Abstract
Ambras syndrome (AS) is a rare form of congenital hypertrichosis with excessive hair on the shoulders, face and ears. Cytogenetic studies have previously implicated an association with rearrangements of chromosome 8. Here we define an 11.5 Mb candidate interval for AS on chromosome 8q based on cytogenetic breakpoints in three patients. TRPS1, a gene within this interval, was deleted in a patient with an 8q23 chromosomal rearrangement, while its expression was significantly downregulated in another patient with an inversion breakpoint 7.3 Mb downstream of TRPS1. Here, we describe the first potential long-range position effect on the expression of TRPS1. To gain insight into the mechanisms by which Trps1 affects the hair follicle, we performed a detailed analysis of the hair abnormalities in Koa mice, a mouse model of hypertrichosis. We found that the proximal breakpoint of the Koa inversion is located 791 kb upstream of Trps1. Quantitative real-time polymerase chain reaction, in situ hybridization and immunofluorescence analysis revealed that Trps1 expression levels are reduced in Koa mutant mice at the sites of pathology for the phenotype. We determined that the Koa inversion creates a new Sp1 binding site and translocates additional Sp1 binding sites within a highly conserved stretch spanning the proximal breakpoint, providing a potential mechanism for the position effect. Collectively, these results describe a position effect that downregulates TRPS1 expression as the probable cause of hypertrichosis in AS in humans and the Koa phenotype in mice.
INTRODUCTION
Ambras syndrome (AS) (OMIM 145701) is a rare form of congenital hypertrichosis in which the body is covered with long, fine, vellus-type hair. The phenotype is accentuated on the shoulders, ears and face, including the forehead, eyelids, cheeks and nose (1). In addition to hypertrichosis, patients display abnormal facial features such as a triangular, coarse face, a bulbous tip of the nose, a broad intercanthal distance and long palpebral fissures (1). Additional facial abnormalities include small and anteverted nares, thick nasal wings, a long, prominent philtrum with a deep groove, a thin vermilion border, a trapezoid mouth and telecanthus, as well as bone abnormalities, such as multiple exostoses, postaxial rudimentary hexadactyly and a low insertion of the first metacarpal (1–4).
Cytogenetic studies have implicated an association with rearrangements of chromosome 8, with a common breakpoint in 8q22–8q24 in three potential AS patients (1–3,5,6). One patient [referred to as ME-1 in Baumeister et al. (1)] was shown to have a pericentric inversion of chromosome 8, inv (8) (p11.2q22) (1). We previously performed a detailed cytogenetic and molecular analysis of this patient and found that the inversion breakpoint in 8q23 did not result in a gene disruption, but instead suggested that the phenotype may be due to a position effect on a nearby gene (6). A second patient [referred to as SS-1 in Balducci et al. (2)] was reported with a paracentric inversion of chromosome 8, inv (8) (q12q22) (2). We performed additional cytogenetic analysis and demonstrated that the chromosomal rearrangements were more complex than originally reported, and involved an insertion of 8q23–8q24 to a more proximal location on 8q, and a large deletion within 8q23, 46, XX, rea (8) (8pter→ 8q13::8q23.2→8q24.1::8q13→8q23.1::8q24.1→8qter) (5). Finally, a third patient (BN-1) with features similar to AS was reported with a deletion in chromosome 8q24 (3). While the diagnosis of patient SS-1 as an AS patient was earlier questioned in the literature because of phenotypic variation from patient ME-1 (7–9), and patient BN-1 was not originally reported to have AS, we postulate that the similar pattern of hair distribution between the three patients mentioned, their associated facial anomalies and the proximity of their cytogenetic rearrangements provide convincing evidence that these three patients unequivocally have AS. Given the rarity of the disorder, and the fact that all three patients have breakpoints in the 8q22–8q24 region, we hypothesized that a position effect on a gene within this region plays a causative role in AS.
