Abstract
Background
Mycoplasma pneumoniae has previously been characterized as a micro-organism that is genetically highly stable. In spite of this genetic stability, homologous DNA recombination has been hypothesized to lie at the basis of antigenic variation of the major surface protein, P1, of M. pneumoniae. In order to identify the proteins that may be involved in homologous DNA recombination in M. pneumoniae, we set out to characterize the MPN229 open reading frame (ORF), which bears sequence similarity to the gene encoding the single-stranded DNA-binding (SSB) protein of other micro-organisms.
Results
The MPN229 ORF has the capacity to encode a 166-amino acid protein with a calculated molecular mass of 18.4 kDa. The amino acid sequence of this protein (Mpn SSB) is most closely related to that of the protein predicted to be encoded by the MG091 gene from Mycoplasma genitalium (61% identity). The MPN229 ORF was cloned, and different versions of Mpn SSB were expressed in E. coli and purified to > 95% homogeneity. The purified protein was found to exist primarily as a homo-tetramer in solution, and to strongly and selectively bind single-stranded DNA (ssDNA) in a divalent cation- and DNA substrate sequence-independent manner. Mpn SSB was found to bind with a higher affinity to ssDNA substrates larger than 20 nucleotides than to smaller substrates. In addition, the protein strongly stimulated E. coli Recombinase A (RecA)-promoted DNA strand exchange, which indicated that Mpn SSB may play an important role in DNA recombination processes in M. pneumoniae.
Conclusion
The M. pneumoniae MPN229 gene encodes a protein, Mpn SSB, which selectively and efficiently binds ssDNA, and stimulates E. coli RecA-promoted homologous DNA recombination. Consequently, the Mpn SSB protein may play a crucial role in DNA recombinatorial pathways in M. pneumoniae. The results from this study will pave the way for unraveling these pathways and assess their role in antigenic variation of M. pneumoniae.
Background
Mycoplasma pneumoniae is a human pathogen that causes a range of respiratory infections, such as tracheobronchitis, pharyngitis, and atypical pneumonia. M. pneumoniae causes up to 40% of community-acquired pneumonias and as many as 18% of cases requiring hospitalization in children (for a review, see [1]). It has previously been reported that M. pneumoniae-induced pneumonia may be observed most frequently among children 5–15 years of age. However, later studies have indicated that M. pneumoniae may also occur endemically and sometimes epidemically in older persons as well as in children less than 5 years of age.
Mycoplasmas are among the smallest self-replicating organisms, in both cellular dimensions and genome size, that are capable of free existence [2]. The genome of M. pneumoniae (strain M129) was found to have a length of 816,394 base pairs (bp), containing 688 open reading frames (ORFs) [3,4]. As would be expected on the basis of the relatively small size of its genome, being at least 5 times smaller than the Escherichia coli genome, M. pneumoniae possesses limited metabolic and biosynthetic capacities in comparison to 'classical' bacteria [5]. These limitations necessitate a parasitic lifestyle, and a close association of the bacterium with the respiratory epithelium of its human host.
A crucial step in the initiation of infection by M. pneumoniae is its attachment to the respiratory epithelium (cytadherence) of the host. This process is essential to pathogenesis since mutants that are unable to adhere are also avirulent [6]. Cytadherence is mediated by a specialized and complex attachment organelle. This organelle is localized at the tip of the bacterium and consists of a network of adhesins and accessory proteins. The major adhesion protein (cytadhesin) that is concentrated in the attachment organelle is the surface-exposed, 170-kDa P1 protein. This protein, which is encoded by the MPN141 gene, was demonstrated to be essential for attachment [6].
Apart from its function in cytadherence, the P1 protein is known to elicit a strong humoral immune response during infection [7]. In relation to this immunodominance, the P1 protein was hypothesized to undergo antigenic variation [8]. At the basis of this hypothesis is the finding within the P1 gene of two sequence blocks of which multiple variants exist throughout the M. pneumoniae genome [9,10]. These sequences (repetitive elements) were designated RepMP4, which is located near the 5' end of the P1 gene, and RepMP2/3, which is located at the 3' end [11]. Within the M. pneumoniae M129 genome, a total of 8 variants of RepMP4 and 10 variants of RepMP2/3 were identified [3]. Because the different variants of a given RepMP element are closely related in sequence, but not identical, it is possible that recombination between the P1 gene and RepMP sequences elsewhere in the genome could generate significant sequence variation within the P1 gene, resulting in amino acid changes in the P1 protein at the bacterial surface.
In order to demonstrate that recombination between RepMP elements lies at the heart of antigenic variation of M. pneumoniae, we initiated a study aimed at the identification of proteins that may be involved in this process. On the basis of sequence comparison, at least four M. pneumoniae ORFs were proposed to play a role in DNA recombination: MPN229 (or G07_orf166), MPN490 (or P02_orf336), MPN535 (or G12_orf206) and MPN536 (or G12_orf307), which may encode homologs of the E. coli single-stranded DNA binding protein (SSB), RecA, RuvA and RuvB, respectively [3]. In E. coli, SSB is a protein that primarily binds single-stranded DNA (ssDNA) and plays a central role in most aspects of DNA metabolism, including DNA replication and recombination. E. coli RecA is responsible for promoting pairing between a single-stranded donor DNA molecule and a homologous, double-stranded recipient DNA molecule, while RuvA and RuvB interact to form RuvAB, which possesses DNA helicase activity and catalyzes branch formation during recombination of two DNA strands.
Of the M. pneumoniae ORFs that were predicted to encode proteins involved in DNA recombination, only MPN535 has been studied in detail [12]. This protein (MpRuvA) was demonstrated to bind Holliday junctions and other branched DNA substrates in a similar fashion as E. coli RuvA [12]. In order to initiate the analysis of the complete set of putative DNA recombination enzymes from M. pneumoniae, we have examined the characteristics of the MPN229 protein product, which was designated Mpn single-stranded DNA-binding protein (Mpn SSB). We here report that Mpn SSB has a strong and selective ssDNA-binding activity, and promotes E. coli RecA-dependent DNA strand transfer.
Methods
Strains
M. pneumoniae strain MAC (ATCC® no. 15492™) was cultured in Mycoplasma medium containing 1.4% Difco™ PPLO broth (Becton Dickinson), 0.15% Difco™ TC Yeastolate, UF (Becton Dickinson), 1.4% glucose, 20% horse serum, 1,000 U/ml Penicillin G, 500 U/ml Polymyxine B, 3 μg/ml Fungizone, 5 μg/ml Voriconazol and 0.02 mg/ml phenol red. The pH of the medium was adjusted to 7.8–8.0 using a solution of 2 N NaOH. Finally, the medium was filter-sterilized. Cells were harvested upon color change of the medium (from red/orange to yellow).
