Abstract
Melanocortin-4 receptor (MC4R) plays critical roles in regulating food intake and energy balance. Recent genome wide scans found common variants near MC4R were related to obesity and insulin resistance. We examined the associations of the reported variants rs17782313 (T>C) and rs17700633 (G>A) with dietary intakes, weight change and diabetes risk in 5724 women (1533 with type 2 diabetes) from a prospective cohort. Under an additive inheritance model, SNP rs17782313 was significantly associated with high intakes of total energy (P = 0.028), total fat (P = 0.008) and protein (P = 0.003). Adjustment for age, BMI, diabetes status and other covariates did not appreciably change the associations. The SNP was also associated with significantly increasing trend of percentage of energy from total fat (P for trend = 0.037). The associations between SNP rs17782313 and higher BMI (P = 0.002) were independent of dietary intakes. In addition, carriers of allele-C had 0.2 kg/m2 greater 10-year increase in BMI from cohort baseline 1976 to 1986 (P = 0.028) compared with the non-carriers. Moreover, per allele-C of rs17782313 was associated with 14% (2–32%) increased risk of type 2 diabetes, adjusting for BMI and other covariates. SNP rs1770833 was not significantly associated with either dietary intakes or obesity traits. In conclusion, the common SNP rs17782313 near MC4R gene was significantly associated with higher intakes of total energy and dietary fat. In addition, the SNP was related to greater long-term weight change and increased risk of diabetes in women.
INTRODUCTION
Melanocortin-4 receptor (MC4R) gene is expressed in several sites in the brain and has been implicated in mediating most of the effects of melanocortin on food intake and energy expenditure (1). Rare mutations in the MC4R gene were previously associated with binge eating, excessive hunger, hyperphagia and food-seeking behavior (2–4). In two recent genome-wide association studies, the common genetic variants near MC4R gene (rs17782313 and rs17700633) were associated with increased obesity risk and insulin resistance (5,6). No study has examined the associations between the common MC4R variants and dietary intakes. In addition, it is unclear whether dietary factors influence the relation between MC4R variants and obesity. Further, the genetic effect on the longitudinal changes in adiposity has yet to be tested.
In this study, we examined the associations of the reported variants near MC4R gene with dietary intakes of energy and the energy-dense macronutrients, the long-term change in adiposity and diabetes risk among women from a prospective cohort. As evidence has shown that MC4R gene may affect linear growth (7,8), we also tested the interaction between MC4R variants and body shape (height) in relation to adiposity.
RESULTS
The minor allele frequencies of the two MC4R SNPs rs17782313 and rs17700633 were 25 and 30%, respectively, in NHS cohort, comparable to the reported distribution in European Caucasians (6). The correlation (r2) between the two SNPs was 0.14 and D′ was 0.44. Table 1 presents the characteristics of the participants (N = 5724) at 1986, when both BMI and waist circumference were measured. The mean age of the study sample was 54.1 years.
Table 1.
Clinic characteristics of the participants
| Variables | |
|---|---|
| n of participants | 5724 |
| Age (years) | 54.1 ± 6.7 |
| Physical activity, MET hours/week | 15.7 ± 21.8 |
| Alcohol consumption (g/day) | 5.9 ± 10.4 |
| Current smoker (%) | 15.0 |
| Postmenopausal status (%) | 85.9 |
| Total energy (Kcal/day) | 1787 ± 524 |
| Total fat (g/day) | 65.5 ± 23.5 |
| Saturated fat (g/day) | 23.2 ± 9.0 |
| Polyunsaturated fat (g/day) | 12.4 ± 5.1 |
| Carbohydrate (g/day) | 215 ± 73 |
Data were derived from questionnaires of 1986.
We examined the relation between the two SNPs and the intakes of total energy and energy-dense macronutrients including dietary fat, carbohydrate and protein. The dietary data were derived from the 1986 questionnaire. Significant associations were observed between rs17782313 and total energy intake under an additive model (P for trend = 0.028) (Table 2). Among the energy-dense nutrients tested, rs17782313 was significantly associated with higher intakes of total fat (P for trend = 0.008) and protein (P for trend = 0.003), but was not related to carbohydrate intake. Women with CC genotype of rs17782313 had significantly higher intakes of total energy (84 kcal/day), total fat (4.6 g/day) and protein (4.4 g/day) compared with those carrying TT genotype (P-values for comparisons between the two genotypes were 0.009, 0.001 and 0.007, respectively). Among dietary fats, the SNP was associated with high intake of saturated fat (P for trend = 0.007). Adjustment for age, BMI, diabetes status, smoking, alcohol consumption, physical activity and postmenopausal status did not appreciably change the associations. SNP rs17700633 was not significantly associated with dietary intakes. In the sensitivity analyses by restricting to the non-diabetic women, we found similar associations (data not shown).