A position effect is defined as an alteration in gene expression caused by a change in the position of a gene relative to its native chromosomal surroundings (10). Gene expression can be affected by a variety of mechanisms, for example, disruption of transcriptional regulation in cis and/or modification of the surrounding chromatin structure (10,11). Position effects have classically been demonstrated in instances where the chromosomal breakpoint occurs at a considerable distance from a disease gene in which intragenic mutations have previously established causality for the disorder (10,11). However, in other instances the position effect may generate a distinct phenotype from the one(s) caused by loss-of-function mutations in the coding region of the same gene (11), as is the case for the role of SHH in preaxial polydactyly (12). These differences in phenotype may be attributed to separation of the gene from a tissue- or temporal-specific modifier of gene expression, such as an enhancer or repressor element, in the case of the position effect (10,11). Indeed, the majority of genes affected by position effects that disrupt transcriptional regulation encode transcription factors that are active in a specific subset of cell types during development that require complex regulation of their expression (10,11).
One intriguing candidate gene that maps to 8q23 and fits the description above is trichorhinophalangeal syndrome 1 (TRPS1), a vertebrate transcription factor that contains nine zinc finger motifs, including a GATA-type zinc finger and two Ikaros-like zinc fingers (13,14). We and others have shown that Trps1 localizes to mesenchymal cells during mouse embryogenesis, where it is expressed in the maxilla, mandible, philtrum, nose, muzzle, precartilaginous condensations on the vertebral axis, phalanges and joints of the limbs, and the developing vibrissae and pelage follicles (14–17). Recently, we demonstrated that Trps1 is also expressed in the nuclei of mesenchyme-derived dermal papilla cells and the highly proliferative epithelial cells of adult mouse, rat and human hair follicles (17).
Heterozygous mutations in TRPS1 in humans result in autosomal dominant inheritance of trichorhinophalangeal syndrome types I and III (TRPS I, OMIM 190350; TRPS III, OMIM 190351) (13,18). Patients have sparse and slow-growing scalp hair and exhibit a number of craniofacial and skeletal abnormalities, including a bulbous nose, a long, flat philtrum, a thin upper vermilion border, protruding ears, cone-shaped epiphyses at the phalanges, hip malformations and short stature (19–21). Similarly, homozygous mutant mice in which the GATA-type zinc finger domain of Trps1 was deleted (Trps1tm1.1Shiv/tm1.1Shiv) completely lack vibrissae follicles and have a number of craniofacial and skeletal defects that mirror the phenotypic characteristics of human TRPS patients (16). Furthermore, Trps1−/− mice were recently generated and reported to have defects in hair follicle development and chondrocyte proliferation. Heterozygous Trps1+/− mice are viable with sparse pelage follicles, whereas homozygous Trps1−/− mice die around birth and have severe hair follicle defects (22). While the affected sites of the Ambras phenotype roughly mimic those of TRPS I, surprisingly, the hair phenotypes are essentially opposite. AS is characterized by hair overgrowth, whereas TRPS I is typified by sparse hair. As previously mentioned, one explanation for such findings (11,12) is a position effect that disrupts regulation of TRPS1 as the molecular basis for the Ambras phenotype.
The koala (Koa) mouse has a hypertrichosis phenotype that shows striking parallels to AS in humans. The koala mutation is a semidominant, radiation-induced chromosomal inversion near the mouse ortholog of Trps1. Adult Koa/+ and Koa/Koa mice have excessive hair growth around the muzzle and on both surfaces of the pinnae (23,24). The Koa inversion was mapped to the distal half of chromosome 15, in a region syntenic to human chromosome 8q (24). More recently, the inversion was shown to span approximately 52 Mb of mouse chromosome 15 (25).
Using comparative genetic approaches and morphological analysis between Koa mice and AS patients, we defined a position effect on TRPS1 associated with hypertrichosis in both mice and humans.
