Abstract
Pyrrolysine, the 22nd genetically-encoded amino acid, is charged onto its specific tRNA by PylS, a pyrrolysyl-tRNA synthetase. While PylS is found as a single protein in certain archaeal methanogens, in the Gram-positive bacterium Desulfitobacterium hafniense, PylS is divided into two separate proteins, PylSn and PylSc, corresponding to the N-terminal and C-terminal domains of the single PylS protein found in methanogens. Previous crystallographic studies have provided the structure of a truncated C-terminal portion of the archaeal Methanosarcina mazei PylS associated with catalysis. Here we report the apo 2.1 Å resolution structure of the intact D. hafniense PylSc protein and compare it to structures of the C-terminal truncated PylS from methanogenic species. In PylSc, the hydrophobic pocket binding the ring of pyrrolysine is more constrained than in the archaeal enzyme; other structural differences are also apparent.
Keywords: Pyrrolysine, PylS, pyrrolysyl-tRNA synthetase
INTRODUCTION
Pyrrolysine is the 22nd amino acid found to have entered natural genetic codes [1–3]. To date, this residue has only been documented in methylamine methyltransferases initiating methane formation in Archaea such as Methanosarcina barkeri [2, 4, 5]. The genes encoding these methyltransferases contain an in-frame amber codon (UAG, typically a stop) that is translated [6–8]. Crystallography of the monomethylamine methyltransferase revealed that the UAG codon is decoded as a novel amino acid, pyrrolysine [2]. Mass spectrometry confirmed the presence of pyrrolysine in three nonhomologous methyltransferases, always corresponding to an amber codon in the encoding gene [5].
Organisms whose genomes possess genes for these methylamine methyltransferases additionally have five pyl genes necessary for the genetic encoding and biosynthesis of pyrrolysine. The pylT gene encodes tRNAPyl [3], while the adjacent pylS encodes a novel class II aminoacyl-tRNA synthetase [3]. PylS is a pyrrolysyl-tRNA synthetase which aminoacylates tRNAPyl with chemically synthesized pyrrolysine [4, 9, 10]. E. coli expressing pylS and pylT and given exogenous pyrrolysine translates UAG as pyrrolysine [9]. PylS and tRNAPyl have great potential as an orthologous pair in E. coli for the programmed incorporation of pyrrolysine or analogs into recombinant proteins with novel properties; as tRNAPyl is not aminoacylated by E. coli aminoacyl-tRNA synthetases [9].
The pylBCD genes adjacent to pylS and pylT were proposed as possible candidates for pyrrolysine biosynthetic genes [3, 11] Their role in biosynthesis has been demonstrated by transformation of E. coli with pylTSBCD, which led to UAG translation as endogenously synthesized pyrrolysine [12]. The results to date are thus consistent with the synthesis of pyrrolysine in a tRNA independent manner. PylS is the first synthetase found beyond those enzymes ligating the common twenty amino acids to their cognate tRNA species [13].
The pyl genes are found in a handful of sequenced genomes, including Methanosarcina and Methanococcoides species [3, 14]. Until recently, Gram-positive Desulfitobacterium hafniense was the only bacterial species known to possess close pyl gene homologs [3]. A recent metagenomic study of gutless marine worm symbiont community uncovered pyl genes in an uncultured δ-proteobacteria [15–17].
Strong distinctions can already be made between archaeal and bacterial pyrrolysyltRNA synthetases. For example, in D. hafniense, the archaeal pylS gene is split into the pylSc and pylSn genes flanking pylBCD [3, 11]. The pylSc gene product averages 60% similarity to the C-terminal catalytic core of archaeal PylS while the pylSn gene product averages 50% similarity to the N-terminal domain of archaeal PylS. In the gutless worm symbiont, the pylSc and pylSn genes are present in a single operon, with the pylBCD genes transcribed separately [15, 16]. The conservation of the N-terminal domain of PylS as a separate polypeptide in bacteria suggests functionality such as tRNA binding [11], and deletion of the 5’ end encoding the N-terminal domain of archaeal PylS renders it non-functional in E. coli [18]. In vitro PylSc ligates a pyrrolysine analog to tRNAPyl, but PylSc binds tRNAPyl with less affinity than intact archaeal PylS [18]. The dichotomy between the bacterial and archaeal domains is striking when considering the limited number of pyrrolysyl-tRNA synthetases known.
