fox JE, Hack AM, Fenton WA, Horwich AL, Rosenberg LE. DNA analysis for ornithine transcarbamylase deficiency. J Inherit Metab Dis. 1986;9 Suppl 1:49–57. doi: 10.1007/BF01800858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR deletion analysis of OTC and neighboring genes. Photograph of agarose gel electrophoresis of PCR products. Each gene and/or exon is indicated at top of each lane. Control DNA demonstrates presence of each amplimer, while DNA from Patient 2 shows absence of amplimers. ‘No DNA’ indicates negative control containing the PCR cocktail mix but no genomic DNA template.