Cloning of the MPN229 gene and generation of Mpn SSB expression constructs
DNA was purified from cultures of M. pneumoniae as follows. Cultures (3 ml) were harvested by centrifugation for 10 min in a microcentrifuge at full speed. The cell pellet was resuspended in 1 ml 0.9% NaCl. After centrifugation, the cell pellet was resuspended in 400 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA pH 8.0). Then, 70 μl of 10% sodium dodecyl sulfate (SDS) and 5 μl of Proteinase K (10 mg/ml) was added. After incubation for 10 min at 65°C, 100 μl of 5 M NaCl was added, followed by the addition of 100 μl of a solution of 10% N-cetyl-N,N,N-trimethylammonium bromide (CTAB) in 0.7 M NaCl. The solution was incubated for 10 min at 65°C. The DNA was subsequently extracted from the solution using 500 μl of chloroform/isoamyl alcohol (24:1, vol:vol). The DNA in the upper, aqueous phase was transferred to a fresh tube and precipitated by the addition of 360 μl of isopropanol. Following incubation at -20°C for 30 min, the DNA was pelleted by centrifugation for 10 min at full speed in a microcentrifuge. The DNA pellet was washed with 70% ethanol, dried, and resuspended in 10 μl of H2O. The MPN229 (or G07_orf166 [3]) ORF was amplified by PCR from the M. pneumoniae MAC genomic DNA. The PCR mixture (50 μl) contained 0.3 μM of oligonucleotide primer SSB_fw (5'-GGTCGTCATATGAACCGCGTTTTTTTATTTG-3'; the sequence in bold indicates a unique NdeI restriction endonuclease recognition site, which overlaps with the translation initiation codon [in italics] of MPN229), 0.3 μM of primer SSB_rv (5'-GCAGCCGGATCCTTATTCATCATCACTCTCCTC-3'; the sequence in bold indicates a unique BamHI restriction site, which is adjacent to the translation termination codon [anticodon in italics] of MPN229), 10 ng of M. pneumoniae genomic DNA, 0.2 mM of each dNTP, 1 unit of Pfu DNA polymerase (Fermentas), and 1 × Pfu buffer containing MgSO4 (20 mM Tris-HCl [pH 8.8], 10 mM (NH4)2SO4, 10 mM KCl, 0.1% Triton X-100, 0.1 mg/ml BSA, and 2 mM MgSO4). PCR was performed using the following conditions: 3 min at 95°C, followed by 30 cycles of 30 sec at 95°C, 30 sec at 50°C, and 2 min at 72°C. The resulting 522-base pairs (bp) PCR fragment was cloned into the pJET1/blunt cloning vector (Fermentas) using the GeneJET™ PCR cloning kit (Fermentas). From the generated plasmid, pJET1-MpnSSB, the MPN229 ORF was excised by digestion with NdeI and BamHI, and cloned into NdeI- and BamHI-digested expression vectors pET-11c and pET-16b (Novagen), resulting in plasmid pET-11c-MpnSSB and pET-16b-MpnSSB, respectively. In plasmid pET-11c-MpnSSB, the MPN229 ORF is cloned such as to express Mpn SSB in its natural, non-tagged form. In plasmid pET-16b-MpnSSB, the MPN229 ORF is fused at its 5' end to a sequence encoding a polyhistidine tag (MG(H)10SSGHIEGRH). This polyhistidine sequence allows one-step affinity purification of the MPN229-encoded protein by Ni2+-affinity chromatography (see below). For the generation of a glutathione S-transferase (GST)-Mpn SSB fusion protein expression construct, the NdeI-BamHI fragment containing MPN229 was first made blunt-ended by incubation with dNTPs and 'Klenow fragment' (Fermentas) and then cloned into SmaI-digested and Shrimp alkaline phosphatase (Promega)-treated vector pRP265 (NCCB Plasmids database, element No. PC-V3271; http://www.cbs.knaw.nl/scripts/Plasmids.dll/ShowPlasmid?Nr=3271). Plasmid pRP265 is a derivative of the expression plasmid pGEX-2T in which the polylinker 5'-GGATCCCCGGGAATTC-3' has been replaced by the sequence 5'-GGATCCCCATGGTACCCGGGTCGACTAGTATGCATAAGCTTGAATTC-3'. The resulting plasmid was termed pRP265-MpnSSB. Vector pRP265 was used for the production of native GST. The integrity of all DNA constructs used in this study was verified by DNA sequencing.
Expression and purification of Mpn SSB, Mpn H10-SSB and GST(-SSB)
Constructs pET-11c-MpnSSB, pET-16b-MpnSSB, pRP265-MpnSSB and pRP265 were introduced into E. coli BL21(DE3) and the resulting strains were grown overnight at 37°C in LB medium containing 100 μg/ml ampicillin. The cultures were diluted 1:100 in 100–150 ml LB medium with ampicillin and grown at 37°C to an optical density at 600 nm of 0.6. Protein expression was then induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.3 mM. After 3 hr, the bacteria were harvested by centrifugation and stored at -20°C.
Non-tagged Mpn SSB was purifed as follows. Frozen, bacterial pellets corresponding to 100 ml of culture were resuspended in 5 ml of buffer A (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT). The suspension was sonicated on ice and clarified by centrifugation for 20 min at 12,000 × g (4°C). To the supernatant, 5 ml of saturated ammonium sulphate was added, followed by 15 min of incubation on ice. After centrifugation for 10 min at 10,000 × g (4°C), the protein pellet, which contained the Mpn SSB protein, was dissolved in 10 ml of buffer A. Subsequently, 1 ml of a 50% suspension of DNA-cellulose (Worthington biochemical corp., Lakewood NJ), equilibrated in buffer A, was added, and the suspension was left on a rotating mixer for 1 hour at 4°C. The DNA-cellulose was poured into a column, and protein was eluted stepwise by the addition of 0.5 ml of a buffer (20 mM Tris-HCl pH 7.5, 1 mM DTT) containing increasing concentrations of NaCl (from 200 to 1500 mM). The fractions containing Mpn SSB were pooled and dialyzed against 20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1% Tween 20, 50% glycerol. The protein was stored at -20°C.
The polyhistidine-tagged Mpn SSB protein (Mpn H10-SSB) was purified as follows. Bacterial pellets corresponding to 150 ml of culture were resuspended in 10 ml of buffer B (20 mM Tris-HCl pH 8.0, 0.3 M NaCl) plus 5 mM imidazole. The suspension was sonicated on ice and clarified by centrifugation for 20 min at 12,000 × g (4°C). To the supernatant, 1 ml was added of a 50% slurry of Ni2+-nitroloacetic acid (Ni-NTA)-agarose (Qiagen), equilibrated previously in buffer B containing 5 mM imidazole. The suspension was stirred for 1 hr at 4°C and subsequently poured into a column. Non-specifically bound proteins were washed from the Ni-NTA agarose by two subsequent washes (of 4 ml each) with buffer B containing 5 mM, 10 mM and 20 mM imidazole, respectively. The bound Mpn SSB protein was eluted from the column with 2.5 ml of buffer B containing 250 mM imidazole. Fractions of 0.5 ml were collected, analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), pooled, and dialyzed against buffer B plus 1 mM DTT and 50% glycerol. Aliquots of purified protein were stored at -20°C.
Both GST and GST-tagged Mpn SSB protein (GST-SSB) were purified as follows. Bacterial pellets corresponding to 100 ml of culture were resuspended on ice in 5 ml of PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3) and sonicated. Then, Triton X-100 was added to a final concentration of 1%, and the suspension was left on a rotating mixer for 30 min at 4°C. Subsequently, the suspension was clarified by centrifugation for 20 min at 10,000 × g (4°C). To the supernatant, 0.8 ml was added of a 50% slurry of Glutathione-Agarose (Sigma, Saint Louis, MO, USA), equilibrated previously in PBS. The suspension was incubated on a rotating mixer for 30 min at 4°C and subsequently poured into a column. The column was washed consecutively with 3 ml of PBS containing 1% Triton X-100 and with 5 ml of PBS. Specifically bound protein was eluted from the column by addition of 3 ml of 50 mM Tris-HCl pH 8.0 containing 10 mM reduced glutathione (Sigma). Fractions of 0.5 ml were collected, analyzed by SDS-PAGE, pooled, and dialyzed against 20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1% Tween 20, 50% glycerol. Aliquots of purified protein were stored at -20°C. Protein concentrations were determined using the BC Assay protein quantitation kit (Interchim) as well as by SDS-PAGE analysis in conjunction with bovine serum albumin standards. All purified proteins were obtained at concentrations of at least 1 mg/ml, having an estimated homogeneity of 95% or greater. The concentrations of all proteins used in this study refer to monomeric protein concentrations throughout the manuscript.