Table 2.
Association of MC4R variants with intakes of total energy, fat, carbohydrates and protein
| Dietary intakes | rs17782313 |
Beta | SE |
P-value |
|||
|---|---|---|---|---|---|---|---|
| TT (N = 2790) | TC (N = 1849) | CC (N = 284) | CC versus TT | Trend | |||
| Total energy | |||||||
| Absolute intake (kcal/day) | 1777 (1758–1796) | 1787 (1763–1811) | 1861 (1800–1922) | 27.3 | 12.4 | 0.009 | 0.028 |
| Total fat | |||||||
| Absolute intake (g/day) | 64.8 (63.9–65.7) | 65.6 (64.5–66.7) | 69.4 (66.7–72.1) | 1.47 | 0.56 | 0.001 | 0.008 |
| Percentage of energy (%) | 32.5 (32.3–32.7) | 32.9 (32.6–33.2) | 33.2 (32.6–33.8) | 0.28 | 0.13 | 0.14 | 0.037 |
| Carbohydrates | |||||||
| Absolute intake (g/day) | 215 (212.3–217.7) | 215 (211.7–218.3) | 222 (213.6–230.4) | 2.56 | 1.71 | 0.11 | 0.13 |
| Percentage of energy (%) | 48.3 (48.0–48.6) | 48.0 (47.7–48.3) | 47.9 (47.1–48.9) | −0.29 | 0.18 | 0.45 | 0.11 |
| Protein | |||||||
| Absolute intake (g/day) | 82.8 (81.8–83.8) | 84.3 (83.1–85.5) | 87.2 (84.2–90.2) | 1.85 | 0.61 | 0.007 | 0.003 |
| Percentage of energy (%) | 18.9 (18.8–19.0) | 19.1 (18.9–19.3) | 18.9 (18.5–19.3) | 0.11 | 0.08 | 0.84 | 0.14 |
Analyses were adjusted for age, BMI and diabetes status; Beta, beta coefficient; SE, standard error.
To further test whether the polymorphism was related to specific nutrient preference, we analyzed the associations between SNP rs17782313 and the percentage of energy from various nutrients (Table 3). The SNP was significantly associated with increasing trend of percentage of energy from total fat (P for trend = 0.037). The associations between the SNP and the percentage of energy from carbohydrate and protein were not statistically significant.
Table 3.
Association of MC4R variant rs17782313 with height, BMI and waist circumference
| rs17782313 |
Model 1 |
Model 2 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | beta | SE | P-value | beta | SE | P-value | |
| BMI, age 18 (kg/m2, N = 5314) | 21.5 (21.4–21.6) | 21.8 (21.7–21.9) | 21.9 (21.5–22.3) | 0.17 | 0.07 | 0.01 | – | – | – |
| BMI (kg/m2, N = 5138) | 26.2 (26.0–26.4) | 26.7 (26.5–26.9) | 26.6 (26.0–27.2) | 0.34 | 0.12 | 0.002 | 0.39 | 0.11 | 3E–04 |
| Waist (cm, N = 3837) | 81.3 (80.8–81.8) | 82.5 (81.9–83.1) | 81.6 (80.1–83.1) | 0.73 | 0.31 | 0.01 | 0.77 | 0.3 | 0.005 |
| Height (cm, N = 5588) | 163.9 (163.6–164.2) | 163.7 (163.4–164.0) | 163.8 (163.0–164.6) | −0.11 | 0.16 | 0.24 | −0.14 | 0.16 | 0.19 |
Model 1: Analyses were adjusted for age and diabetes status; Model 2: analyses were further adjusted for total energy (in quartiles), dietary fat (in quartiles), smoking status, alcohol consumption, physical activity and postmenopausal status.