The archaeal PylS is relatively unstable; and the structure of an intact pyrrolysyl-tRNA synthetase has not been obtained. This problem was partially solved by crystallization of Methanosarcina mazei PylS (MmzPylS) lacking the N-terminal 185 residues which yielded the structure of the catalytic domain of archaeal PylS [19]. Recently we purified PylSn and PylSc from D. hafniense, and set the goal of forming a complete structure of a PylRS from the structures of the two bacterial proteins. These structures will further inform the evolutionary history underlying the acquisition of pyrrolysine by the genetic code, the understanding of the interaction of PylRS enzymes and tRNAPyl, and directed evolution experiments to exploit PylRS for incorporation of novel amino acids into recombinant proteins. Here, we report the structure of the first of the gene products encoding bacterial pyrrolysyl-tRNA synthetase, the intact PylSc protein from D. hafniense (DhaPylSc).
MATERIALS AND METHODS
Cloning, overexpression, and purification of DhaPylSc
The D. hafniense pylSc gene was amplified by polymerase chain reaction (PCR) from genomic DNA using Ex-taq (Takara Mirus Bio, Madison, WI) with primers PylScNdeI (CATATGTTTTTAACAAGGAGGGACCCACCCTTGAGC) and PylScNotI (GCGGCCGCATTGATATTTAAGCGTACTCCATCAAGATAGC). PCR-amplified pylSc gene was subsequently ligated into PCR2.1-Topo (Invitrogen, Carlsbad, CA), then inserted as a NdeI and NotI fragment into pET22b(+) (Novagen, Madison, Wisconsin) so as to produce sequence encoding PylSc with a C-terminal hexahistidine tag (PylSc-His6) under the control of an inducible T7 promoter. The plasmid, pPylScHis, was then transformed into E. coli BL21 (DE3) (Stratagene, La Jolla, California).
Cells were grown at 37°C in Miller’s LB Broth medium (10g/L tryptone, 5g/L yeast extract and 10g/L sodium chloride) containing 50 mg/L kanamycin. When the culture had 6 grown to OD600 of ~0.5, 80 µM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce pylSc. The cell culture was centrifuged 3.5 hours post-induction and cells lysed in 500 mM NaCl, 10 mM imidazole, and 20 mM sodium phosphate buffer, pH 7.4. The cell extract was applied to a 1-mL bed volume Ni-activated HisTrap HP column (GE Healthcare Bio-Sciences Corp., Piscataway, New Jersey). Bound protein was then eluted with a gradient of 10 – 500 mM imidazole in 500 mM NaCl and 20 mM sodium phosphate, pH 7.4. PylSc eluted at 250 mM imidazole and was 95% pure by SDS-PAGE and Coomassie staining.
Reductive alkylation of DhaPylSc
All methylation reaction steps to reduce DhaPylSc were carried out at 4°C. 520 µL of the native PylSc (5 mg/mL) in 50 mM Hepes pH 7.5, 500 mM NaCl and 10% glycerol was placed in a glass tube and incubated with 10.4 µL of freshly prepared 1 M dimethylamine-borane complex (ABC) and 20.8 µL 1 M formaldehyde for 2 hours. This was followed by a second addition of 10.4 µL 1 M ABC and 20.8 µL 1 M formaldehyde, and the reaction was incubated for a further 2 hours. A final addition, consisted of only 10.4 µL 1 M ABC was added to the reaction mixture, which was then left at 4°C overnight to ensure complete reduction of the protein. The reaction was quenched using 59.3 µL 2 M ammonium sulfate. The methylated protein was buffer-exchanged into 20 mM PBS pH 7.4, 300 mM imidazole, 500 mM NaCl, 10% glycerol and 1 mM pyrrolysine analog 2-amino-6-((R)-tetrahydrofuran-2-carboxamido)hexanoic acid (2THF-lys), followed by concentration using a BioMax centrifugal filter unit (Millipore). Methylation of the protein had no significant effect on the activity of protein (15 min−1) in mediating 32P-pyrophosphate: ATP exchange dependent upon the addition of amino acid, as assayed after the procedure described by Blight et al. [9].
Crystallization of DhaPylSc
Crystals of methylated DhaPylSc were grown at room temperature by mixing 2 µL of protein (7 mg/mL) in 20 mM PBS pH 7.4, 300 mM imidazole, 500 mM NaCl, 10% glycerol and 1 mM 2THF-lys with 1 µL of precipitant (1.2 M potassium sodium tartrate, 0.1 M Tris pH 8.5, 10% glycerol) using the hanging drop vapor diffusion method. The crystals were flash-cooled in liquid nitrogen for data collection.
