Abstract
Cleavage of proteins within their membrane-spanning segments is an ancient regulatory mechanism that has evolved to control a myriad of cellular processes in all forms of life. Although three mechanistic families of enzymes have been discovered that catalyze hydrolysis within the water-excluding environment of the membrane, how they achieve this improbable reaction has been both a point of controversy and skepticism. The crystal structures of rhomboid and site-2 protease, two different classes of intramembrane proteases, have been solved recently. Combined with current biochemical analyses, this advance provides an unprecedented view of how nature has solved the problem of facilitating hydrolysis within membranes in two independent instances. We focus on detailing the similarities between these unrelated enzymes to define core biochemical principles that govern this conserved regulatory mechanism.
Keywords: membrane, protease, Alzheimer’s disease, cell signalling, malaria
INTRODUCTION
Studies focused on identifying molecular mechanisms of cellular regulation and disease over the past decade uncovered a new class of enzymes: the intramembrane proteases [1]. While soluble proteases use ambient water molecules to facilitate cleavage of peptide bonds, intramembrane proteases evolved independently and had to solve the problem of catalyzing hydrolysis within the water-excluding environment of the membrane. Discovery of intramembrane protelysis thus defined a new frontier in enzymology, but mechanistic insights into these membrane-embedded enzymes have remained rudimentary until recently.
Three mechanistic classes of intramembrane proteases have been discovered. Site-2 protease (S2P) was discovered to be responsible for releasing transcription factors from the membrane to regulate cholesterol and fatty acid biosynthesis [2]. Sequence and topology analysis of S2P suggested four transmembrane (TM) segments with several membrane-inserted loops, and a transmembrane HExxH metalloprotease motif [3]. This motif had been characterized structurally in soluble metalloproteases and functions by coordinating a zinc ion [4]. Accordingly, mutagenesis of any of these residues in various S2P family members abolishes activity [2,5].
Efforts to understand the molecular events leading to Alzheimer’s disease identified gamma-secretase, an aspartyl protease whose catalytic centre is the presenilin protein [6], but requires at least three other proteins that assemble into a large complex [7,8]. Gamma-secretase contains two conserved aspartates essential for activity, and has been termed ‘proteasome of the membrane’ because it cleaves virtually any transmembrane protein of type I orientation that lacks an ectodomain [9]. A simpler version is signal peptide peptidase (SPP), a presenilin-type aspartyl protease that removes remnant signal peptides from membranes; some of the released peptides have regulatory activity [10].
The serine intramembrane protease rhomboid functions to release epidermal growth factor domains from their transmembrane anchors to initiate signalling during Drosophila development [11]. Mutagenesis of conserved residues shared by rhomboid family members suggested a mechanism: proteolysis was found to depend on residues known to form a serine protease active site [12]. Three of these were transmembrane: the catalytic serine in TM4, histidine in TM6, and a glycine preceding the catalytic serine (GxS), which in some serine proteases functions in oxyanion stabilization [13]. The function of the fourth essential residue, an arginine within the conserved WR motif in the long extracellular loop connecting TM1 and 2, remained ambiguous. Rhomboid activity is sensitive to only serine protease inhibitors, further strengthening the intramembrane serine protease hypothesis [12,14].
Although these are the major roles that led to the discovery of each intramembrane protease family, parallel work in other fields broadened the role of these unusual enzymes to dozens of processes, including eukaryotic cell signalling [15–17], apoptosis [18], and stress responses [19,20], bacterial quorum sensing [21], mating [22], sporulation [5] and stress responses [23], and yeast mitochondrial dynamics [24]. Recent work has also placed intramembrane proteolysis at the core of a diverse array of pathogenic processes, including rhomboid in malaria parasite invasion of host cells [25,26], S2P in tuberculosis [27]. and Cryptococcus virulence [28], and SPP in viral assembly [29,30]. These discoveries now add a potential therapeutic application to understanding the detailed biochemistry of these enzymes.
The central question of how intramembrane proteases catalyze hydrolysis within cellular membranes has lagged behind cell biological advances. But in the last few years the tide has turned: defined reconstitution systems have been developed for the study of these enzymes [14,31–34], and more recently crystal structures have been solved for both rhomboid and site-2 proteases [35–40].