In our earlier analysis, SNP rs17782313 has been associated with higher BMI and obesity risk in 2265 women from NHS, which was a subset of the present study sample (6). In this updated analysis of 5724 women, the associations between rs17782313 and the adiposity measures (BMI and waist circumference) remained significant (P = 0.002 and P = 0.01, respectively) (Table 3). Each C-allele was associated with 0.34 kg/m2 higher BMI, 0.73 cm higher waist circumference and 12% (1–26%) increased risk of obesity (BMI ≥ 30 kg/m2). In addition, the SNP was significantly associated with BMI at age 18 (P = 0.01). SNP rs17700633 was not significantly related to these adiposity measures. We did not find a significant association between the two SNPs and height. Adjustment for dietary intakes and other lifestyle covariates did not materially change the associations. Similar associations were observed when the analyses were restricted to the non-diabetic women.
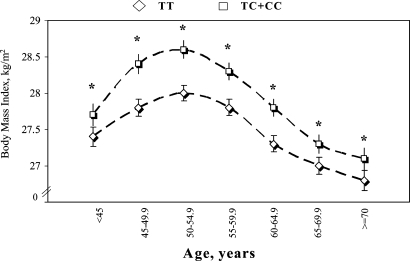
We examined the associations between SNP rs17782313 and BMI in different age groups (<45, 45–49.5, 50–54.9, 55–59.5, 60–64.9, 65–69.9 and ≥70 years). Because the heterozygotes and minor homozygotes were comparably associated with BMI, we pooled these two genotypes together in the stratified analyses. The difference in BMI between the carriers of C-allele and the non-carriers was consistently significant in all age groups (Fig. 1). In addition, SNP rs17782313 was significantly associated with 10-year (1976–1986, corresponding to the population mean age of 44–54 years) change in BMI and body weight, adjusting for baseline age and BMI (Table 4). The increase in BMI was greater in risk-allele carriers compared to the non-risk-allele homozygotes. The SNP was not associated with the change in adiposity at the later stage of follow-up through 2004.
Figure 1.
The geometric means (± standard errors) of BMI (at 1986) by the genotypes of SNP rs17782313 in different age groups among women, <45, 45–49.5, 50–54.9, 55–59.5, 60–64.9, 65–69.9 and ≥70 years old. The analyses were adjusted for diabetes status. *P < 0.05.
Table 4.
Change in BMI according to the presence of C-allele of SNP rs17782313
| Period of change | Mean age (years) | Adiposity measures | Without C-allele | With C-allele | P-value |
|---|---|---|---|---|---|
| 1976–1986 | 44–54 | BMI (kg/m2) | 1.62 (1.52, 1.72) | 1.79 (1.67, 1.91) | 0.028 |
| Weight (kg) | 4.35 (4.08–4.62) | 4.84 (4.54–5.14) | 0.018 | ||
| 1986–1996 | 54–64 | BMI (kg/m2) | 0.97 (0.87, 1.07) | 0.97 (0.85, 1.09) | 0.99 |
| Weight (kg) | 2.62 (2.35–2.88) | 2.63 (2.32–2.94) | 0.95 | ||
| 1996–2004 | 64–72 | BMI (kg/m2) | −0.28 (−0.38, −0.18) | −0.22 (−0.34, −0.10) | 0.45 |
| Weight (kg) | −0.77 (−1.06, −0.49) | −0.59 (−0.92, −0.26) | 0.42 |
Analyses were adjusted for age and body mass index at the start point of each period examined.
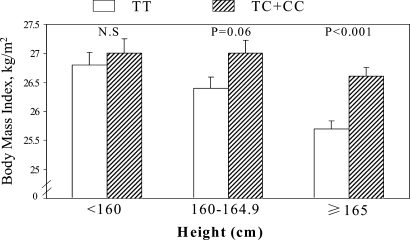
We assessed the modification effects of dietary intakes (total energy and macronutrients) and body size (height) on the associations of MC4R variant. A significant interaction (P = 0.03) was observed between rs17782313 and height in relation to BMI (Fig. 2). The differences in BMI between the carriers of allele-C and the non-carriers were 0.9 (P < 0.001), 0.6 (P = 0.06) and 0.2 kg/m2 (P = 0.66), respectively, in women with the height of greater than 165 cm, 160–164.9 cm, and less than 160 cm. No significant interactions were found between dietary intakes and the genotype in determining BMI.
Figure 2.