SDS-PAGE
Proteins were separated on SDS-polyacrylamide gels, essentially as described by Laemmli [13]. Following electrophoresis, gels were stained with Coomassie brilliant blue (CBB), destained in 40% methanol/10% acetic acid, and photographed using a GelDoc XR system (Bio-Rad). Digital images were processed using Quantity One® 1-D Analysis Software (Bio-Rad).
Gel filtration chromatography
Gel filtration chromatography was carried out by applying a sample of 500 μl of Mpn SSB (at 0.3 mg/ml in 50 mM Tris-HCl (pH 7.5), 135 mM NaCl) to a Sephadex G-150 column with a length of 1 m and an inner diameter of 1.0 cm. The column was eluted with 50 mM Tris-HCl (pH 7.5), 135 mM NaCl at an elution rate of 4 ml/h. The column was calibrated with bovine serum albumin (BSA, 66.4 kDa), ovalbumin (42.9 kDa), and cytochrome C (12.3 kDa). Void volume was determined with blue dextran (2,000 kDa). Fractions of 1.0 ml were collected and analyzed by measuring the optical density at 280 nm (OD280). Since a relatively low concentration of Mpn SSB was loaded on the column, the eluted fractions were precipitated with trichloroacetic acid, and separated on 14% SDS-PAGE gels. Gels were stained with CBB and recorded using the GelDoc XR system (Bio-Rad). In fractions that contained Mpn SSB, the amount of protein was determined semi-quantitatively using BioNumerics Version 3.0 software (Applied Maths).
DNA substrates
The sequences of the oligonucleotide substrates that were used in this study are listed in Table 1. Oligonucleotides were purchased from Eurogentec and were used either unlabeled or 5'-end labeled with 32P. Bacteriophage M13mp18 circular, single-stranded DNA was purified from cultures of infected E. coli XL-Blue cells (Stratagene), essentially as described previously [14]. Bacteriophage φX174 RF1 DNA was purchased from Fermentas. The DNA was linearized by digestion with PstI (Invitrogen), followed by inactivation of the restriction enzyme by incubation for 20 min at 80°C. Viron DNA from φX174 was purchased from New England Biolabs. The concentration of bacteriophage-derived DNAs was determined using the NanoDrop 1000 (Thermo Scientific). The DNA concentrations are indicated for DNA molecules and not for nucleotides for all substrates throughout the manuscript.
Table 1.
Sequences of the oligonucleotide substrates used in this study
| Name | Sequence |
| Oligo 1 (48-mer) | 5'-GGAACAGCTACAGCTGATCATCACCATCACCATCACTAGGATCCGCAT-3' |
| Oligo 2 (49-mer) | 5'-CACGTGCTAGCCATCACCATCACCATCACGTGAAGACAACAGTCTATCC-3' |
| Oligo 3 (50-mer) | 5'-CAGGTGCACCAGAACAACCACATCACCATCACCATCACTAGGATCCGCAT-3' |
| (A)50 (50-mer) | 5'-AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA-3' |
| (T)50 (50-mer) | 5'-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3' |
| (C)50 (50-mer) | 5'-CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC-3' |
| (G)50 (50-mer) | 5'-GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG-3' |
| 15 nt (15-mer) | 5'-CCCTGTTAGCACTGT-3' |
| 20 nt (20-mer) | 5'-CCCTGTTAGCACTGTTGTAT-3' |
| 30 nt (30-mer) | 5'-CCCTGTTAGCACTGTTGTATCCAGCATCCT-3' |
| 40 nt (40-mer) | 5'-CCCTGTTAGCACTGTTGTATCCAGCATCCTGTCTTGCAGA-3' |
| 50 nt (50-mer) | 5'-CCCTGTTAGCACTGTTGTATCCAGCATCCTGTCTTGCAGAGCATTCCGAC-3' |
DNA-binding assays
DNA-binding experiments were carried out with either oligonucleotide substrates (Table 1), bacteriophage M13mp18 DNA, or bacteriophage φX174 DNA (both single- and double-stranded). Reactions were performed in volumes of 20 μl with various concentrations of purified protein in 25 mM Tris-OAc pH 7.5, 10 mM Mg(OAc)2, 1 mM DTT, 5% glycerol, and either a single-stranded (ss) unlabeled oligonucleotide (5 μM), 5'-32P-labeled oligonucleotide (1 μM), double-stranded (ds) supercoiled bacteriophage DNA (M13mp18 or φX174 DNA) (1 nM), or ss bacteriophage DNA (M13mp18 or φX174 DNA) (2 nM). After incubation for 15 min at 37°C, 2 μl was added of a solution containing 40% glycerol and 0.25% bromophenol blue. Subsequently, the samples were electrophoresed in 0.5 × TBE buffer, either through 1.0% agarose gels (when using unlabeled oligonucleotides), 0.6% agarose gels (when using bacteriophage DNA) or 5% polyacrylamide gels (when using 32P-labeled oligonucleotides). Agarose gels were stained with ethidium bromide and photographed. The polyacrylamide gels were subjected to autoradiography.
Three-strand transfer assay
E. coli RecA-promoted DNA strand transfer reactions were carried out using φX174 DNA, in a similar fashion as described previously [15,16]. Reactions (30 μl) contained 70 mM Tris-HCl pH 7.6, 10 mM MgCl2, 5 mM DTT, 5 mM ATP, 0.08 U/μl Pyruvate Kinase Preparation (Type VII, from rabbit muscle [Sigma]), 3.2 mM phospho(enol)pyruvic acid (Sigma), 1 nM ss circular φX174 DNA, 2 nM PstI-digested, ds φX174 DNA, 0.2 μg/μl E. coli Rec A (New England Biolabs), and 3 ng/μl of either E. coli SSB (Epicentre Biotechnologies), or 6.5 ng/μl of either Mpn SSB, GST-SSB or GST. Samples of 10 μl were taken at 0, 30 and 60 min of incubation at 37°C, and reactions were terminated by the addition of 1 μl of a solution of 5% SDS and 50 mM EDTA pH 8.0, followed by incubation for 10 min at 55°C. After the addition of loading dye, the samples were separated on 0.6% agarose gels in 0.5 × TBE. The gels were stained with ethidium bromide and photographed.