WHAT THE BIOCHEMISTRY TOLD US
The main objection to the intramembrane proteolysis hypothesis was the lack of biochemical support. The first cell-free assay was developed for gamma-secretase [41], but gamma-secretase functions as a complicated multi-subunit enzyme complex [7,8]. What was needed to test whether these membrane proteins are themselves proteases was to reconstitute intramembrane proteolysis with a single catalytic polypeptide. Such assays were soon developed for both site-2 protease and rhomboid, which had bacterial homologs to serve as model enzymes, unlike gamma-secretase.
These advances demonstrated that S2P and rhomboid polypeptides could catalyze intramembrane proteolysis in isolation, without the requirement for other protein cofactors [14,31–33]. The requirement for zinc was also finally demonstrated for S2P activity [33]. Moreover, these studies have also started to uncover unique properties of these proteases, including modulation of activity by different membrane lipids [14,34], although the mechanistic basis of which are not yet clear. Further enzymatic and chemical probe analyses using these defined pure-enzyme systems promise to reveal a detailed understanding of the catalytic function of these unusual enzymes.
Throughout these studies, the catalytic residues and thus reaction chemistry at the core of these presumed membrane-embedded enzymes strikingly conformed to what was known for soluble proteases [4,13]. However, a structural approach was required to begin addressing how hydrolysis is catalyzed within membranes. An added implication of the reconstitution systems was that they showed recombinant intramembrane proteases are sufficiently robust for structural analysis.
THE FLOODGATES OPEN
Although it was the last mechanistic class to be discovered, rhomboid provided the first structural glimpse of an intramembrane protease, and did so in a dramatic fashion. Four groups solved the structure of two bacterial rhomboid homologs, GlpG from Escherichia coli and Haemophilus influenzae, as 6 TM core domains in the detergent-solubilized state [35–38]. Although they were solved in four different space groups (R32, P31, P21, C2), the structures overlay with remarkable similarity (the greatest difference is an RMSD of 1.24Å over 164 Cα atoms with GlpGs from the two bacterial species), giving a high degree of confidence that the overall structure is not compromised to a large extent by environmental or crystal effects. The structure of a core domain from an Archaeal S2P, the N-terminal two thirds of the Methanocaldococcus jannaschii S2P enzyme (MjS2P) was solved a year later [39]. These structures of proteolytically active core domains now provide an unprecedented opportunity to compare the features of two unrelated intramembrane proteases.
INTRAMEMBRANE PROTEASE ARCHITECTURE
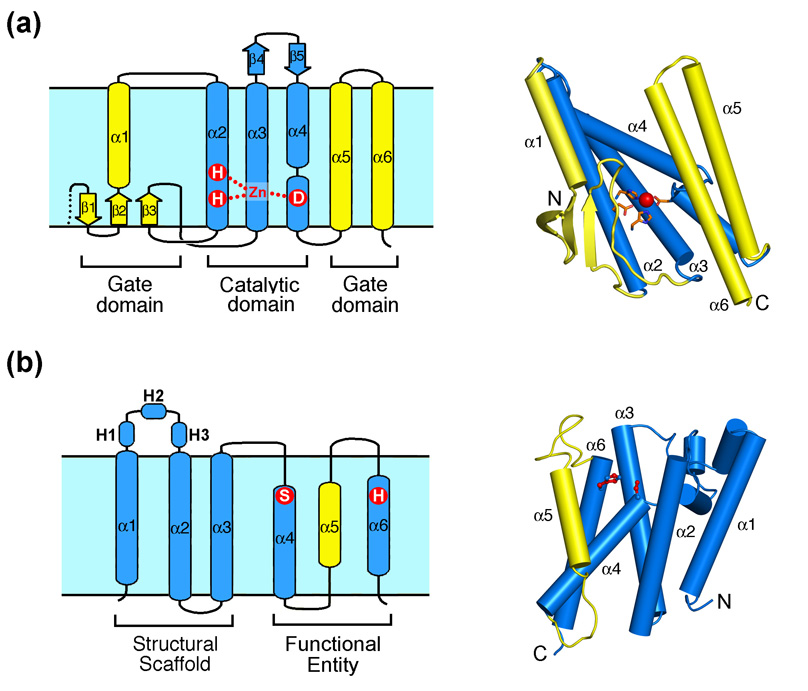
One dramatic feature of the rhomboid and S2P structures is that, apart from having 6 TM helices, they share no common structural features whatsoever. S2P family members share a central core of three TMs, with great variety of inserted TM and nonTM sequences before, after, and/or between these three TMs [42]. The structure of MjS2P shows that the three-TM core provides the structural scaffold for the enzyme, as well as the active site (Figure 1a). MjS2P TM2 contains the HExxH motif, while the TM4 helix is interrupted in its middle by a loop that contributes the Asp residue for Zn coordination [39]. TM3 makes a series of close contacts with both TM 2 and 4 to provide structural support. The preceding TM1, and following TM 5 and 6 of MjS2P are variable among S2P members, as are the several short, membrane-inserted beta-strands in MjS2P.