The geometric means (standard errors) of BMI (at 1986) of carriers and non-carriers of the risk allele-C of MC4R SNP rs17782313 by height (<160, 160–149 and ≥165 cm). The analyses were adjusted for age and diabetes status. P for interaction between the genotype and height is 0.04.
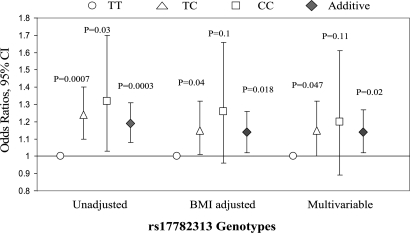
We found that rs17782313 genotypes TC and CC were significantly associated with increased risk of type 2 diabetes (OR = 1.24, 95% CI 1.10–1.40; and 1.32, 95% CI 1.03–1.70, respectively) compared with TT genotype (Fig. 3). Adjustment for BMI, waist circumference and other covariates slightly attenuated the associations (OR = 1.15, 95% CI 1.00–1.32; and 1.20, 95% CI 0.89–1.61, respectively). Under an additive model, per allele-C was significantly associated with 14% (2–27%; P = 0.02) increased diabetes risk, after adjustment for the covariates. We performed a meta-analysis combining our data with data from DIAGRAM consortium including Diabetes Genetics Initiative (DGI), Wellcome Trust Case Control Consortium (WTCCC) and Finland-United States Investigation of NIDDM Genetics (FUSION) studies (9). In total, 6082 diabetes cases and 9770 controls were included. The summary OR (additive model) was 1.07 (95% CI 1.01–1.14; fixed effect; P for heterogeneity test = 0.07).
Figure 3.
Odds ratios (P-values) of type 2 diabetes associated with rs17782313 genotypes. Multivariable analysis adjusted for age, BMI, waist circumference, smoking, alcohol consumption, physical activity and menopausal status. Error bars denote 95 percent confidence intervals.
DISCUSSION
The melanocortin-4 receptor (MC4R) is expressed in brain and is part of the melanocortin pathway controlling food intake and energy homeostasis (3). MC4R−/− mice display hyperphagia and maturity onset obesity characteristics (10). For the first time, we found that the common obesity associated MC4R variant rs17782313 was associated with high intake of total energy, and elevated intakes of dietary fat and protein. These findings are in line with the biological function of MC4R in regulating food intake (3) and the previous observations that rare mutations in MC4R gene were related to binge-eating disorder, hyperphagia and energy intakes in humans (2,4,11). SNP rs17700633, which showed a low correlation with rs17782313 (r2 = 0.14), was not related to dietary intakes. It is likely that the observed genetic effect may be driven by rs17782313 or a causal variant in LD with it.
We observed that the MC4R variant was associated with higher intake of energy and also higher percentage of energy from diet fat. These findings are in concord with the experimental data that MC4-R play a role in controlling fat preference (12). MC4R knockout mice were not hyperphagic when they were fed low-fat diets but became hyperphagia after the introduction of high-fat diets. In addition, it was observed that the treatment by selective MC4-R agonist specifically reduced the intake of a high-fat diet (97% energy was from fat), but did not affect the intakes of a high-protein diet (93% energy was from protein) or a high-carbohydrate diet (100% energy was from carbohydrates). This effect was absent in MC4R−/− mice (12). Likewise, agouti-related protein, an inverse agonist of MC4-R, was found to specifically enhance the intake of high fat diets in rats (13).
A wealth of evidence has shown that dietary intake is affected by genetic components (14,15). Similarly, genetic variants may also determine other behaviors such as physical activity, alcohol consumption and smoking (16–18). All these behavioral or environmental factors, which are closely related to human health, are modifiable. Therefore, research on identifying genetic determinants for the behavioral factors would have important implications for improving interventions to reduce unhealthy behaviors especially in high-risk populations with a specific genetic background.
We further confirmed significant associations between SNP rs17782313 and higher BMI and waist circumference and greater obesity risk. The effect size was comparable to that from a previous study (6). SNP rs17700633 was not significantly associated with adiposity measures in NHS. The reason for the discrepancy between our study and the previous study is not clear but may be partly due to the difference in the population characteristics, including age, gender composition and environmental exposures.