Results
Coding capacity of the MPN229 ORF
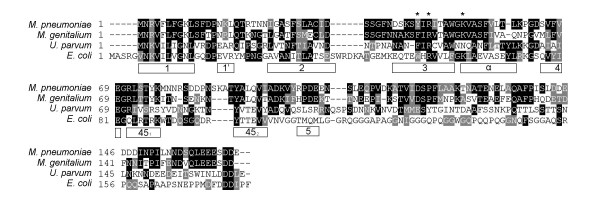
The M. pneumoniae MPN229 ORF was recognized as an ORF putatively encoding a homolog of single-stranded DNA binding proteins after determination of the complete genome sequence of M. pneumoniae strain M129 [3]. MPN229 has the capacity to encode a 166-amino acid protein (termed Mpn SSB) with a theoretical molecular mass of 18.4 kDa. The predicted amino acid sequence of Mpn SSB displays only limited similarity with the protein encoded by the SSB gene from E. coli (17% identity). A significantly higher similarity was seen with the protein that is putatively encoded by the MG091 gene [17] from Mycoplasma genitalium (61% identity). The residues that are shared among the predicted amino acid sequences of the (putative) SSB proteins from M. pneumoniae, M. genitalium, Ureaplasma parvum (all belonging to the family of Mycoplasmataceae) as well as from E. coli, are depicted in a multiple sequence alignment in Fig. 1. Most bacterial SSB proteins that have been studied to date possess a so-called oligonucleotide/oligosaccharide-binding (OB) fold and function as homotetramers [18,19]. By contrast, the SSB proteins from Thermus aquaticus, Thermus thermophilus [20] and Deinococcus radiodurans [21], which have a significantly higher molecular weight than the E. coli (-like) SSB protein(s), were reported to contain two OB folds per monomer and function as homodimers. As shown in Fig. 1, Mpn SSB, which is 12 amino acids smaller than the 178-amino acid E. coli SSB, may contain a single OB fold. The amino acid motifs that are typical of the OB fold are indicated by the open boxes below the aligned sequences in Fig. 1.
Figure 1.
Multiple alignment of the amino acid sequences predicted to be encoded by the M. pneumoniae MPN229 [3], M. genitalium MG091[17], U. parvum ssb [39], and E. coli ssb [40] genes. The amino acid motifs that are characteristic for the OB DNA-binding fold are indicated by the open boxes below the aligned sequences [18]. The asterisk above Y44 of the M. pneumoniae sequence indicates the position of a Trp residue (W55) in E. coli SSB that was shown to play a role in DNA-binding. The other two asterisks indicate conserved arginine (R) and lysine (K) residues (R46 and K52, respectively, in the M. pneumoniae sequence); for E. coli SSB these residues were shown to be oriented toward the DNA binding cleft [32]. The multiple aligment was performed using Clustal W http://www.ebi.ac.uk/Tools/clustalw/index.html. The program BOXSHADE 3.21 http://www.ch.embnet.org/software/BOX_form.html was used to generate white letters on black boxes (for residues that are identical in at least two out of four sequences) and white letters on grey boxes (for residues that are similar in at least two out of four sequences).
Salient features of the Mpn SSB sequence
The C-terminus of the Mpn SSB sequence contains a high proportion of acidic amino acid residues: 13 of the 24 C-terminal amino acids are aspartic acid (D) or glutamic acid (E) residues. Albeit less extensive, acidic residues are also concentrated near the C-terminus of other SSB proteins. In E. coli, these residues have been demonstrated to be involved in the interaction of SSB with other proteins, such as exonuclease I, uracil DNA glycosylase, and DNA polymerase III [22-30]. Several residues of the E. coli SSB protein have been demonstrated to play a role in DNA-binding. These include three tryptophan (W) residues at position 41 (adjacent to OB box '2' in Fig. 1), 55 (within box '3') and 89 (adjacent to box '451') [31]. However, none of these residues appear to be conserved with the SSB proteins from the Mycoplasmataceae. Only W55 seems to have a clear counterpart in the mycoplasma proteins, i.e. a tyrosine (Y) residue in M. pneumoniae (Y44) and a phenylalanine (F) in both M. genitalium (F44) and U. parvum (F43). Other residues that were found to be implicated in E. coli SSB DNA-binding, however, do have a counterpart in the mycoplasma SSBs. The arginine (R) and lysine (K) residues at positions 57 and 63, respectively, of E. coli SSB, which were shown to be oriented toward the DNA binding cleft [32], are conserved in the M. pneumoniae and M. genitalium SSB proteins (R46 and K52, respectively, in both proteins). These residues are located in the predicted OB fold motifs '3' and 'α' (Fig. 1). Interestingly, the histidine (H) residue that was described to be important for tetramerization of E. coli SSB (H56, central to OB fold box '3') [33] is not conserved among the proteins listed in Fig. 1; an isoleucine (I) is present at a position congruent to that of H56 in the mycoplasma proteins (residue I45 in M. pneumoniae SSB). The lack of conservation of this apparently crucial amino acid residue, however, can be reconciled with the finding that substitution of residue H56 of E. coli SSB for either an F or I residue, did not change the properties of the protein measurably [33].
Expression and purification of the MPN229-encoded protein
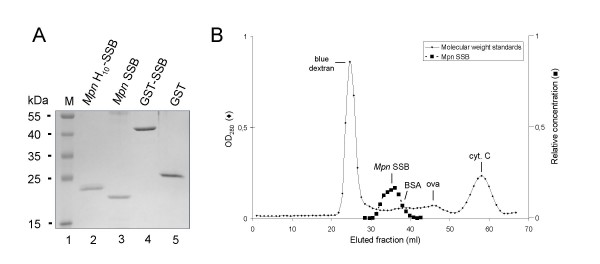
The MPN229 gene was cloned into different expression vectors in order to express Mpn SSB either in its native form (Mpn SSB), as an N-terminal polyhistidine-tagged polypeptide (Mpn H10-SSB) or as a glutathione S-transferase (GST)-tagged fusion protein (GST-SSB). Each of these proteins was expressed to high levels in E. coli and was purified to near homogeneity (Fig. 2A). The estimated molecular weights of the purified proteins corresponded to their theoretical molecular weights of 18.4 kDa, 20.9 kD and 45.0 kDa for Mpn SSB, Mpn H10-SSB, and GST-SSB, respectively. As a control, we also purified non-fused GST (Fig. 2A, lane 5). While most experiments were performed using Mpn H10-SSB, the activities of the three different SSB variants were found to be indistinguishable.
Figure 2.
Purification of recombinant Mpn SSB proteins. (A) Samples of purified recombinant Mpn SSB proteins, i.e. Mpn H10-SSB (lane 2), Mpn SSB (lane 3), GST-SSB (lane 4), and purified GST (lane 5), were analyzed by SDS-PAGE (12%) and Coomassie brilliant blue (CBB)-staining. The sizes of protein markers (lane 1; PageRuler™ Prestained Protein Ladder [Fermentas]) are shown on the left-hand side of the gel. (B) Gel filtration analysis of Mpn SSB. Gel filtration chromatography was performed by applying Mpn SSB to a Sephadex G-150 column. The column was calibrated with blue dextran (2,000 kDa), bovine serum albumin (BSA, 66.4 kDa), ovalbumin (ova, 42.9 kDa), and cytochrome C (cyt. C, 12.3 kDa). Fractions of 1.0 ml were collected and monitored by measuring the optical density at 280 nm (OD280, Y-axis at the left-hand side of the graph). The fractions eluted from the subsequent run containing Mpn SSB were precipitated with trichloroacetic acid, and separated on 14% SDS-PAGE gels. Gels were stained with CBB and recorded using the GelDoc XR system (Bio-Rad). In fractions that contained Mpn SSB, the amount of protein was determined semi-quantitatively using BioNumerics Version 3.0 software (Applied Maths). The relative concentration of Mpn SSB (Y-axis on the right-hand side, in arbitrary units) in these fractions is plotted. In all other fractions, Mpn SSB was not detected by SDS-PAGE analysis and Coomassie brilliant blue-staining.