RESULTS
Genetic rearrangements in human chromosome 8q that result in Ambras syndrome
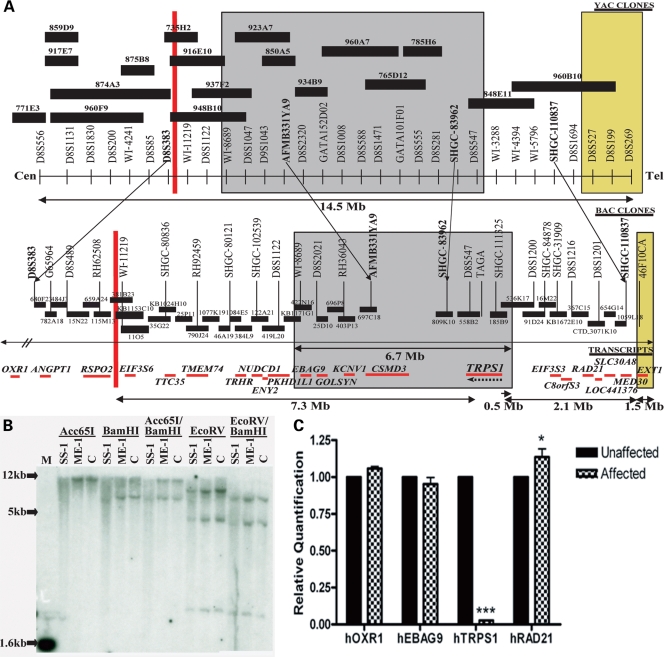
Detailed cytogenetic analysis of patient ME-1 mapped the inversion breakpoint to a region of 8q23 between markers RH62508 and WI-11219 (Fig. 1A). Cloning and sequencing of the breakpoint indicated that it occurs between RSPO2 and EIF3S6 but does not disrupt either gene (6). We also performed loss of heterozygosity (LOH) mapping with markers from 8q22–23 in this patient and found no microdeletions (data not shown).
Figure 1.
Genetic rearrangements in human chromosome 8q that result in Ambras syndrome. (A) Patient ME-1 has a pericentric inversion with a breakpoint in 8q23.1 that lies 7.3 Mb downstream of TRPS1 (red line). Patient SS-1 has a 6.7 Mb deletion in 8q23 that encompasses TRPS1 (gray box). Patient BN-1 has a 1.5 Mb deletion in 8q24, 2.1 Mb upstream of TRPS1 (yellow box). (B) Southern blot analysis using a TRPS1 cDNA probe revealed less intense hybridization signals in patient SS-1 than in a control individual. No differences were detected between hybridization signals in patient ME-1 and a control individual. (C) A bar graph depicting quantitative reverse transcriptase-polymerase chain reaction values for OXR1, EBAG9, TRPS1 and RAD21 in the lymphoblasts of patient ME-1 and an unaffected parent. Data are represented as mean ± SD. Patient ME-1 had a significant reduction in TRPS1 expression.
In order to determine the borders of the deleted interval in the second patient, SS-1, at higher resolution, fluorescence in situ hybridization (FISH) analysis was performed with BAC clones that map to 8q23. Eleven BAC clones from this interval were tested, and four of these were deleted on the rearranged chromosome 8. The borders of the deleted interval were mapped between BAC clones RP11–419L20 and RP11–536K17, corresponding to an interval of approximately 6.7 Mb, and encompassing EBAG9, GOLSYN, KCNV1, CSMD3 and TRPS1 (Fig. 1A).
To more precisely define the borders of the deleted interval in patient SS-1, we performed extensive deletion mapping analysis using polymorphic markers from 8q22–24. Eighty-four markers from chromosome 8 were tested, completely saturating the interval between the genes EIF3S6 and TRPS1. All of the available markers from the Marshfield sex-average based map between 118 and 127 cM (26) were used, in addition to microsatellite markers that were generated on the basis of the known sequence data. In agreement with the cytogenetic data, a deletion was detected within chromosomal band 8q23. The borders of the deleted interval were established between markers D8S1122 and D8S1694, which corresponds to a region of approximately 7.9 Mb. This distance overestimates the size of the deleted interval however, as there are no informative markers between D8S1122, which is still present on the rearranged allele, and D8S1047, which is the first marker with which a deletion can be observed. A deletion was detected for markers D8S1047, D8S2320, CHLC.GATA152D02, D8S588, CHLC.GATA101F01 and TAGA on the paternal allele (Fig. 1A).