Data collection of DhaPylSc
Diffraction data were collected at 100 K using the BLu-ICE interface [20] and a MarMosaic-325 CCD detector on beamline 9–2 at the Stanford Synchrotron Radiation Laboratory (SSRL). Data reduction and scaling were performed with the programs MOSFLM and SCALA, respectively [21, 22]. The crystallographic data collection and refinement statistics are shown in Supporting Table 1.
Structure determination and refinement of DhaPylSc
The 2.1 Å resolution structure of DhaPylSc was determined by molecular replacement (MR) using a single subunit of the MmzPylS complexed with Pyl-AMP and pyrophosphate (PDB: 2Q7H) as the search model. The initial MR solution was identified using the program Phaser [23], and was improved through iterative model building and refinement using Coot and CNS, respectively [24, 25]. During the refinement process, a flat bulk solvent correction was applied and 5% of the data was omitted for the Rfree calculation. Metal ions, ligands, and waters were added in the latter stages of the rebuilding process. The quality of the final model was verified using PROCHECK [26]. 99.2% of the residues were in either the most favored regions (92.3%) or the additional allowed regions (6.9%) of the Ramachandran plot while 0.8% of the residues were in the generously allowed regions. All of the stereochemical parameters were better than expected. The coordinates have been deposited into the Protein Data Bank (PDB) with the accession ID code: pending.
RESULTS
Crystal structure of DhaPylSc
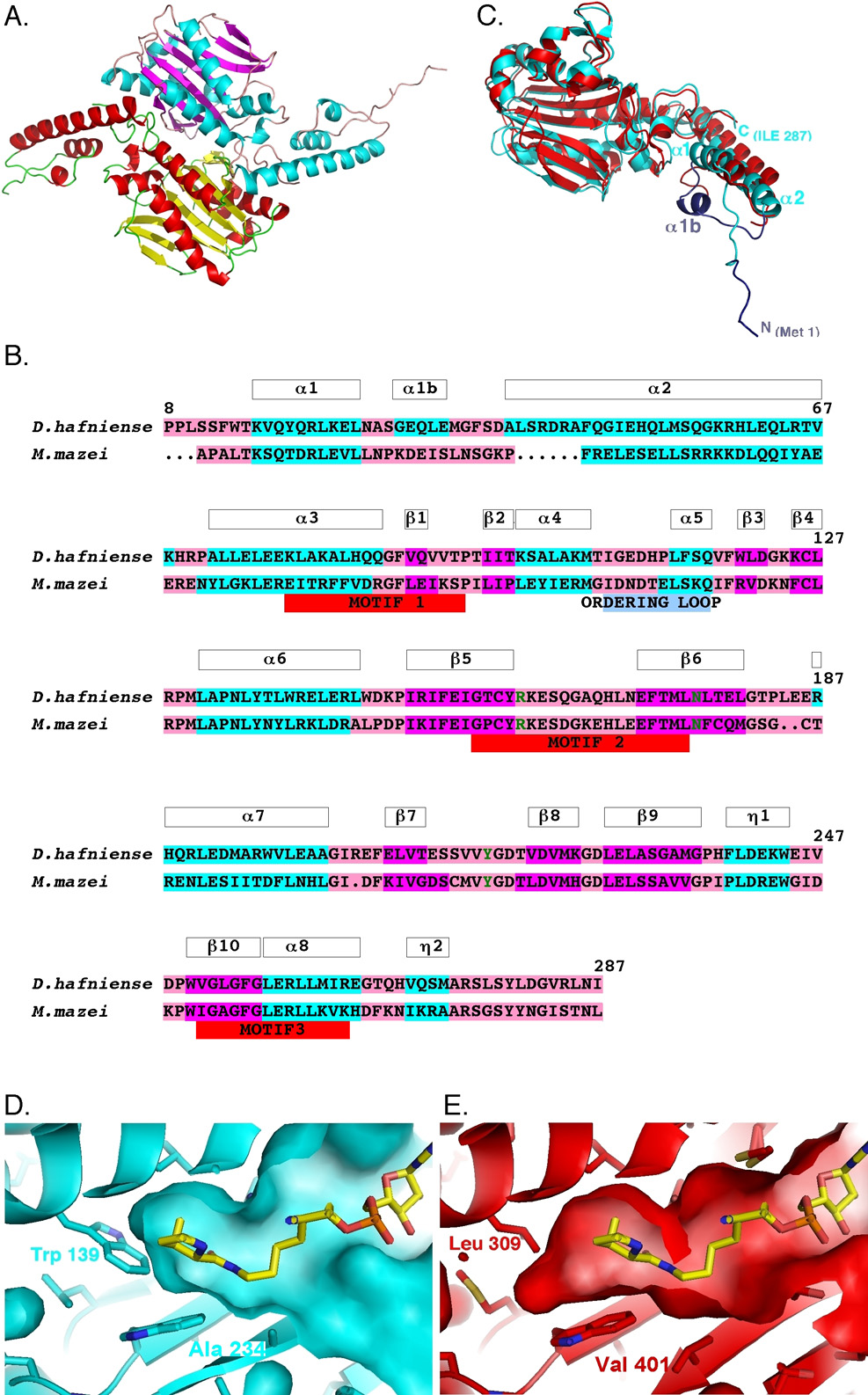
The crystal structure of the apo DhaPylSc (Fig. 1A) reveals a homodimer with an architecture that is typical of the catalytic domains of class II aminoacyl-tRNA synthetases (aaRSs) [27]. Each subunit is made up of nine α-helices surrounding a β-sheet core comprised of seven anti-parallel β-strands that contain specific motifs (motifs 2 and 3) which are involved in recognizing the nucleotide, and an eighth β-strand that lies off-side to the seven β-strands and which harbors another motif (motif 1) that participates in dimerization. In general, the three motifs are canonical, followed faithfully the signature sequence layouts as observed in other class II aaRSs [28]. Invariant residues are also highly conserved in each motif. However, there is a slight deviation from the general pattern for motif 3 (λXφGφGφERφφφφφ; where λ=small amino acids P,G,S,T; φ=hydrophobic residues; underlined Arg(R) is invariant) [28]. Instead of five consecutive hydrophobic residues following the invariant Arg residue that are the hallmark of motif 3, two charged residues replace two hydrophobic residues within this hydrophobic patch. Similar replacement was also observed in the structures of the truncated C-terminal domain of MmzPylS [19], although the substituting residues in this protein are different from those of DhaPylSc.