The oligomeric form of Mpn SSB in solution
In order to determine the oligomeric state of Mpn SSB in solution, the untagged, purified protein was subjected to size-exclusion chromatography. As shown in Fig. 2B, Mpn SSB eluted from the column as a single, major protein species with an estimated molecular weight of ~75 kDa. This molecular weight corresponds to the theoretical molecular weight of the tetrameric form of Mpn SSB (73.6 kDa). These data therefore suggests that Mpn SSB exists predominantly as a homo-tetramer in solution. A similar oligomeric composition has previously also been found for SSB proteins from other bacterial species, such as E. coli [32].
Oligonucleotide binding by Mpn SSB
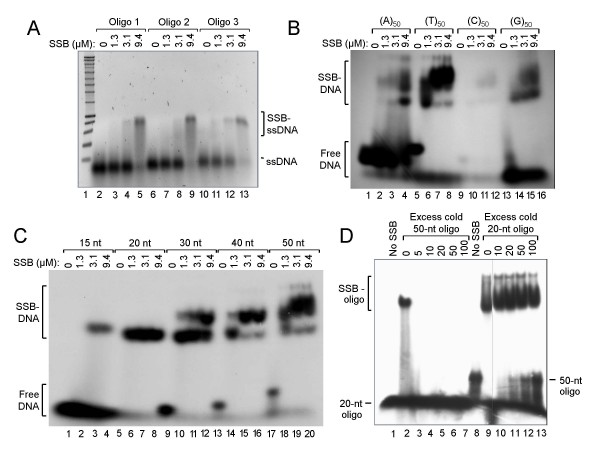
To investigate the potential of Mpn H10-SSB to bind DNA, the protein was incubated at various concentrations with three different single-stranded oligonucleotide (ssDNA) substrates. The resulting complexes were analyzed by non-denaturing agarose gel electrophoresis, followed by staining with ethidium bromide. As shown in Fig. 3A, an increasing amount of product with a lower mobility than that of the free oligonucleotides was generated with increasing Mpn H10-SSB concentrations. At the highest protein concentration tested (9.4 μM; lanes 5, 9 and 13), virtually all of the ssDNA formed part of the lower mobility complex, which was assumed to represent Mpn H10-SSB-ssDNA complexes. DNA-binding by Mpn H10-SSB was found to be rapid, as the DNA-protein complexes shown in Fig. 3A were already produced in less than 1 min of incubation at 37°C (data not shown). Similar results were obtained with the other recombinant SSB proteins (data not shown).
Figure 3.
The Mpn SSB protein binds oligonucleotide substrates in a DNA sequence-independent fashion. (A) Binding of Mpn H10-SSB to three different oligonucleotide substrates. Reactions were performed in volumes of 20 μl and contained either 0 μM (lanes 2, 6 and 10), 1.3 μM (lanes 3, 7 and 11), 3.1 μM (lanes 4, 8 and 12) or 9.4 μM (lanes 5, 9 and 13) of Mpn H10-SSB, and 5 μM of either of three different single-stranded oligonucleotides (Oligo 1, 2 or 3; Table 1). After incubation for 15 min at 37°C, the samples were electrophoresed on a 1.0% agarose gel in 0.5 × TBE buffer. A black/white inverted image of a typical ethidium bromide-stained gel is shown. (B) Binding of Mpn H10-SSB (at 0, 1.3, 3.1 or 9.4 μM in lanes 1–4, 5–8, 9–12 and 13–16, respectively) to 5'-32P-labeled homooligomeric DNA substrates (at 1 μM; Table 1). The samples were separated on 5% polyacrylamide gels in 0.5 × TBE buffer. An autoradiograph is shown. (C) Binding of Mpn H10-SSB (at 0, 1.3, 3.1 or 9.4 μM) to a series of 15- to 50-mer 5'-32P-labeled oligonucleotides (at 1 μM), each containing the same 15-nucleotide core sequence (Table 1). The samples were processed as described above in (B). (D) A DNA-binding competition experiment in which Mpn H10-SSB (at 3.1 μM) was incubated with a constant amount (1 μM) of either the 5'-32P-labeled '20 nt' oligonucleotide (lanes 1–7) or '50-nt' oligonucleotide (lanes 8–13) (Table 1), and increasing amounts of the other, unlabeled ('cold') oligonucleotide. A molar excess of 5 to 100 times unlabeled oligonucleotide over labeled oligonucleotide was tested, as indicated above the lanes. From the samples loaded in lanes 1 and 8, Mpn H10-SSB was omitted; the gel was processed similarly as in (B).
Because protein-DNA complexes were generated with each of the three different ssDNA substrates tested, we hypothesized that Mpn SSB may bind ssDNA in a sequence-independent manner. To further investigate this, we tested the ability of Mpn H10-SSB to bind 32P-labeled homooligomers (50-mers) of dA, dT, dC and dG (Table 1). Although the (T)50 substrate seemed to be bound most efficiently, each of the homooligomers was bound by the protein (Fig. 3B). The (C)50 substrate, despite being labeled inefficiently, most likely due to the formation of inaccessible secondary structures, was also bound by Mpn H10-SSB. These data show that the protein indeed binds ssDNA irrespective of the DNA substrate sequence. It is interesting to note that binding of Mpn H10-SSB to the 50-nucleotide homooligomer substrates resulted in the formation of two different DNA-protein complexes (Fig. 3B). As the formation of the larger of the two complexes was favored at higher protein concentrations, it is likely that this larger species represents DNA-protein complexes in which a higher number of SSB DNA-binding units (eg., dimers or tetramers) are present than in the smaller species. Thus, a 50-mer DNA substrate may be bound by a single binding unit at relatively low protein concentrations, whereas two or more binding units may occupy the substrate at higher protein concentrations. To study this further, we tested binding of Mpn H10-SSB to a series of oligonucleotides with lengths ranging from 15 to 50 nucleotides, and each containing the same 15-nucleotide 'core sequence', which was randomly chosen (Table 1). As shown in Fig. 3C, only the 15-nucleotide substrate was bound inefficiently by the protein, whereas the other substrates were bound efficiently, each being completely complexed at higher protein concentrations. Interestingly, a second, larger DNA-protein complex was seen exclusively with substrates with a length equal to or larger than 30 nucleotides. The efficiency of formation of these larger complexes was even higher with the 40- or 50-mer substrates (lanes 14–20). Thus, whereas a 20-mer substrate may accommodate binding of a single SSB DNA-binding unit, longer substrates may allow binding of two or more units, generating protein-DNA complexes with a lower electrophoretic mobility than do single unit-bound complexes.