Figure 1. Topology and structure of intramembrane proteases.
(a) Topology and overall structure of mjS2P (PDB code 3B4R), a zinc metalloprotease from M. jannaschii. The zinc-binding residues are indicated in the topology diagram (left panel) and the structure (right panel). The conserved three-TM core is in blue, and the structural elements that may constitute a lateral gate are highlighted in yellow. In all diagrams, extracellular is upwards and cytosol is downwards. (b) Topology and overall structure of GlpG (PDB code 2NRF), a serine protease from E. coli. The approximate positions of the active site residues Ser201 and His254 are indicated in the topology diagram (left panel). These two residues are colored red in the structure (right panel). The putative lateral gate TM5 is highlighted in yellow.
All rhomboid proteases contain a core region of 6 TM segments and a long loop between TM 1 and 2 [12,43,44]. This basic structure is elaborated by the addition of cytosolic domains (and a TM for mitochondrial enzymes) preceding the 6TM core, and/or a seventh TM and a C terminal extension following the 6TM core. The structures of GlpG contained the 6TM core only: the 86 residue N terminal domain of E. coli GlpG was experimentally removed, while H. influenzae GlpG naturally lacks an N terminal domain. The structures revealed a ring of TM segments, with the surprise that the long L1 loop is partially submerged as a sideways hairpin between TMs 1 and 3, with the WR motif stabilizing the hairpin (Figure 1b) [35–38]. The longer helices of TM 1–3 and the L1 loop likely provide the structural scaffold of the enzyme. But unlike MjS2P, the functional regions are contributed by shorter helices on the other side of the enzyme. TM4 helix enters the core of the enzymes at an oblique angle, bringing the catalytic serine into the centre, but the helix stops abruptly and continues in an extended conformation above the serine. TM6 provides the catalytic histidine and the GxxxG transmembrane dimerization motif that solidifies the TM4:6 interaction. TM5 bears no sequence conservation, and exhibits the greatest flexibility of all TM segments (discussed later).
THE PROOF: …INTRAMEMBRANE…AND PROTEASE
The first dramatic similarity between the structures is the location of the catalytic residues: in both cases the active site residues are indeed submerged beneath the surface of the membrane (Fig 2), proving that proteolysis occurs within the plane of the membrane [35–39]. Moreover, the position reflects the site of substrate cleavage and the direction of their release. For rhomboid, the catalytic serine is recessed 10Å into the outer leaflet of the membrane (Figure 2a). This is the region where rhomboid substrates are known to be cleaved, and in the majority of cases the function is to release domains to the outside of the cell [11]. For MjS2P, the zinc ion, coordinated by the histidines in the HExxH motif on TM2 and an Asp residue on TM4, is sunk ~14Å into the membrane from the cytosolic face (Figure 2b) [39]. Again, S2P substrates are cleaved within the first few residues of their transmembrane segment, and released into the cytosol [1].
Figure 2. Active sites of intramembrane proteases.
(a) Active site in GlpG (PDB code 2NRF). The left panel shows that the active site residues Ser201 and His254 are located below the presumed lipid membrane surface. The right panel compares the active site of GlpG (top) to that of trypsin (bottom). GlpG has a catalytic dyad, compared to a catalytic triad in trypsin. Asn154 of GlpG is invariant in rhomboid proteases but cannot serve as the third catalytic residue. (b) Active site in mjS2P (PDB code 3B4R). The left panel shows that the catalytic zinc ion is located below the presumed lipid membrane surface. The right panel compares the active site of mjS2P (grey) to that of thermolysin (yellow).
Secondly, the structures provide the first picture of the catalytic centres of both enzymes, and show that they are indeed proteases. The catalytic serine in TM4 of rhomboid proteases is hydrogen bonded to a histidine in TM6 (Figure 2a). This observation indicates that rhomboid enzymes use a catalytic dyad instead of a triad. The issue of triad versus dyad has been a point of debate ever since the original observation that mutation of a transmembrane asparagine, proposed to be the third catalytic residue, abolished the activity of Drosophila rhomboid-1, while the asparagine mutant had mild effects in human RHBDL2 [12]. Similar discrepancies have been observed for bacterial rhomboid enzymes in vivo [31,45,46] and in vitro [32,47]. Therefore, the structures unequivocally show that no other residue is hydrogen-bonded to the histidine, and the candidate asparagine on TM2 lies on the opposite side of the serine such that it could not bond with the histidine. However, the histidine may enjoy some support; a conserved tyrosine one helical turn below the serine was observed to stack onto the imidazole ring [35], and mutation of this tyrosine decreases proteolytic activity [47].