Fig. 1. Structure of the D. hafniense PylSc and its comparison to that of the M. mazei PylS C-terminal catalytic domain.
(A) Ribbons diagram of the DhaPylSc homodimer colored by subunit and secondary structure. Subunit A is colored with α-helix in red, β-sheet in yellow, and random coil in green. Subunit B is colored with α-helix in cyan, β-sheet in magenta, and random coil in pink. (B) Structure-based sequence alignment of DhaPylSc and MmzPylS C-terminal domain. The amino acids are highlighted by secondary structure (same color scheme as Subunit B in Fig. 1A) and critical residues associated with binding pyrrolysine are colored in green. (C) Structural overlap of the B-subunit of DhaPylSc colored in red and MmzPylS C-terminal domain colored in cyan. The novel regions in the DhaPylSc structure are colored in slate. (D) Pyrrolysine binding pocket in DhaPylSc. Adenylated pyrrolysine analog colored yellow is modeled into the binding pocket according to its structurally equivalent position in MmzPylS. Trp139 residue shown in stick leads to a much tighter pyrrolysine pocket. (E) Pyrrolysine binding pocket in MmzPylS. Leu309 in structurally equivalent position as Trp139 in DhaPylSc is shown as stick.
The two subunits of DhaPylSc are generally similar with the exception of the electron density for the N-terminal residues 1–7 (MFLTRRD), which is visible for the B-subunit, but absent in the A-subunit. The additional N-terminal 1–7 amino acids of the B-subunit form a random coil whose orientations appear to be stabilized by interactions with neighboring packing molecules. Given the disorder of the corresponding amino acids of the A-subunit, it appears likely that these residues are generally disordered in the holoenzyme. Whether these residues form a more ordered conformation in the presence of tRNA, as might be suggested by the high Arg content, or by interaction with the DhaPylSn, which it is linked to in the methanogenic PylS enzymes, will require further study. In either case, such an interaction would be unique as sequence alignments with the methanogenic PylS synthetases indicate that these residues are found only in DhaPylSc.