To investigate whether Mpn SSB binds with similar affinities to 20- and 50-mer substrates, a DNA-binding competition experiment was performed in which Mpn H10-SSB was incubated with a constant amount of radiolabeled oligonucleotide and increasing amounts of the other, unlabeled oligonucleotide (Fig. 3D). Complexes with 20-mer substrates were no longer observed when a 5-fold molar excess of the 50-mer substrate was present in the DNA-binding reaction (compare lanes 2 and 3). By contrast, the formation of complexes with the 50-mer substrate was only partially inhibited in the presence of a 20-100-fold excess of unlabeled 20-mer oligonucleotide (Fig. 3D, lanes 11–13). These results indicate that Mpn H10-SSB binds with a higher affinity to the 50-mer substrate than to the 20-mer substrate.
dsDNA- versus ssDNA-binding of SSB and divalent cation-dependence of DNA-binding
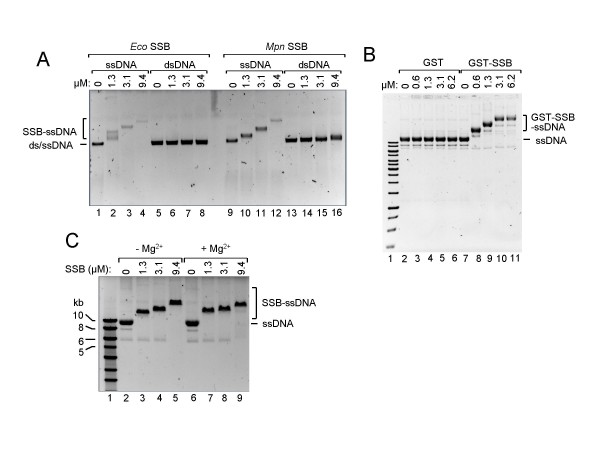
To study the ability of Mpn SSB to also bind dsDNA, the protein was incubated with either single- or double-stranded φX174 DNA (Fig. 4A). While almost all ssDNA was shifted towards a lower mobility form at the lowest concentration of Mpn SSB tested (1.3 μM; lane 10), significant binding to dsDNA could not be observed at any protein concentration used (lanes 14–16). These results demonstrate that Mpn SSB preferentially binds ssDNA. Fig. 4A further shows that the gel mobility of Mpn SSB-ssDNA complexes becomes lower when Mpn SSB concentrations are increased (lanes 10–12). Similarly as described above for the oligonucleotide substrates, it is likely that increasing numbers of Mpn SSB molecules accumulate on the ssDNA substrates when higher concentrations of protein are present. Consequently, the size of the DNA-protein complexes will increase when the protein concentration becomes higher, resulting in lowering of the gel mobility of these complexes. Similar DNA-binding results were obtained for each of the purified, recombinant Mpn SSB proteins, including GST-SSB (Fig. 4B, lanes 7–11), as well as for E. coli SSB (Fig. 4A, lanes 2–4). Interestingly, the E. coli SSB protein may assemble on DNA substrates in a different fashion than does Mpn SSB, as more heterogeneously sized protein-DNA complexes were formed by the E. coli protein than by the M. pneumoniae protein, in particular at relatively low protein concentrations (compare lanes 2 and 10 in Fig. 4A).
Figure 4.
Mpn SSB binding to long DNA substrates and Mg2+-dependence of DNA-binding. (A) Binding of E. coli SSB (Eco SSB, lanes 1–8) and Mpn SSB (lanes 9–16) to either 2 nM of ss φX174 DNA (ssDNA, lanes 1–4 and lanes 9–12) or 1 nM of ds φX174 DNA (dsDNA, lanes 5–8 and lanes 13–16). Reactions were performed in volumes of 20 μl and contained various concentrations of protein, as indicated above the lanes. The samples were separated on a 0.6% agarose gel in 0.5 × TBE buffer. A black/white inverted image of a typical ethidium bromide-stained gel is shown. (B) Binding of GST-SSB to circular, single-stranded M13mp18 DNA (ssDNA). Binding reactions were carried out with various concentrations of either GST-SSB (lanes 8–11) or GST (lanes 3–6), as indicated above the lanes, and 2 nM of DNA. The samples were separated on 0.6% agarose gels in 0.5 × TBE buffer and processed as described above. (C) DNA-binding by Mpn SSB is Mg2+-independent. Reactions with Mpn H10-SSB were executed similarly as described in (B), except for the omission of Mg(OAc)2 in the reaction mixtures of the samples loaded in lanes 2–5. Complexes of Mpn SSB bound to ssDNA (SSB-ssDNA), and the position of unbound DNA (ssDNA and dsDNA), are indicated alongside the gel. The marker DNA loaded in lane 1 of both (B) and (C) is the SmartLadder (Eurogentec).
To determine whether Mpn SSB is dependent upon divalent cations for its activity, a DNA-binding experiment was conducted in either the presence or absence of Mg2+. Fig. 4C shows that similar DNA-protein complexes are generated irrespective of the presence of Mg2+ in the binding reaction (compare lanes 3–5 to lanes 7–9), which indicates that the protein does not require divalent cations for DNA-binding. Similarly, the SSB proteins from E. coli and S. pneumoniae were previously also shown to bind DNA irrespective of the presence of Mg2+ in the binding reaction [22,34]. However, these proteins do display different binding modes in either the presence or absence of divalent cations [22,34].
Mpn SSB stimulates E. coli RecA-promoted three-strand DNA exchange
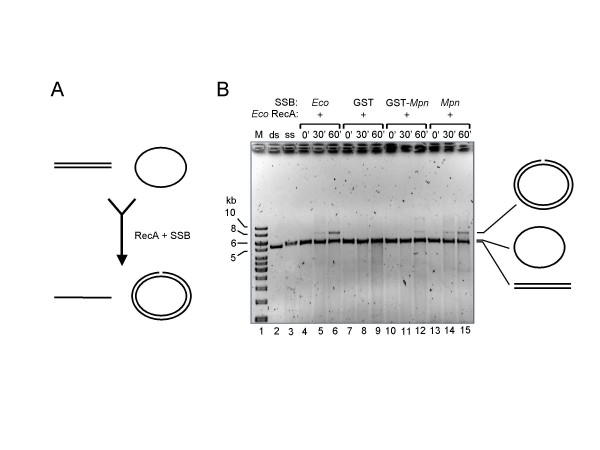
Single-stranded DNA-binding proteins from several micro-organisms have previously been shown to stimulate in vitro DNA strand exchange reactions catalyzed by (homologs of) RecA. Also, the SSB proteins from S. pneumoniae and D. radiodurans were found to stimulate the activity of RecA from E. coli [16,21]. To study the potential of Mpn SSB to stimulate E. coli RecA-catalyzed DNA recombination, so-called three-strand exchange reactions were performed. In these reactions, RecA promotes the transfer of one of the strands of a linear, dsDNA molecule (donor) to a complementary, circular ssDNA molecule (acceptor), resulting in a double-stranded, circular product and a linear, single-stranded product (Fig. 5A).
Figure 5.
Mpn SSB promotes E. coli RecA-catalyzed three-strand DNA transfer. (A) Schematic representation of the three-strand transfer reaction. E. coli RecA catalyzes the transfer of a single strand from a linear, double-stranded DNA donor molecule (at the top left) to a complementary, single-stranded, circular acceptor molecule (at the top right), resulting in a linear, single-stranded product (bottom, left) and a (nicked) circular, double-stranded product (bottom, right). This reaction is strongly promoted by the E. coli SSB protein. (B) E. coli RecA-promoted DNA strand transfer reactions using φX174 DNA. Reactions were performed at 37°C in the presence of either E. coli SSB (Eco SSB, lanes 4–6), GST (lanes 7–9), GST-SSB (GST-Mpn, lanes 10–12) or Mpn SSB (lanes 13–15). DNA concentrations used were 1 nM and 2 nM for the ssDNA and dsDNA, respectively. Reactions were terminated at either 0, 30 or 60 min of incubation (0', 30' and 60', respectively, above the lanes). The samples were separated on 0.6% agarose gels in 0.5 × TBE buffer. A black/white inverted image of an ethidium bromide-stained gel is shown. A schematic representation of the major DNA products is indicated at the right-hand side of the gel. The DNA marker (M) is the SmartLadder (Eurogentec). ds (lane 2), linear, dsDNA donor; ss (lane 3), circular, ssDNA acceptor.