Moreover, we found that SNP rs17782313 was significantly associated with long-term change in BMI at middle age. Our data indicate that the genetic effect increases with age at least before early middle age (<55 years). In the study of Loos et al. (6), this SNP was associated with a greater difference of BMI in children than in adults, suggesting a decreasing trend of the genetic effect with age. Of note, the genetic associations observed among adults in Loos’ study varied significantly across different populations. We suspect the discrepancy between our observation and Loos’ data may be partly due to population variation in the genetic effect. The lack of association between MC4R SNP and BMI change in the later stage of follow-up may be partly explained by cessation of weight gain in older participants. In addition, environmental influences and co-morbidities accumulate with age and likely exert stronger influences on later BMI.
The relation between MC4R SNP and dietary intakes did not account for the associations with adiposity. It has been documented that MC4Rs on different neurons may influence both food intakes and energy expenditure through distinct pathways (19). We suspect that MC4R genetic variants may affect not only dietary intakes but also energy expenditure. In addition, underreporting of dietary intakes that are commonly observed in obese subjects (20) may affect our ability to accurately adjust for these variables in the model. We found that body size (height) significantly modified the relation between rs17782313 and BMI. The association was stronger in women with greater height (≥165 cm) than those who were shorter. The mechanisms underlying these observations need to be further elucidated.
We found a significant association between rs17782313 and increased diabetes risk. The association was independent of adiposity (BMI and waist circumference). Our results were in line with the observation that MC4R SNP was significantly related to insulin resistance in the study of Chambers et al. (5). SNP rs17782313 was also related to increased diabetes risk in the study of Loos et al. (6), albeit the association became marginally significant after adjusting for BMI. Our data suggest that the association of MC4R variant and diabetes may be more evident in women.
The major strengths of our study include the detailed information on the dietary intakes, the prospective design that allows assessing the genetic effects on the longitudinal changes in adiposity and the large sample size. Several limitations need to be acknowledged. Population stratification, mostly arising from ethic admixture, may cause spurious associations. However, population stratification is less likely to explain the associations observed in the present study. The study population is highly homogeneous including only European whites. Underreporting of dietary intake especially in the obese may cause misclassifications in dietary variables. However, such misclassifications would tend to lead to an underestimate of the true effect. Adiposity measures were self-reported in our cohorts. However, self-reported information has been reliably validated with a high correlation with technician-measured measures (21,22). In addition, previous analyses in our cohorts have demonstrated that self-reported BMI and waist circumference strongly predicted various chronic diseases (23–25). The findings of the present study were restricted to women and need to be replicated in other populations.
In summary, we found a common SNP rs17782313 near MC4R gene was associated with high intakes of total energy especially from dietary fat. We demonstrated that the SNP was significantly related to long-term changes in adiposity at middle age. Our data suggest that the association between MC4R and obesity risk is not entirely explained by the changes in dietary intakes. In addition, we found that SNP rs17782313 was associated with increased diabetes risk in women. Comprehensive re-sequencing, fine-mapping and functional test are warranted to identify the causal variant(s) for the observed associations and elucidate the underlying molecular changes.
MATERIALS AND METHODS
Study population
The Nurses’ Health Study was established in 1976 when 121 700 female registered nurses aged 30–55 years and residing in 11 large US states completed a mailed questionnaire on their medical history and lifestyle (26). The lifestyle factors, including smoking, menopausal status and postmenopausal hormone therapy, and body weight, have been updated by validated questionnaires every 2 years. A total of 32 826 women provided blood samples between 1989 and 1990. Information about health and disease is assessed biennially. Samples for the present study were selected from a subcohort of 32 826 women who provided a blood sample between 1989 and 1990 (27,28). We included in total 5724 women. Among them, 2265 non-diabetic women were included in the paper by Loos et al. (6). In the present analysis (primarily on the associations with dietary intakes, BMI changes and diabetes risk), we additionally included 3459 women, 1533 of those were diabetic patients. Diabetes cases were defined as self-reported diabetes confirmed by a validated supplementary questionnaire. For cases before 1998, diagnosis was made using criteria consistent with those proposed by the National Diabetes Data Group (NDDG) (29). We used the American Diabetes Association diagnostic criteria for diagnosis of diabetes cases after 1998 (30). All participants were Caucasians of European ancestry.