In order to determine whether the third patient, BN-1, has any cytogenetic anomalies in the same chromosomal regions as patients ME-1 and SS-1, FISH analysis was performed using BAC clones that were found to either encompass the breakpoint in patient ME-1 (BAC clone KB115353C10) or to be deleted in patient SS-1 (BAC clones RP11–403P13, RP11–697C18, RP11–558B2, RP11–185B9). No abnormalities were detected using these FISH probes.
To ascertain whether the deletion in patient BN-1 overlaps with the deleted interval in patient SS-1, deletion analysis was performed using markers spanning 8q23–8q24. No deletions were detected for markers D8S1047, D8S1043 and CHLC.GATA152D02, all of which were deleted in patient SS-1. Deletions were confirmed for markers 46F10CA, D8S199 and D8S269 (Fig. 1A), in agreement with the results reported by Wuyts et al. (3). In each case, the deletion was detected on the paternally-derived allele.
In summary, the genetic aberrations of the three AS patients map within 11.5 Mb of one another on chromosome 8q, in and around TRPS1. None of the breakpoints in these chromosomal rearrangements disrupt the coding sequence of a gene. Patient ME-1 has a pericentric inversion with a breakpoint in 8q23.1 that lies 7.3 Mb centromeric of TRPS1. Patient SS-1 has a large deletion in 8q23, mapping to a 6.7 Mb interval between markers D8S1122 and D8S1200. TRPS1 falls within this deletion interval, 0.5 Mb from the telomeric border. Finally, patient BN-1 carries a 1.5 Mb deletion in 8q24, the centromeric end of which lies 2.1 Mb upstream of TRPS1 (Fig. 1A). As there is no overlap between the breakpoints in the three patients, the entire 11.5 Mb interval between markers RH62508 (the first marker centromeric to the breakpoint in patient ME-1) and D8S269 (the most distal marker deleted in patient BN-1) was defined as the candidate interval. There are 20 genes present within this region of chromosome 8q, including TRPS1.
Reduced expression of TRPS1 in an Ambras patient with an inversion breakpoint in 8q23
In order to ascertain the genomic status of TRPS1 in AS patients ME-1 and SS-1, we performed Southern blot analysis on genomic DNA obtained from the two patients and control individuals using TRPS1 cDNA as a probe. As expected, hybridization signals for patient ME-1 were roughly equivalent to those of control individuals, indicating that no large-scale genomic anomalies occurred in TRPS1 in this patient. However, Southern blot probe hybridization signals were noticeably less intense than in control individuals for patient SS-1, suggesting a deletion of one copy of TRPS1 (Fig. 1B).
To determine the effect of the inversion in patient ME-1 on TRPS1 transcript expression, RNA was isolated from lymphoblast cell lines derived from the blood of the patient and an unaffected parent. qRT-PCR analysis was performed to examine the expression of TRPS1 and additional transcripts spanning 8q23–q24, including OXR1 (centromeric of the inversion breakpoint in patient ME-1), EBAG9 (within the deletion interval of patient SS-1) and RAD21 (telomeric of the deletion in patient SS-1).
Strikingly, patient ME-1 had a 97.35% reduction (±0.16%; p < 0.0001) in TRPS1 expression when compared with an unaffected parent (Fig. 1C). Importantly, there were no significant changes in the expression of the nearby genes OXR1 and EBAG9 in patient ME-1, and a modest 13.60% increase (±4.57%; p < 0.05) in RAD21 expression (Fig. 1C). No RNA sample was available for patient BN-1, hence qRT-PCR analysis was not possible in this proband. This result demonstrates that an inversion breakpoint 7.3 Mb downstream of TRPS1 significantly reduces the expression of the gene, consistent with a position effect.