Comparison of apo DhaPylSc structure to the MmzPylS structures
Due to the highly limited distribution of PylS in nature to a relatively small number of archaeal and bacterial species, a comparison of their structural features is useful for understanding the diversity of the family as well as identifying common features relevant to its mechanism. Previous sequence alignments of DhaPylSc and MmzPylS (40% identity; 61% similarity in their overlapping regions) have been performed using standard sequence alignment algorithms [19, 29]. With the structure of the DhaPylSc in hand, we could now perform a more accurate structure-based sequence alignment of DhaPylSc with MmzPylS. Our alignment result (Fig. 1B) was most similar to that performed by Steitz and colleagues and verified the correctness of their residue-to-residue alignment for the MmzPylS's loop segment (residue 208–211 ISLN) connecting α-helix 1 and 2 to the DhaPylSc's short α-helical turn (residue 30–33 EQLE). In their MmzPylS structure, this loop segment was disordered and was not resolved as a result of high thermal factor [19].
Structural alignment of DhaPylSc and MmzPylS reveals specific differences in the structural features of the two proteins (Fig. 1C). In the MmzPylS structure [19], the loop between helices α1 and α2 (residues 202–214), or α1-α2 loop, is disordered between residues 208–211. Based on the structural alignment, the corresponding loop in the DhaPylSc (residues 26–38) is of similar length, but adopts a slightly different orientation relative to helices α1 and α2. The α1-α2 loop contains a short α-helical turn (residues 30–33) that may help to contribute to a fairly ordered region based on the electron density maps. In addition to these differences, the length of the α2-helix of DhaPylSc is longer than that of MmzPylS. This impacts the orientation of the N-terminal loop which is directed further away due to potential steric constraints.
A second loop belonging to the region of the ordering loop (residues 278–287 in MmzPylS and 108–117 in DhaPylS) also exhibits disorder in both DhaPylSc and MmzPylS structures [19, 29]. In DhaPylSc, residue 109–112 (GEDH) are disordered and omitted from the model, while in the MmzPylS structures, the disorder in the loop depends on the ligand state. In all three MmzPylS structures by Steitz and colleagues [19], residues 280–283 that correspond to the peptide NDTE could not be observed, resulting in breakage within the ordering loop. In the structures published by Yokoyama and colleagues [29], although this loop could be modeled, the aforementioned segments have high thermal factor. In particular, the ordering loop, from residue 280 to residue 287 in the apo MmzPylS structure (PDB: 2E3C), has thermal factor in excess of 100. This disorder, in turn, can lead to subtle displacements of the ordering loop. Thus this region appears to be fairly flexible in both proteins – though the significance of this feature is unclear.
DISCUSSION
Support for a conformational change of the β7-β8 loop in PylS enzymes
Previous crystallographic studies of apo and various liganded structures of the trunctated MmzPylS C-terminal domain have been determined [19, 29]. These structures, most notably those of apo and pyrrolysyl-adenylate bound MmzPylS, have led to the suggestion that although the overall main chain remains relatively unchanged, recognition of the bound pyrrolysine substrate leads to changes in the orientations of a conserved Asn and Tyr (Asn346 and Tyr384 in MmzPylS; Asn176 and Tyr217 in DhaPylSc) which form hydrogen bonding interactions with the bound ligand. For the Asn residue this change involves an alteration in a rotomer conformation, while for the conserved Tyr, the change is associated with a shift of the entire β7-β8 loop. Notably, in the apo MmzPylS C-terminal domain, the β7-β8 loop and the associated Tyr residue (Tyr384) are positioned away from the active site pocket, while in the pyrrolysyl-adenylate bound complex, Tyr384 is positioned near the active site in an orientation that allows the Tyr hydroxyl to form a hydrogen bond to the pyrroline imine nitrogen of pyrrolysine.
Analysis of the apo DhaPylSc structure reveals that as in the apo MmzPylS C-terminal domain structure [29], the β7-β8 loop and the associated Tyr residue (Tyr217) are located far away from the active site. These observations are suggestive of a general conformational change of β7-β8 loop for the entire family of enzymes. Elucidating whether this conformation change is utilized by PylS to promote catalysis, enhance the substrate specificity or both, will require further study.