In MjS2P, the coordination of zinc by the two histidines of the conserved HExxH metalloprotease motif in TM2 and an aspartate within TM4 overlayed well with the active site from the soluble metalloprotease thermolysin (Figure 2b) [39]. Zinc coordination is likely to be tetrahedral, with the fourth coordinating ligand coming from a water molecule. The glutamate in HExxH is thus thought to function during catalysis to activate the zinc-bound water, as occurs in other metalloproteases [4].
Apart from the basic chemistry underlying catalysis, many of the other stabilizing interactions remain completely obscure. In both cases, peptide bond attack results in a negatively-charged oxygen in the transition state that should be stabilized. For MjS2P, the oxyanion stabilization may be provided by an invariant asparagine that sits above the zinc-coordinating aspartate, in a similar position as an arginine that serves the same function in thermolysin (Figure 2b) [39]. Other interactions with the peptide chain as it lies across the active site are unknown. Even less is known about secondary contacts in the rhomboid active site, but could involve the conserved HxxxxHxxxN motif on TM2, and/or the backbone of the GxS motif [37,38].
THE OLD PROBLEM SOLVED: WATER DELIVERY
The historical objection to intramembrane proteolysis has been the question of how water enters and/or is held within the membrane long enough for catalysis to occur. The structures showed that membrane-embedded active sites are indeed hydrated, and illustrated the means with which this occurs (Figure 3). In the case of rhomboid, the catalytic serine sits at the bottom of a funnel-shaped cavity that is lined with polar residues and opens to the outside of the cell [35–38,48]. The cavity is large, providing ample opportunity for water entry, and indeed all structures, in both closed and open conformations, contain multiple water molecules. In this case, it seems that blocking water entry is more difficult that facilitating it.
Figure 3.
Water delivery in intramembrane proteases. The left panel shows the V-shaped hydrophilic cavity in GlpG (PDB code 2NRF) that converges on the active site (red). The two water molecules are represented by yellow spheres. The right panel shows a narrow channel in mjS2P (PDB code 3B4R) that is connected to the active site zinc ion. This channel is large enough to allow water passage.
MjS2P may also have a dedicated system for water delivery, but in this case the path may be through a narrow channel rather than a large funnel [39]. The channel runs from the cytosolic side of MjS2P to the zinc ion (Figure 3), and is lined with polar residues including backbone carbonyls and acidic and basic side chains. Although the path constricts are several points, the space is sufficient for water to pass. This hypothesis is also consistent with accessibility studies, which showed that while many residues in the GlpG active site were accessible to modifying agents [49], active site modification could only be achieved with the E. coli S2P when a chaotrophic agent was included [50]. However, other routes of water entry cannot necessarily be excluded, and could come from structural vibration of the enzyme, or if larger conformation changes occurred during catalysis.
A NEW PROBLEM RAISED: SUBSTRATE GATING
The structures therefore reformulated the central problem of intramembrane proteolysis: the issue isn’t how water gets into the membrane-embedded active site, but how water within the hydrated active site, and a hydrophobic substrate within the membrane lipid, meet. Unusual structural features, and tantalizing differences between structures, provided several alternative hypotheses.
The first structure of GlpG revealed a ring of transmembrane segments that delimit the hydrated active site from membrane lipid. However, a gap was observed between TM1 and 3 that was plugged by membrane insertion of the L1 loop (Figure 4a). It was proposed that displacement of the L1 loop, which is satisfyingly conserved in all rhomboid enzymes, served a gating function [35]. In support of this model, overlay of the active sites of Heamophlus GlpG with chymotrypsin revealed that substrate entering from the L1 direction would lie across the active site in the same direction as substrates are known to traverse the active sites of soluble serine proteases [38].
Figure 4. Substrate gating models for intramembrane proteases.