Either alone (data not shown) or in the presence of GST (Fig. 5B, lanes 7–9), E. coli RecA did not detectably catalyze φX174 DNA strand exchange. However, in the presence of E. coli SSB, strand transfer was readily detectable after 60 min of incubation at 37°C (lane 6), while a faint DNA species at the position of the double-stranded, open circular recombination product was already visible after 30 min (lane 5). Similar results were obtained when E. coli SSB was replaced by either Mpn SSB (lanes 13–15), Mpn H10-SSB (data not shown) or GST-SSB (lanes 10–12), albeit that the latter protein was somewhat less efficient than the other recombinant proteins in stimulating RecA activity.
Taken together, these data show that Mpn SSB, despite having a relatively low level of sequence similarity with E. coli SSB, is capable of stimulating DNA strand transfer activity of E. coli RecA. Consequently, Mpn SSB may play a similar, crucial role as E. coli SSB in DNA recombination processes. The elucidation of the activities of Mpn SSB can therefore be considered an important step in unraveling the recombinatorial events that may occur in the genome of M. pneumoniae, and which may play a pivotal role in antigenic variation of this bacterium.
Discussion
In order to assess whether recombination between RepMP elements within the genome of M. pneumoniae lies at the heart of antigenic variation of this bacterium, we set out to identify the proteins that may be involved in DNA recombination. Previously, four M. pneumoniae ORFs were proposed to code for proteins that could play a role in DNA recombination: MPN229, MPN490, MPN535 and MPN536 [3]. These ORFs show sequence similarity with the E. coli genes encoding SSB, RecA, RuvA and RuvB, respectively. Up until now, only one of these four ORFs, i.e. MPN535, has been subjected to a detailed study. This ORF was found to encode a RuvA homolog (MpRuvA) which is capable of binding Holliday junctions and other branched DNA structures in a manner similar to E. coli RuvA [12]. In contrast to E. coli RuvA, however, MpRuvA did not support branch migration mediated by E. coli RuvB. MpRuvA was also found to differ from E. coli RuvA in its inability to allow E. coli RuvC to stably bind a MpRuvA-bound DNA junction complex [12]. In addition, MpRuvA was unable to promote DNA repair in E. coli ruvA mutants [12]. These findings clearly illustrated that the DNA recombination machinery in M. pneumoniae may differ considerably from that in E. coli. In agreement with this notion, we found the Mpn SSB protein to differ in several characteristics from its E. coli counterpart. First, the sequence similarity between these proteins is rather low (17% identity). Second, Mpn SSB and E. coli SSB appear to diverge in the way by which they assemble on long, single-stranded DNA substrates. This was illustrated by the heterogeneity of the DNA-protein complexes formed by E. coli SSB, whereas complexes formed by Mpn SSB appeared rather homogeneous in size (Fig. 4A). The heterogeneous size of these E. coli SSB-DNA complexes may be indicative of the non-random distribution of the protein on DNA substrates, reflecting the cooperative DNA-binding activity of E. coli SSB (see Lohman and Ferrari for a review)[22]. Nevertheless, E. coli SSB can interact with ssDNA in different binding modes, each having different characteristics, including cooperativity [22]. These binding modes were found to be dependent on the solution conditions, such as protein to DNA ratio, salt concentration and nucleotide sequence of the DNA substrates. Although Mpn SSB appears to bind with a significantly lower level of cooperativity than its E. coli counterpart in Fig. 4A, it is possible that cooperative binding by Mpn SSB is also strongly dependent on the binding conditions used. In our study, these conditions may not have been optimal for Mpn SSB cooperativity.
Another factor that plays an eminent role in protein-DNA complex formation is the multimeric state of the protein. E. coli SSB was reported to form homotetramers in solution [35,36]. The Mpn SSB protein also appears to exist as a tetramer in solution. With regard to the oligomeric status of Mpn SSB, is interesting to note that the bacterial SSBs that have previously been analyzed can be divided in separate groups on the basis of their oligomeric status and number of OB DNA-binding domains per monomer. The most prominent group consists of proteins that contain a single OB fold per monomer and function as homotetramers [18,19]. Among the members of this group are the SSB proteins from E. coli [32], Streptococcus pneumoniae [16,34] and Herbaspirillum seropedicae [37] and M. pneumoniae (this study). Another group of bacterial SSB proteins appears to possess two OB folds and function as dimers. Members of this group are the SSBs from T. aquaticus [20], T. thermophilus [20] and D. radiodurans [21]. These proteins are considerably larger in size than those from the first group. A common feature of both groups of SSBs is that they contain four OB folds per functional protein oligomer [21].
In contrast to the differences between the SSB proteins from E. coli and M. pneumoniae in primary sequence as well as DNA-binding characteristics, these proteins were found to be indistinguishable in their capacity to promote E. coli RecA-catalyzed DNA strand exchange reactions. In previous studies it has been shown that the SSBs from D. radiodurans and S. pneumoniae were also able to stimulate E. coli RecA protein-promoted DNA three-strand exchange reactions with at least the same efficiency as E. coli SSB [16,21]. However, these activities of the D. radiodurans and S. pneumoniae SSB proteins may not be regarded as unexpected, as they show, unlike Mpn SSB, a relatively high level of sequence similarity with their E. coli counterpart (38% and 31%, respectively) [16,21].
Conclusion
We conclude that Mpn SSB represents the M. pneumoniae counterpart of the E. coli SSB protein. As a consequence, it is likely that this protein may be involved in various aspects of the DNA metabolism of M. pneumoniae. In a study on the complete proteome of M. pneumoniae by two-dimensional gel electrophoresis followed by mass spectrometry, the Mpn SSB protein was the only protein detected of the four proteins that were proposed to be involved in DNA recombination [38]. Studies aimed at the detection and characterization of the other three proteins, i.e. the RecA, RuvA and RuvB homologs, are currently underway. It will be important to determine whether these proteins interact and/or cooperate to catalyze homologous DNA recombination in M. pneumoniae and whether these proteins are necessary and sufficient for the recombination between RepMP elements in the bacterial genome. However, as homologs of SSB, RecA, RuvA and RuvB are found in all living organisms from bacteria to humans, it is tempting to speculate that the site-specific recombination of RepMP elements may require the activity of one or more specialized, yet unidentified proteins. The identification of such proteins and the elucidation of the DNA recombinatorial pathways in M. pneumoniae will be the main challenge of our future studies.
Authors' contributions
MS cloned and sequenced the Mpn SSB gene, generated most of the recombinant plasmid constructs, purified the majority of the recombinant proteins, and performed most of the DNA-binding and strand transfer experiments. TH cultured M. pneumoniae and verified the identity of the purified recombinant proteins. CV generated some of the plasmid constructs, performed several protein purifications and did part of the DNA-binding experiments. NGH and CV designed the study and drafted the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Prof. Dr. J.S. Jensen (Statens Serum Institut, Copenhagen, Denmark) for permission to use M. pneumoniae strains that were originally isolated in his laboratory. We thank Dr. S.A.J. Zaat (Academic Medical Center Amsterdam, The Netherlands) for supplying these strains. Prof. Dr. E. Jacobs and Dr. R. Dumke (Medical Faculty Carl Gustav Carus, Institute of Medical Microbiology and Hygiene, Dresden, Germany) are thanked for their help with setting up the M. pneumoniae culturing facility at our laboratory. We thank T. Roelofse and K.-Y. Liu for technical assistance.