Hair abnormalities in Koa mutant mice
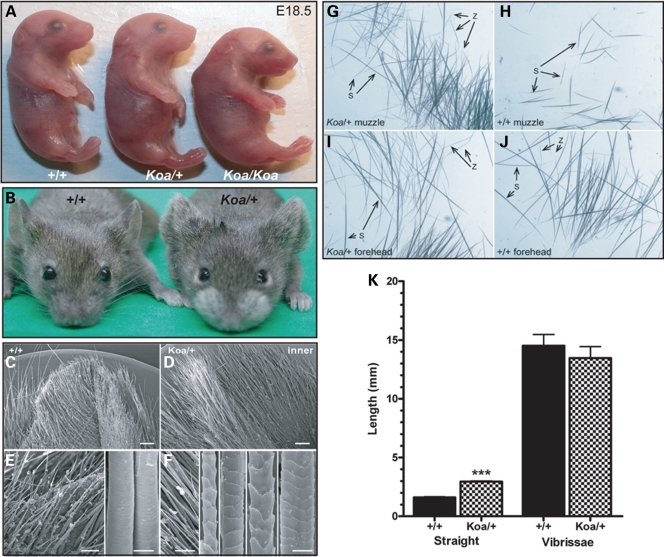
Koa/Koa embryos are smaller than their Koa/+ and wild-type (+/+) littermates, and have a curved back and a flatter, wider head (Fig. 2A). As previously reported, Koa/Koa embryos often have open eyelids at birth (23,24). Adult Koa mice have long hair on both surfaces of the pinnae and around the muzzle (Fig. 2B) (23,24). In wild-type mice, the outer surface of the pinna is covered by a uniform group of short, broad, widely-spaced hair fibers and the epidermal surface is visible, whereas the inner surface of the ear is concealed by a relatively dense population of uniformly-sized, broad, elongated hair fibers, with a visible epidermal surface (Fig. 2C and E). In contrast, in Koa/+, both the outer and inner surfaces of the pinna are obscured by a dense population of long, heterogeneous hair fibers (Fig. 2D). Scanning electron microscopy (SEM) images revealed that the fibers on the inner surface of the Koa/+ pinna resembled those of truncal (pelage) hair types, specifically auchene, awl, guard and zigzag fibers (Fig. 2F).
Figure 2.
Hair abnormalities in Koa mutant mice. (A) Koa/Koa embryos are smaller than their Koa/+ and wild-type littermates at E18.5, with a curved back and a flatter, wider head. (B) Koa/+ mice have long hair on both surfaces of the pinnae of the ears and around the muzzle. (C–F) Scanning electron microscopy revealed that the inner surface of the Koa/+ pinna is obscured by a dense population of long, heterogeneous hair fibers that resemble pelage hair types, specifically auchene, awl, guard and zigzag. (G–J) Whereas only straight hairs were detected in a wild-type muzzle (excluding vibrissae), both straight and zigzag hairs were found in a Koa/+ muzzle. Both straight and zigzag hairs were present in the foreheads of wild-type and Koa/+ mice. (K) A bar graph illustrating that the straight hairs on the Koa/+ muzzle were significantly longer than the straight hairs on the wild-type muzzle, whereas the difference in length of vibrissae hairs was not statistically significant. Data are represented as mean ± SD.
The hair fibers covering the muzzle of Koa/+ mice also included pelage hair types. Whereas only straight hairs were detected in a wild-type muzzle (excluding vibrissae), both straight and zigzag hairs were found in a Koa/+ muzzle (Fig. 2G–J), where the zigzag hairs made up nearly 50% of the muzzle hair fibers. Furthermore, the straight hairs on the Koa/+ muzzle were significantly longer (2.95 ± 0.07 mm) than the straight hairs on the wild-type muzzle (1.61 ± 0.04 mm; p < 0.0001) (Fig. 2K). No differences were observed in SEM images of wild-type versus Koa/+ vibrissae fibers (data not shown), and the difference in length of vibrissae hairs in wild-type (14.51 ± 0.97 mm) and Koa/+ (13.46 ± 0.98 mm) mice was not statistically significant (Fig. 2K). The dorsal skin and pelage follicles of Koa/+ and Koa/Koa mice were also indistinguishable from wild-type (data not shown). The Koa mouse therefore represents a rodent hypertrichosis model in which the ears and muzzle are predominantly affected, identical to the pattern of excessive hair growth in Ambras patients.