Assessment of adiposity
At baseline, participants were asked to report their height and current body weight; weight was then updated during the biennial follow-up. To assess the adiposity in early adulthood, the 1980 NHS questionnaire asked about weight at 18 years of age. We calculated BMI as weight in kilograms divided by height squared in meters. In 1986–1987, participants reported direct measurements of their waists (at the umbilicus) and hips (at the largest circumference) to the nearest quarter of an inch, using a paper tape and detailed measuring directions. The validity of self reported adiposity measures were assessed in a random sample living in the greater Boston area, with high correlation with measured weight (r ≥ 0.96) and waist (r = 0.95) (21,22). We defined obesity as BMI ≥ 30 kg/m2.
Assessment of dietary intakes
Detailed dietary information was obtained through the use of semi-quantitative food-frequency questionnaires (FFQ), which included 116 food items with specified serving sizes that were described by using natural portions or standard weight and volume measures of the servings commonly consumed in this study population. Participants were asked to report their average frequency of consumption of selected foods and beverages with a specified commonly used unit or portion size during the previous year. The reproducibility and validity of the dietary questionnaires were previously evaluated by comparing with the multiple 1-week dietary records. High correlation coefficients were observed for dietary fats and carbohydrate-rich food items and foods (31,32). When corrected for week-to-week variations in diet records that were used to assess validity, the correlations between the FFQ and two 1-week diet records were 0.67, 0.65, 0.67, 0.70 and 0.56 for total energy, carbohydrate, total fat, saturated fat and protein, respectively (32). We used the dietary intakes from 1986 questionnaire, when both BMI and waist circumference were measured, in the primary analysis. The SNPs showed similar associations with dietary intakes from questionnaires of other years.
Genotype determination
DNA was extracted from the buffy coat fraction of centrifuged blood using the QIAmp Blood Kit (Qiagen, Chatsworth, CA, USA). Two reported SNPs near MC4R gene, rs17782313 and rs17700633 (6), were genotyped using the OpenArray™ SNP Genotyping System (BioTrove, Woburn, MA, USA). Another reported SNP rs12970134 (5) was in strong LD with rs17782313 (D′ = 0.96 and r2 = 0.81; HapMap CEU) and therefore was not genotyped. Replicate quality control samples (10%) were included and genotyped with >99% concordance. The call rate was higher than 95% and genotype distribution was in Hardy–Weinberg equilibrium (P > 0.05).
Statistical analyses
The geometric means of dietary factors were compared among the genotypes using general linear models. In the multivariable analyses, we adjusted for age, BMI, diabetes status, physical activity (quintiles), smoking (never, past and current), alcohol intake [non-drinker and drinker (0.1–4.9, 5–10, or >10 g/day)], and menopausal status [pre- or postmenopausal (never, past or current hormone use)]. When adiposity measures were examined, we also adjusted for intakes of total energy (in quartiles). In the longitudinal analysis, changes in BMI were defined as the difference between BMI assessed at different time points (1976–1986; 1986–1996; and 1996–2004). We adjusted for age and BMI at each start point in general linear model. Interaction was examined by introducing the cross-product term of the variables tested in the regression model. Associations between genotype and dichotomous outcomes (obesity and diabetes risk) were tested using unconditional logistic regression, adjusting for covariates. The SAS statistical package was used for the analyses (SAS, Version 8.2 for UNIX). Statistical significance was set at the 0.05 level. The replication tests for the effects of MC4R variants on adiposity were one-tailed (higher adiposity associated with the risk alleles) and other tests were two-tailed.
We performed a meta-analysis on the genetic association (additive model) with diabetes risk, by combining our data with data from DIAGRAM consortium (available at http://www.well.ox.ac.uk/DIAGRAM/) (9). Chi-square test indicated there was no significant between-study heterogeneity (P = 0.07). We, therefore, reported the summary odds ratio (OR) derived from fixed-effect model, in which the summary OR was obtained by averaging the natural logarithms of the OR from individual studies, weighted by the inverses of their variances. Meta analyses were performed with Stata (version 8.2, Stata Corp.).
FUNDING
Supported by grants (DK58845 and CA87969) from the National Institutes of Health. L.Q.’s research is partly supported by the American Heart Association Scientist Development Award and the Boston Obesity Nutrition Research Center (DK46200); F.B.H.’s research is partly supported by the American Heart Association Established Investigator Award.