(a) A gating model of GlpG that relies on the L1 loop (PDB code 2IC8). The amphipathic L1 loop (yellow) was thought to serve as the gate that regulates access of a substrate protein to the active site (left panel). The right panel shows the myriad of packing interactions among residues in L1 loop and between residues in L1 loop and α3 helix. The WR motif is labelled. (b) A gating model of GlpG that relies on TM5 and the L5 Cap loop. The left panel shows GlpG structure (PDB code 2NRF) in the open conformation (TM5 and Cap in yellow; active site in red; the rest in blue). The right panel shows interactions between TM5 and TM2. (c) A proposed gating model for S2P. In the left panel shows mjS2P (PDB code 3B4R) in the open conformation (gating elements in yellow; active site in red; the core elements in blue). The right panel shows interactions between α1 and α6.
But other structures of GlpG revealed differences in the position of TM5, which in the first structure formed close contacts with neighbouring helix 2 through a series of interdigitated, large, hydrophobic residues (Figure 4b). In the most open structure, the top of helix 5 was observed to tilt outwards by 35 degrees [36]. In another structure, a lipid was bound between the outwardly tilted TM5 and the neighbouring TM2 [37]. Thus, tilting of TM5 was alternatively proposed to serve a lateral substrate gating function.
While the wealth of structural information has provided a number of alternative models for substrate gating, enzymatic activity analysis served as an initial functional test. Mutational analysis of residue interactions in GlpG revealed that loosening the interdigitating contacts between TM5 and TM2, or kinking TM5 outwards with a proline substitution, resulted in an increase of enzymatic activity up to 10-fold in vitro depending on the mutation [47]. This observation served both as functional evidence that TM5, rather than the L1 loop, is the lateral substrate gate, and the first identification of a rate-limiting step for intramembrane proteolysis. Interestingly, these gating mutants also dramatically enhance GlpG activity in live E. coli cells, suggesting that gate opening is rate-limiting in natural cell membranes [51].
The current substrate gating model via TM5 tilting is consistent with substrate requirements: rhomboid enzymes rely on helix disrupting residues within the top region of their transmembrane domain, presumably to facilitate unwinding of the substrate helix into the active site through the space provided by tilting of the top of TM5 [25,52,53]. However, definitive proof of this model awaits the substrate:enzyme complex structure.
In the case of S2P, detailed scrutiny of the differences between two MjS2P molecules in the asymmetric unit suggested a gating model that is similar in principle but more complex than for GlpG. The gating again appears to derive from breaking of interdigitating interactions between two TM segments [39]. But in this case, the result pushes TM1 and TM6 10–12Å away from each other, opening as ‘French doors’ to expose the active site (Figure 4c). The result generates a cleft running down the length of the enzyme that could accommodate a TM substrate. Intriguingly, the cleft reveals a new surface that could provide specificity in substrate binding, and a narrowing at its bottom that could force the substrate to unwind near the site of cleavage. Other possibilities for gating exist, and these tantalizing observations provide a specific and detailed framework for future biochemical study; it might be possible to generate activating mutants that open the gate, as proved to be informative for GlpG [47].
In both GlpG and MjS2P, substrate gating appears to be provided by TM segments that do not have conserved sequence elements. While this may seem surprising at first, it’s precisely what should be expected: gating occurs as a result of knob-hole interactions for packing between TM segments, thus congruent surfaces can be reached by a myriad of different residue combinations. Moreover, in this way, the interface can evolve to fine-tune activity and selectivity.
EXTRAMEMBRANE REGIONS MAY PROVIDE PROTECTION
While the structures of both rhomboid and site-2 protease revealed their transmembrane core domains, function of intramembrane proteases may also involve extramembranous regions.
Site-2 proteases usually require a prior site-1 cleavage by a different protease to remove the protein ectodomain and convert proteins into S2P substrates. A PDZ domain inserted between TM1 and 2 of the E. coli S2P RseP functions to limit cleavage of full-length proteins. Elegant work showed that removal of the PDZ domain, which is thought to bind the C-terminus of proteins, obviated the need for site-1 cleavage and resulted in unregulated intramembrane protelysis of full-length proteins [54]. While the MjS2P does not have any domain inserted between its analogous TMs (MjS2P TM3 and 4), a noticeable crater exists in the structure near this region (Figure 5). It’s tempting to speculate that this would be the region where a PDZ domain of bacterial enzymes, and the cysteine-rich region of the human enzyme, would normally sit, above the active site core of the enzyme.
Figure 5.
Roles of extracellular domains. The Cap (magenta) sits above the membrane surface in GlpG (PDB code 2NRF). The PDZ domain of an S2P protease may dock into the open cleft at the extracellular side of mjS2P (PDB code 3B4R).