Characterization of the Koa inversion breakpoint
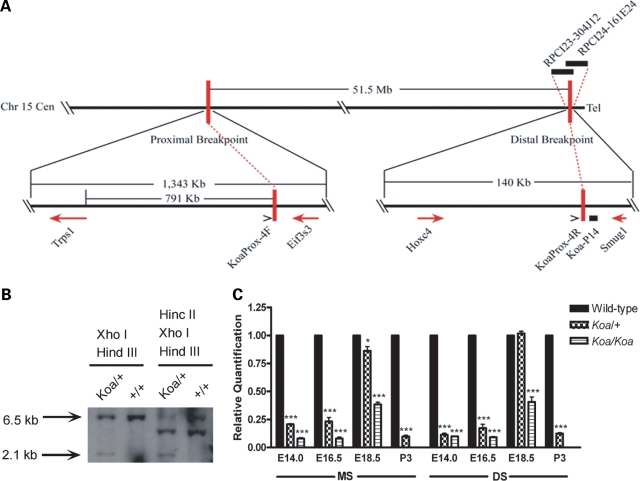
The Koa inversion was previously mapped to the distal half of chromosome 15 (24), in a region syntenic to human chromosome 8q. To determine the precise sequences of the Koa inversion breakpoints, the distal breakpoint was further mapped by FISH analysis on metaphase chromosome spreads from a Koa/+ mouse. The Koa distal inversion breakpoint was localized between two overlapping BAC clones, RPCI23–304J12 and RPCI24–161E24 (Fig. 3A). A series of 0.5–1 kb Southern blot probes of unique sequence were then amplified by PCR from BAC DNA templates and used to analyze a region of approximately 90 kb using DNA from a Koa/+mouse and its C3H/HeH+/+littermate. A 2.1 kb band of altered size was identified using Southern blot probe Koa-P14 (Fig. 3B) and subsequently cloned into a pBlueScript plasmid, allowing for identification of the Koa distal inversion breakpoint. The proximal breakpoint sequence was determined by direct sequencing of a 1.8 kb PCR product amplified using primers flanking the breakpoint. Mapping of both breakpoint sequences to the current mouse genome assembly using the UCSC Genome Browser (http://genome.ucsc.edu/) revealed that neither breakpoint interrupts a gene. The distal breakpoint lies between Hoxc4 and Smug1, while the proximal breakpoint lies between Trps1 and Eif3s3, and is located 791 kb upstream of Trps1 (Fig. 3A). Analogous to what we observed in the Ambras patient with an inversion mutation (patient ME-1), the Koa proximal inversion breakpoint likewise does not disrupt a gene, and lies very near to Trps1.
Figure 3.
Characterization of the Koa inversion mutation. (A) The Koa phenotype is due to a 51.5 Mb inversion on mouse chromosome 15. The proximal Koa inversion breakpoint was mapped between Trps1 and Eif3s3, 791 kb upstream of Trps1. The distal inversion breakpoint falls between Hoxc4 and Smug1. (B) Southern blot analysis using probe Koa-P14 identified a 2.1 kb band of altered size in a Koa/+ mouse that was not present in its +/+ littermate. (C) A bar graph depicting quantitative real time-polymerase chain reaction values for Trps1 expression in the muzzle skin (MS) and dorsal skin (DS) at E14.0, E16.5, E18.5 and P3 in wild-type, Koa/+ and Koa/Koa mice. Data are represented as mean ± SD. Trps1 expression was significantly reduced in both Koa/+ and Koa/Koa MS at all timepoints examined, with lower expression in the homozygous mutants than in heterozygous mice. Trps1 expression was significantly reduced in both Koa/+ and Koa/Koa samples at E14.0, E16.5 and P3 in the DS. Trps1 expression differences were less extreme in the MS and DS at E18.5.