Analysis of the hydrophobic binding pocket that binds the pyrroline ring
In addition to the recognition of pyrrolysine via hydrogen bonding to the conserved Asn and Tyr residues, a hydrophobic pocket that binds the pyrroline ring of pyrrolysine likely plays an important role in dictating the specificity of PylS for its targeted amino acid [19, 29]. Previous analysis of the structures of MmzPylS highlighted four residues in forming this hydrophobic binding pocket - Tyr306, Trp417, Cys348, and Val401 [19]. In DhaPylSc, these positions are filled by Tyr136, Trp250, Thr178 and Ala234, respectively. Thus only the former two residues are conserved. Perhaps the largest change in the pyrrolysine binding pocket is the substitution of a fifth relatively distant residue, Leu309, at the back of the pocket in MmzPylS for Trp139 in DhaPylSc. This much larger Trp139 residue now interacts directly with the pyrroline ring through van der Waals interactions. The presence of the larger Trp139 and Thr178 residues lead to a much tighter pyrrolysine pocket (Fig. 1D). While the substitution of the Val401 in MmzPylS by an Ala234 residue in DhaPylSc might initially be thought to relieve this compression, it in fact adds to this compression because the smaller side chain is compensated by the movement of the main chain to fill the otherwise open space. The net consequence of these changes suggests that the specificity of DhaPylSc pyrrolysine could be much higher than MmzPylSc, and the incorporation of substituted pyrrolysine analogs more difficult without substantial mutagenesis. Nevertheless, the application of DhaPylSc, for the incorporation of smaller analogs, such as acetylated lysine [30], could be worth further investigation.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grant GM 061796.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Atkins JF, Gesteland R. The 22nd amino acid. Science. 2002;296:1409–1411. doi: 10.1126/science.1073339. [DOI] [PubMed] [Google Scholar]
- 2.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 3.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 4.Hao B, Zhao G, Kang P, Soares J, Ferguson T, Gallucci J, Krzycki J, Chan MK. Reactivity and chemical synthesis of L-pyrrolysine— the 22nd genetically encoded amino acid. Chem. Biol. 2004;11:1317–1324. doi: 10.1016/j.chembiol.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Soares JA, Zhang L, Pitsch RL, Kleinholz NM, Jones RB, Wolff JJ, Amster J, Green-Church KB, Krzycki JA. The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases. J. Biol. Chem. 2005;280:36962–36969. doi: 10.1074/jbc.M506402200. [DOI] [PubMed] [Google Scholar]
- 6.Burke SA, Lo SL, Krzycki JA. Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine. J.Bacteriol. 1998;180:3432–3440. doi: 10.1128/jb.180.13.3432-3440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul L, Ferguson DJ, Krzycki JA. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 2000;182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James CM, Ferguson TK, Leykam JF, Krzycki JA. The amber codon in the gene encoding the monomethylamine methyltransferase isolated from Methanosarcina barkeri is translated as a sense codon. J. Biol. Chem. 2001;276:34252–34258. doi: 10.1074/jbc.M102929200. [DOI] [PubMed] [Google Scholar]
- 9.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 10.Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Soll D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. USA. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzycki JA. The direct genetic encoding of pyrrolysine. Curr. Opin. Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Longstaff DG, Larue RC, Faust JE, Mahapatra A, Zhang L, Green-Church KB, Krzycki JA. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc. Natl. Acad. Sci. USA. 2007;104:1021–1026. doi: 10.1073/pnas.0610294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schimmel P, Beebe K. Molecular biology: genetic code seizes pyrrolysine. Nature. 2004;431:257–258. doi: 10.1038/431257a. [DOI] [PubMed] [Google Scholar]
- 14.Goodchild A, Saunders NF, Ertan H, Raftery M, Guilhaus M, Curmi PMG, Cavicchioli R. A proteomic determination of cold adaptation in the Antarctic archaeon, Methanococcoides burtonii. Mol. Microbiol. 2004;53:309–321. doi: 10.1111/j.1365-2958.2004.04130.x. [DOI] [PubMed] [Google Scholar]
- 15.Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, Boffelli D, Anderson IJ, Barry KW, Shapiro HJ, Szeto E, Kyrpides NC, Mussmann M, Amann R, Bergin C, Ruehland C, Rubin EM, Dubilier N. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Gladyshev VN. High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis. Nucleic Acids Res. 2007;35:4952–4963. doi: 10.1093/nar/gkm514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atkins JF, Baranov PV. Translation: duality in the genetic code. Nature. 2007;448:1004–1005. doi: 10.1038/4481004a. [DOI] [PubMed] [Google Scholar]
- 18.Herring S, Ambrogelly A, Gundllapalli S, O'Donoghue P, Polycarpo CR, Soll D. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007;581:3197–3203. doi: 10.1016/j.febslet.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kavran JM, Gundllapalli S, O'Donoghue P, Englert M, Soll D, Steitz TA. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. USA. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Rad. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 21.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data, Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26. 1992. [Google Scholar]
- 22.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Cusack S, Berthet-Colominas C, Hartlein M, Nassar N, Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 28.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 29.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J. Mol. Biol. 2008;378:634–652. doi: 10.1016/j.jmb.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 30.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.