ACKNOWLEDGEMENT
We thank Dr Zeggini and DIAGRAM consortium for the kind help in meta-analysis.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Tao Y.X. Molecular mechanisms of the neural melanocortin receptor dysfunction in severe early onset obesity. Mol. Cell. Endocrinol. 2005;239:1–14. doi: 10.1016/j.mce.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Branson R., Potoczna N., Kral J.G., Lentes K.U., Hoehe M.R., Horber F.F. Binge eating as a major phenotype of melanocortin 4 receptor gene mutations. N. Engl. J. Med. 2003;348:1096–1103. doi: 10.1056/NEJMoa021971. [DOI] [PubMed] [Google Scholar]
- 3.Adan R.A., Tiesjema B., Hillebrand J.J., la Fleur S.E., Kas M.J., de Krom M. The MC4 receptor and control of appetite. Br. J. Pharmacol. 2006;149:815–827. doi: 10.1038/sj.bjp.0706929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mergen M., Mergen H., Ozata M., Oner R., Oner C. A novel melanocortin 4 receptor (MC4R) gene mutation associated with morbid obesity. J. Clin. Endocrinol. Metab. 2001;86:3448. doi: 10.1210/jcem.86.7.7809. [DOI] [PubMed] [Google Scholar]
- 5.Chambers J.C., Elliott P., Zabaneh D., Zhang W., Li Y., Froguel P., Balding D., Scott J., Kooner J.S. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 6.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santoro N., Rankinen T., Perusse L., Loos R.J., Bouchard C. MC4R marker associated with stature in children and young adults: a longitudinal study. J. Pediatr. Endocrinol. Metab. 2005;18:859–863. doi: 10.1515/jpem.2005.18.9.859. [DOI] [PubMed] [Google Scholar]
- 8.Huszar D., Lynch C.A., Fairchild-Huntress V., Dunmore J.H., Fang Q., Berkemeier L.R., Gu W., Kesterson R.A., Boston B.A., Cone R.D., et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 9.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen A.S., Metzger J.M., Trumbauer M.E., Guan X.M., Yu H., Frazier E.G., Marsh D.J., Forrest M.J., Gopal-Truter S., Fisher J., et al. Role of the melanocortin-4 receptor in metabolic rate and food intake in mice. Transgenic Res. 2000;9:145–154. doi: 10.1023/a:1008983615045. [DOI] [PubMed] [Google Scholar]
- 11.Heid I.M., Vollmert C., Kronenberg F., Huth C., Ankerst D.P., Luchner A., Hinney A., Bronner G., Wichmann H.E., Illig T., et al. Association of the MC4R V103I polymorphism with the metabolic syndrome: The KORA Study. Obesity (Silver Spring) 2008;16:369–376. doi: 10.1038/oby.2007.21. [DOI] [PubMed] [Google Scholar]
- 12.Samama P., Rumennik L., Grippo J.F. The melanocortin receptor MCR4 controls fat consumption. Regul. Pept. 2003;113:85–88. doi: 10.1016/s0167-0115(02)00299-9. [DOI] [PubMed] [Google Scholar]
- 13.Hagan M.M., Rushing P.A., Benoit S.C., Woods S.C., Seeley R.J. Opioid receptor involvement in the effect of AgRP- (83–132) on food intake and food selection. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001;280:R814–R821. doi: 10.1152/ajpregu.2001.280.3.R814. [DOI] [PubMed] [Google Scholar]
- 14.Cai G., Cole S.A., Bastarrachea R.A., Maccluer J.W., Blangero J., Comuzzie A.G. Quantitative trait locus determining dietary macronutrient intakes is located on human chromosome 2p22. Am. J. Clin. Nutr. 2004;80:1410–1414. doi: 10.1093/ajcn/80.5.1410. [DOI] [PubMed] [Google Scholar]
- 15.Perusse L., Tremblay A., Leblanc C., Cloninger C.R., Reich T., Rice J., Bouchard C. Familial resemblance in energy intake: contribution of genetic and environmental factors. Am. J. Clin. Nutr. 1988;47:629–635. doi: 10.1093/ajcn/47.4.629. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell B.D., Rainwater D.L., Hsueh W.C., Kennedy A.J., Stern M.P., Maccluer J.W. Familial aggregation of nutrient intake and physical activity: results from the San Antonio Family Heart Study. Ann. Epidemiol. 2003;13:128–135. doi: 10.1016/s1047-2797(02)00255-7. [DOI] [PubMed] [Google Scholar]