Reduced expression of Trps1 in Koa mutant mice
The Koa proximal breakpoint is within 1 Mb from the Trps1 coding sequence, and as such, it potentially disrupts the regulatory region of this gene. We next compared Trps1 expression between wild-type and Koa mutant mice. qRT-PCR was performed on whole muzzle and dorsal skin from wild-type, Koa/+ and Koa/Koa mice at E14.0, E16.5, E18.5 and postnatal day 3 (P3). Trps1 expression was significantly reduced in Koa muzzle skin at all timepoints examined, with lower expression in the homozygous mutants than in heterozygous mice (Fig. 3C). Strikingly, Trps1 expression was reduced by as much as 91.93% in E14.0 (±0.79%; p < 0.0001) and E16.5 (±1.26%; p < 0.0001) Koa/Koa muzzle skin. Similar trends were observed in the dorsal skin, where expression was significantly reduced in Koa samples at E14.0, E16.5 and P3 (Fig. 3C). Of note, Trps1 expression differences were less extreme in the muzzle and dorsal skin at E18.5, when transcript expression was reduced by 61.47% (±1.98%; p < 0.0001) in Koa/Koa muzzle skin and 59.30% (±4.42%; p = 0.0002) in Koa/Koa dorsal skin (Fig. 3C).
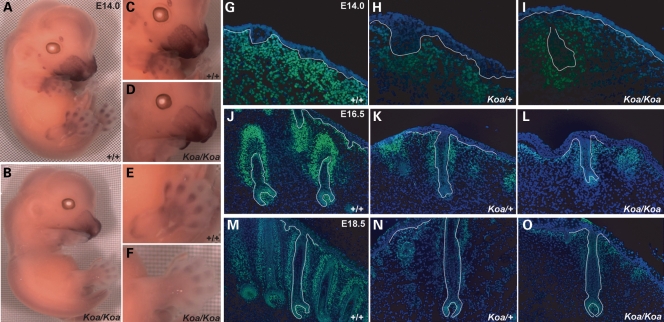
We next assessed differences in Trps1 expression levels between wild-type and Koa/Koa mice during embryogenesis by whole-mount in situ hybridization. At E14.0, Trps1 expression was drastically reduced at all sites of transcript localization in the Koa/Koa mice (Fig. 4A and B), including the maxillary region, the anlagen of the rows of vibrissae follicles, the three tactile follicles located in the sinus and supraorbital regions (Fig. 4C and D), and the prospective phalanges and joints of the limbs (Fig. 4E and F). Of note, wild-type embryos have five rows of developing vibrissae follicles (Fig. 4C), whereas Koa/Koa embryos have only four rows (Fig. 4D).
Figure 4.
Reduced expression of Trps1 in Koa mutant mice. (A, C, E) Trps1 transcripts were detected in the maxilla, mandible, philtrum, nose, muzzle skin, the mesenchyme surrounding the developing vibrissae and tactile follicles, and the prospective phalanges and joints of the limbs at E14.0 in a wild-type mouse by whole mount in situ hybridization. (B, D, F) Trps1 expression was reduced at all sites of transcript localization in a Koa/Koa mutant. (G–I) As detected by immunofluorescence, Trps1 expression is drastically reduced in the nuclei of dermal cells underlying the developing vibrissae follicles of Koa/+ and Koa/Koa mice at E14.0. (J–O) By E16.5 and E18.5, Trps1 expression is notably decreased in the nuclei of mesenchymal cells surrounding the vibrissae follicles, the dermal papillae cells, the germinative matrix and the precortex of Koa/+ and Koa/Koa mice. Trps1, green; DAPI, blue. Dotted line indicates basement membrane.
We then performed immunofluorescence analysis to examine differences in Trps1 protein expression between wild-type, Koa/+ and Koa/Koa mice during embryogenesis. At E14.0, Trps1 expression was considerably reduced from wild-type expression levels (Fig. 4G) in the nuclei of dermal cells underlying the developing vibrissae follicles of Koa mice (Fig. 4H and I). By E16.5 and E18.5, we observed notable decreases from wild-type Trps1 expression levels (Fig. 4J and M) in the nuclei of the mesenchymal cells surrounding the vibrissae follicles, the mesenchyme-derived dermal papillae cells, and the proliferating epithelial cells of the lower follicle, including the germinative matrix and precortex, in Koa mice (Fig. 4K, L, N and O). Interestingly, the vibrissae follicles of Koa/+ (Fig. 4H, K and N) and Koa/Koa (Fig. 4I, L and O) mutant mice were reduced in number and were more widely spaced than in their wild-type counterparts (Fig. 4G, J and M).