In GlpG, both the L1 loop and the L5 Cap (that emerges from TM5) cover the active site cavity from the top (Figure 5) [48]. Since rhomboid proteases cleave full-length proteins without requiring a prior activating cleavage, the extracellular regions do not function to limit substrate access normally [52]. In fact, movement of TM5 necessarily results in L5 Cap displacement, thus both need to move to facilitate substrate entry into the active site. However, given the size of the funnel-shaped cavity, the primary function of the Cap may be to limit inappropriate cleavage of soluble proteins (present in the periplasm of bacteria, or lumen of the secretory pathway in eukaryotic cells). S2P does not appear to have a large enough cavity that would require similar protection [39]. Rhomboid proteases also often contain highly-divergent cytosolic N-terminal domains, the function of which remains unknown [12,43,44].
FUTURE DIRECTIONS
The recent flurry of intramembrane protease structures have provided major initial insights, but this information has more set the stage for understanding this process in detail rather than solved its mysteries. Indeed, the available structures provide a key framework with which to study the process in detail, and both further structural and functional studies are required.
To understand the catalytic reaction, structure of the protease:substrate complex is required to address important outstanding questions that are currently under intense debate. Such a structure would pinpoint exactly where and how substrate entry occurs. More interestingly, interactions revealed at the active site should provide information regarding the role of conserved residues in stabilizing the peptide chain as a whole, as well as the transition state intermediate. These structural advances would, in turn, provide new information that can be used to design selective inhibitors, which do not exist for either rhomboid or site-2 proteases.
The available structures represent the first glimpse of an intramembrane protease prototype for two classes, but each family consists of many members with great diversity between them [42–44]. In particular, structures of eukaryotic intramembrane proteases would greatly advance our understanding of their specific functions. Moreover, all of the structures have been analyzed with the widely-held assumption that it is possible to predict the way in which these enzymes are positioned into biological membranes simply by observing the hydrophobic belt that circumnavigates each enzyme. The reality is unlikely to be this simple.
Lastly, although we now have first example structures of intramembrane serine and metalloproteases, no high-resolution information exists for the asparyl intramembrane proteases. Low-resolution EM reconstruction of gamma-secretase [55,56], and elegant biochemical analysis with accessibility and substrate probes [57–59], hint that similar principles as observed with rhomboid and site-2 protease may apply: an internal active site cavity protected laterally from membrane lipid with channels for water delivery and gating have been postulated. The recent biochemical advance showing model SPPs can be expressed in bacteria and purified in active form provides new hope for the high-resolution structure of an aspartyl intramembrane protease [34]. It will be exciting to re-evaluate the currently-defined core principles of intramembrane proteolysis with the third class of intramembrane proteases.
ACKNOWLEDGEMENTS
We sincerely apologize to those authors whose work could not be cited or described due to space limitations. SU is grateful to Chelsea Newhouse for expert administrative assistance. Work in the Urban lab is supported by NIH grant R01AI066025 (to SU), a career award from the Burroughs-Wellcome Fund (to SU), a Packard Foundation Fellowship for Science and Engineering (to SU). Work in the Shi lab is supported by Princeton University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 3.Zelenski NG, Rawson RB, Brown MS, Goldstein JL. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274:21973–21980. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]
- 4.Bode W, Grams F, Reinemer P, Gomis-Ruth FX, Baumann U, McKay DB, Stocker W. The metzincin-superfamily of zinc-peptidases. Adv Exp Med Biol. 1996;389:1–11. doi: 10.1007/978-1-4613-0335-0_1. [DOI] [PubMed] [Google Scholar]
- 5.Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci U S A. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 7.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 8.Sato T, Diehl TS, Narayanan S, Funamoto S, Ihara Y, De Strooper B, Steiner H, Haass C, Wolfe MS. Active gamma-secretase complexes contain only one of each component. J Biol Chem. 2007;282:33985–33993. doi: 10.1074/jbc.M705248200. [DOI] [PubMed] [Google Scholar]
- 9.Kopan R, Ilagan MX. Gamma-secretase: proteasome of the membrane? Nat Rev Mol Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 10.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide paptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 11.Urban S. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 2006;20:3054–3068. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- 12.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 13.Blow DM, Birktoft JJ, Hartley BS. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969;221:337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- 14.Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci U S A. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedmann E, Hauben E, Maylandt K, Schleeger S, Vreugde S, Lichtenthaler SF, Kuhn PH, Stauffer D, Rovelli G, Martoglio B. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFalpha in activated dendritic cells to trigger IL-12 production. Nat Cell Biol. 2006;8:843–848. doi: 10.1038/ncb1440. [DOI] [PubMed] [Google Scholar]