- 17.Thomasson H.R., Edenberg H.J., Crabb D.W., Mai X.L., Jerome R.E., Li T.K., Wang S.P., Lin Y.T., Lu R.B., Yin S.J. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am. J. Hum. Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- 18.Bierut L.J., Cubells J.F., Iacono W.G., Li M.D., Madden P.A., Nelson E.C., Pollock J.D., Rutter J.L., Swan G.E., Vanyukov M. Genetic research and smoking behavior. JAMA. 2007;297:810. doi: 10.1001/jama.297.8.809. [DOI] [PubMed] [Google Scholar]
- 19.Balthasar N., Dalgaard L.T., Lee C.E., Yu J., Funahashi H., Williams T., Ferreira M., Tang V., McGovern R.A., Kenny C.D., et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 20.Lissner L. Measuring food intake in studies of obesity. Public Health Nutr. 2002;5:889–892. doi: 10.1079/phn2002388. [DOI] [PubMed] [Google Scholar]
- 21.Willett W., Stampfer M.J., Bain C., Lipnick R., Speizer F.E., Rosner B., Cramer D., Hennekens C.H. Cigarette smoking, relative weight, and menopause. Am. J. Epidemiol. 1983;117:651–658. doi: 10.1093/oxfordjournals.aje.a113598. [DOI] [PubMed] [Google Scholar]
- 22.Rimm E.B., Stampfer M.J., Colditz G.A., Chute C.G., Litin L.B., Willett W.C. Validity of self-reported waist and hip circumferences in men and women. Epidemiology. 1990;1:466–473. doi: 10.1097/00001648-199011000-00009. [DOI] [PubMed] [Google Scholar]
- 23.Hu F.B., Willett W.C., Li T., Stampfer M.J., Colditz G.A., Manson J.E. Adiposity as compared with physical activity in predicting mortality among women. N. Engl. J. Med. 2004;351:2694–2703. doi: 10.1056/NEJMoa042135. [DOI] [PubMed] [Google Scholar]
- 24.Hu F.B., Manson J.E., Stampfer M.J., Colditz G., Liu S., Solomon C.G., Willett W.C. Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. N. Engl. J. Med. 2001;345:790–797. doi: 10.1056/NEJMoa010492. [DOI] [PubMed] [Google Scholar]
- 25.Manson J.E., Colditz G.A., Stampfer M.J., Willett W.C., Rosner B., Monson R.R., Speizer F.E., Hennekens C.H. A prospective study of obesity and risk of coronary heart disease in women. N. Engl. J. Med. 1990;322:882–889. doi: 10.1056/NEJM199003293221303. [DOI] [PubMed] [Google Scholar]
- 26.Colditz G.A., Manson J.E., Hankinson S.E. The Nurses’ Health Study: 20-year contribution to the understanding of health among women. J. Womens Health. 1997;6:49–62. doi: 10.1089/jwh.1997.6.49. [DOI] [PubMed] [Google Scholar]
- 27.Qi L., Meigs J., Manson J.E., Ma J., Hunter D., Rifai N., Hu F.B. HFE genetic variability, body iron stores, and the risk of type 2 diabetes in U.S. women. Diabetes. 2005;54:3567–3572. doi: 10.2337/diabetes.54.12.3567. [DOI] [PubMed] [Google Scholar]
- 28.Qi L., van Dam R.M., Meigs J.B., Manson J.E., Hunter D., Hu F.B. Genetic variation in IL6 gene and type 2 diabetes: tagging-SNP haplotype analysis in large-scale case–control study and meta-analysis. Hum. Mol. Genet. 2006;15:1914–1920. doi: 10.1093/hmg/ddl113. [DOI] [PubMed] [Google Scholar]
- 29.Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. National Diabetes Data Group. Diabetes. 1979;28:1039–1057. doi: 10.2337/diab.28.12.1039. [DOI] [PubMed] [Google Scholar]
- 30.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 31.Hu F.B., Stampfer M.J., Manson J.E., Rimm E., Colditz G.A., Rosner B.A., Hennekens C.H., Willett W.C. Dietary fat intake and the risk of coronary heart disease in women. N. Engl. J. Med. 1997;337:1491–1499. doi: 10.1056/NEJM199711203372102. [DOI] [PubMed] [Google Scholar]
- 32.Willett W. Nutritional Epidemiology. 2nd edn. New York: Oxford University Press; 1998. [Google Scholar]