Contributor Information
Marcel Sluijter, Email: m.sluijter@erasmusmc.nl.
Theo Hoogenboezem, Email: t.hoogenboezem@erasmusmc.nl.
Nico G Hartwig, Email: n.hartwig@erasmusmc.nl.
Cornelis Vink, Email: c.vink@erasmusmc.nl.
References
- Waites KB, Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004;17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MH, Collier AM. Ultrastructural study of Mycoplasma pneumoniae in organ culture. J Bacteriol. 1976;125:332–339. doi: 10.1128/jb.125.1.332-339.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, Doerks T, Sanchez-Pulido L, Snel B, Suyama M, et al. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 2000;28:3278–3288. doi: 10.1093/nar/28.17.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T, Sauerborn R. Comparative genome analysis and pathway reconstruction. Pharmacogenomics. 2002;3:245–256. doi: 10.1517/14622416.3.2.245. [DOI] [PubMed] [Google Scholar]
- Baseman JB. The cytadhesins of Mycoplasma pneumoniae and M. genitalium. Subcell Biochem. 1993;20:243–259. doi: 10.1007/978-1-4615-2924-8_9. [DOI] [PubMed] [Google Scholar]
- Leith DK, Trevino LB, Tully JG, Senterfit LB, Baseman JB. Host discrimination of Mycoplasma pneumoniae proteinaceous immunogens. J Exp Med. 1983;157:502–514. doi: 10.1084/jem.157.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenri T, Taniguchi R, Sasaki Y, Okazaki N, Narita M, Izumikawa K, Umetsu M, Sasaki T. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect Immun. 1999;67:4557–4562. doi: 10.1128/iai.67.9.4557-4562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CJ, Chavoya A, Baseman JB. Regions of Mycoplasma pneumoniae cytadhesin P1 structural gene exist as multiple copies. Infect Immun. 1988;56:3157–3161. doi: 10.1128/iai.56.12.3157-3161.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel R, Herrmann R. Repetitive DNA sequences in Mycoplasma pneumoniae. Nucleic Acids Res. 1988;16:8337–8350. doi: 10.1093/nar/16.17.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland K, Wenzel R, Herrmann R. Analysis of three different repeated DNA elements present in the P1 operon of Mycoplasma pneumoniae: size, number and distribution on the genome. Nucleic Acids Res. 1990;18:6311–6317. doi: 10.1093/nar/18.21.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingleston SM, Dickman MJ, Grasby JA, Hornby DP, Sharples GJ, Lloyd RG. Holliday junction binding and processing by the RuvA protein of Mycoplasma pneumoniae. European journal of biochemistry/FEBS. 2002;269:1525–1533. doi: 10.1046/j.1432-1033.2002.02805.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley and Sons, Inc; 2007. [Google Scholar]
- Corrette-Bennett SE, Lovett ST. Enhancement of RecA strand-transfer activity by the RecJ exonuclease of Escherichia coli. The Journal of biological chemistry. 1995;270:6881–6885. doi: 10.1074/jbc.270.12.6881. [DOI] [PubMed] [Google Scholar]
- Steffen SE, Bryant FR. Purification and characterization of the single-stranded DNA binding protein from Streptococcus pneumoniae. Archives of biochemistry and biophysics. 2001;388:165–170. doi: 10.1006/abbi.2001.2286. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. The EMBO journal. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F, Seitz EM, Kowalczykowski SC. Novel homologs of replication protein A in archaea: implications for the evolution of ssDNA-binding proteins. Trends in biochemical sciences. 1998;23:273–277. doi: 10.1016/S0968-0004(98)01243-2. [DOI] [PubMed] [Google Scholar]
- Dabrowski S, Olszewski M, Piatek R, Brillowska-Dabrowska A, Konopa G, Kur J. Identification and characterization of single-stranded-DNA-binding proteins from Thermus thermophilus and Thermus aquaticus – new arrangement of binding domains. Microbiology. 2002;148:3307–3315. doi: 10.1099/00221287-148-10-3307. [DOI] [PubMed] [Google Scholar]
- Eggington JM, Haruta N, Wood EA, Cox MM. The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC microbiology. 2004;4:2. doi: 10.1186/1471-2180-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annual review of biochemistry. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- Curth U, Genschel J, Urbanke C, Greipel J. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–2711. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel J, Curth U, Urbanke C. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biological chemistry. 2000;381:183–192. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. The Journal of biological chemistry. 1994;269:30005–30013. [PubMed] [Google Scholar]
- Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa P, Acharya N, Varshney U. Chimeras between single-stranded DNA-binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C-terminal domains interact with uracil DNA glycosylases. The Journal of biological chemistry. 2001;276:16992–16997. doi: 10.1074/jbc.M100393200. [DOI] [PubMed] [Google Scholar]
- Glover BP, McHenry CS. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. The Journal of biological chemistry. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. The EMBO journal. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Urbanke C, Curth U. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamis MI, Casas-Finet JR, Maki AH, Murphy JB, Chase JW. Investigation of the role of individual tryptophan residues in the binding of Escherichia coli single-stranded DNA binding protein to single-stranded polynucleotides. A study by optical detection of magnetic resonance and site-selected mutagenesis. The Journal of biological chemistry. 1987;262:10938–10945. [PubMed] [Google Scholar]
- Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curth U, Bayer I, Greipel J, Mayer F, Urbanke C, Maass G. Amino acid 55 plays a central role in tetramerization and function of Escherichia coli single-stranded DNA binding protein. European journal of biochemistry/FEBS. 1991;196:87–93. doi: 10.1111/j.1432-1033.1991.tb15789.x. [DOI] [PubMed] [Google Scholar]
- Hedayati MA, Grove DE, Steffen SE, Bryant FR. Expression and purification of the SsbB protein from Streptococcus pneumoniae. Protein expression and purification. 2005;43:133–139. doi: 10.1016/j.pep.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Weiner JH, Bertsch LL, Kornberg A. The deoxyribonucleic acid unwinding protein of Escherichia coli. Properties and functions in replication. The Journal of biological chemistry. 1975;250:1972–1980. [PubMed] [Google Scholar]
- Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nature structural biology. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- Vernal J, Serpa VI, Tavares C, Souza EM, Pedrosa FO, Terenzi H. Expression, purification and biochemical characterization of a single-stranded DNA binding protein from Herbaspirillum seropedicae. Protein expression and purification. 2007;53:195–200. doi: 10.1016/j.pep.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Regula JT, Ueberle B, Boguth G, Gorg A, Schnolzer M, Herrmann R, Frank R. Towards a two-dimensional proteome map of Mycoplasma pneumoniae. Electrophoresis. 2000;21:3765–3780. doi: 10.1002/1522-2683(200011)21:17<3765::AID-ELPS3765>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Glass JI, Lefkowitz EJ, Glass JS, Heiner CR, Chen EY, Cassell GH. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature. 2000;407:757–762. doi: 10.1038/35037619. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]