- 16.Fluhrer R, Grammer G, Israel L, Condron MM, Haffner C, Friedmann E, Bohland C, Imhof A, Martoglio B, Teplow DB, et al. A gamma-secretase-like intramembrane cleavage of TNFalpha by the GxGD aspartyl protease SPPL2b. Nat Cell Biol. 2006;8:894–896. doi: 10.1038/ncb1450. [DOI] [PubMed] [Google Scholar]
- 17.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 18.Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D'Adamio L, et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 19.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 20.Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature. 2006;441:894–897. doi: 10.1038/nature04830. [DOI] [PubMed] [Google Scholar]
- 21.Rather PN, Ding X, Baca-DeLancey RR, Siddiqui S. Providencia stuartii genes activated by cell-to-cell signaling and identification of a gene required for production or activity of an extracellular factor. J Bacteriol. 1999;181:7185–7191. doi: 10.1128/jb.181.23.7185-7191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An FY, Sulavik MC, Clewell DB. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J Bacteriol. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- 25.Baker RP, Wijetilaka R, Urban S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog. 2006;2:e113. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Donnell RA, Hackett F, Howell SA, Treeck M, Struck N, Krnajski Z, Withers-Martinez C, Gilberger TW, Blackman MJ. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J Cell Biol. 2006;174:1023–1033. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makinoshima H, Glickman MS. Regulation of Mycobacterium tuberculosis cell envelope composition and virulence by intramembrane proteolysis. Nature. 2005;436:406–409. doi: 10.1038/nature03713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 29.Heimann M, Roman-Sosa G, Martoglio B, Thiel HJ, Rumenapf T. Core protein of pestiviruses is processed at the C terminus by signal peptide peptidase. J Virol. 2006;80:1915–1921. doi: 10.1128/JVI.80.4.1915-1921.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Targett-Adams P, Schaller T, Hope G, Lanford RE, Lemon SM, Martin A, McLauchlan J. Signal peptide peptidase cleavage of GB virus B core protein is required for productive infection in vivo. J Biol Chem. 2006;281:29221–29227. doi: 10.1074/jbc.M605373200. [DOI] [PubMed] [Google Scholar]
- 31.Maegawa S, Ito K, Akiyama Y. Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry. 2005;44:13543–13552. doi: 10.1021/bi051363k. [DOI] [PubMed] [Google Scholar]
- 32.Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. Embo J. 2005;24:464–472. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. Embo J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narayanan S, Sato T, Wolfe MS. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J Biol Chem. 2007;282:20172–20179. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]
- **35. Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. Reports the first crystal structure of any intramembrane protease, the rhomboid homolog GlpG from Escherichia coli. The overall topology of the enzyme, active site geometry, hydrated cavity above the catalytic serine, and position of conserved residues and interactions are detailed in an exciting discussion. Faced with the problem of how substrates gain access to the active site, the authors argued that the partially-membrane submerged L1 loop functions as the substrate gate.□
- **36. Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. A different crystal form of E. coli GlpG confirmed the overall architecture observed in the first structure, but differences between the two suggested an alternative model for substrate entry: gating by tilting of transmembrane segment 5, which lies on the opposite face of □the enzyme compared to the L1 loop.
- **37. Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci U S A. 2007;104:462–466. doi: 10.1073/pnas.0609773104. Structure of E. coli GlpG solved independently, and with a phospholipid bound between TM2 and TM5. The position of the tetrahedral phosphate head group allowed the authors to propose how the oxyanion might be □stabilized in the active site.
- **38. Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc Natl Acad Sci U S A. 2007;104:750–754. doi: 10.1073/pnas.0609981104. The crystal structure of GlpG from a different bacterial species, H. influenzae was solved, and provides further support that the overall structures are highly similar. These authors also provide a comparison of the active sites of GlpG versus chymotrypsin with bound substrate, and discuss possible mechanistic implications including oxyanion stabilization.
- **39. Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. Ten years after the discovery of the first intramembrane protease, S2P, the crystal structure of an Archaeal homolog was solved. The active site is confirmed to be that of a metalloprotease with bound zinc, and new models for water delivery and gating are proposed.
- 40.Wang Y, Maegawa S, Akiyama Y, Ha Y. The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J Mol Biol. 2007;374:1104–1113. doi: 10.1016/j.jmb.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li YM, Lai MT, Xu M, Huang Q, DiMuzio-Mower J, Sardana MK, Shi XP, Yin KC, Shafer JA, Gardell SJ. Presenilin 1 is linked with gamma-secretase activity in the detergent solubilized state. Proc Natl Acad Sci U S A. 2000;97:6138–6143. doi: 10.1073/pnas.110126897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urban S, Schlieper D, Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in eukaryotic and prokaryotic Rhomboids. Current Biology. 2002;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- 46.Clemmer KM, Sturgill GM, Veenstra A, Rather PN. Functional characterization of Escherichia coli GlpG and additional rhomboid proteins using an aarA mutant of Providencia stuartii. J Bacteriol. 2006;188:3415–3419. doi: 10.1128/JB.188.9.3415-3419.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **47. Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci U S A. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. Enzymatic analysis of E. coli GlpG with a pure enzyme reconstitution assay and an intramembrane-cleaved substrate was used as a functional test to distinguish between the different substrate gating models. □Mutations that help TM5 open result in highly increased GlpG activity, suggesting that TM5 acts as a rate-limiting substrate gate.
- 48.Wang Y, Ha Y. Open-cap conformation of intramembrane protease GlpG. Proc Natl Acad Sci U S A. 2007;104:2098–2102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maegawa S, Koide K, Ito K, Akiyama Y. The intramembrane active site of GlpG, an E. coli rhomboid protease, is accessible to water and hydrolyses an extramembrane peptide bond of substrates. Mol Microbiol. 2007;64:435–447. doi: 10.1111/j.1365-2958.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- 50.Koide K, Maegawa S, Ito K, Akiyama Y. Environment of the active site region of RseP, an Escherichia coli regulated intramembrane proteolysis protease, assessed by site-directed cysteine alkylation. J Biol Chem. 2007;282:4553–4560. doi: 10.1074/jbc.M607339200. [DOI] [PubMed] [Google Scholar]
- 51.Urban S, Baker RP. In vivo analysis reveals substrate gating mutants of a rhomboid intramembrane protease have increased activity in live cells. Biol Chem. 2008 doi: 10.1515/BC.2008.122. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 53.Akiyama Y, Maegawa S. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol Microbiol. 2007;64:1028–1037. doi: 10.1111/j.1365-2958.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 54.Kanehara K, Ito K, Akiyama Y. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. Embo J. 2003;22:6389–6398. doi: 10.1093/emboj/cdg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *55.Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Electron microscopic structure of purified, active gamma-secretase reveals an aqueous intramembrane chamber and two pores. Proc Natl Acad Sci U S A. 2006;103:6889–6894. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *56. Ogura T, Mio K, Hayashi I, Miyashita H, Fukuda R, Kopan R, Kodama T, Hamakubo T, Iwatsubo T, Tomita T, et al. Three-dimensional structure of the gamma-secretase complex. Biochem Biophys Res Commun. 2006;343:525–534. doi: 10.1016/j.bbrc.2006.02.158. These two papers describe EM particle reconstruction of negatively-stained gamma-secretase purified from mammalian or insect cells, providing the first structural glimpse of this complicated □enzyme. Antibody or lectin binding (to the nicastrin subunit) was used to orient the molecules. Although the structures are low resolution and do not fully agree, both observed a roughly spherical protein, □with an irregular inner, hollow chamber, and two or more pores emanating both upwards and downwards.
- *57.Sato C, Morohashi Y, Tomita T, Iwatsubo T. Structure of the catalytic pore of gamma-secretase probed by the accessibility of substituted cysteines. J Neurosci. 2006;26:12081–12088. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *58. Tolia A, Chavez-Gutierrez L, De Strooper B. Contribution of presenilin transmembrane domains 6 and 7 to a water-containing cavity in the gamma -secretase complex. J Biol Chem. 2006 doi: 10.1074/jbc.M604997200. These elegant papers probe water-exposed regions of gamma-secretase by reacting cysteines introduced at various positions in TM6 and 7 with modifying agents that react only in the presence of water. It was found that residues near the active site can be modified, supporting the notion that the gamma-secretase active site is hydrated. Moreover, cysteine substitutions at the catalytic aspartates could be crosslinked with bifunctional reagents with spacer arms of defined length.
- 59.Kornilova AY, Bihel F, Das C, Wolfe MS. The initial substrate-binding site of gamma-secretase is located on presenilin near the active site. Proc Natl Acad Sci U S A. 2005;102:3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]