In summary, Trps1 expression is markedly reduced in Koa mutant mice at the sites of pathology for the phenotype, as a result of an inversion breakpoint near the gene that recapitulates the level of TRPS1 expression in the Ambras inversion patient.
The Koa inversion alters the pattern of transcription factor binding sites upstream of Trps1
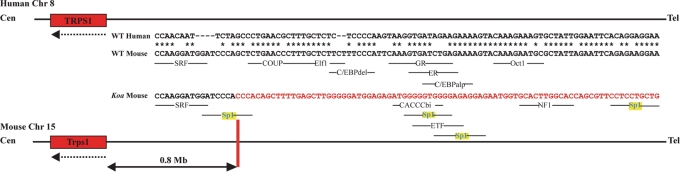
In order to determine the effect of the Koa inversion on the position of transcription factor binding sites in the region of the proximal breakpoint, we compared a 100 bp stretch spanning the breakpoint between wild-type mouse, Koa mouse and human sequences. This region is very well conserved between mouse and human, with 74% of the basepairs identical between the two species (Fig. 5). Using the AliBaba 2.1 prediction program (http://www.gene-regulation.com/pub/programs/alibaba2/index.html), we determined that the wild-type mouse sequence contains eight transcription factor binding sites within this region, including SRF, COUP, Elf1, C/EBPdel, GR, ER, C/EBPalp and Oct1. Interestingly, the inversion creates a new Sp1 site, and moreover, translocates three additional Sp1 binding sites within this 100 bp stretch (Fig. 5), providing a potential mechanism for the observed position effect.
Figure 5.
Altered configuration of transcription factor binding sites due to Koa inversion. A 100 bp stretch spanning the Koa proximal inversion breakpoint is highly conserved between wild-type mouse and human sequences, with 74% of the basepairs identical. The wild-type mouse sequence contains eight transcription factor binding sites within this region, including SRF, COUP, Elf1, C/EBPdel, GR, ER, C/EBPalp and Oct1. The Koa inversion creates a new Sp1 transcription factor binding site and translocates three additional Sp1 binding sites within this 100 bp stretch, in addition to CACCCbi, ETF and NF1 sites. Identical base pairs are indicated by an asterisk. Inverted basepairs in the Koa sequence are in red. Sp1 binding sites are highlighted in yellow.
DISCUSSION
The goal of this study was to define the molecular basis of the AS hypertrichosis phenotype using classical genetics. Our results indicate that underexpression of TRPS1 due to a position effect is likely responsible for the AS phenotype. Intragenic mutations in TRPS1 were previously demonstrated to play a causative role in TRPS I (13). While AS and TRPS type I in humans exhibit opposite hair phenotypes, the affected body sites are essentially the same. Patients affected with both syndromes have hair follicle defects in addition to craniofacial and skeletal abnormalities, including a bulbous tip of the nose, a long philtrum and a thin vermillion border common to both. The nature of these phenotypes suggests that they are likely due to disturbances in the expression of one or more genes active during development. We provide several lines of evidence in humans and mice implicating a position effect on TRPS1 as the cause of hypertrichosis. In support of a position effect, we show that TRPS1 expression is significantly reduced in an Ambras patient with an inversion breakpoint that lies 7.3 Mb from TRPS1. It therefore appears that while loss of TRPS1 function due to intragenic mutations leads to sparse hair, underexpression of the protein owing to a position effect can result in excessive or ectopic hair growth.
Numerous position effect genes have been identified in human diseases with associated breakpoints at a distance of 1 Mb or more from the gene, including FOXC1 in autosomal dominant iridogoniodysgenesis (27), MAF associated with cataract, ocular anterior segment dysgenesis and coloboma (28), SHH in preaxial polydactyly (12) and SOX9 in campomelic dysplasia (29). Furthermore, position effects have been shown to act on either the upstream (5′) or downstream (3′) sides of a gene, and in some instances, on both sides, as is the case for SRY in sex reversal (30,31), SHFM1 in split-hand/split-foot malformation (32), and SOX9 in campomelic dysplasia (29).
Exploiting the comparative genomics approach, we also mapped the Koa inversion breakpoints and found that the proximal breakpoint lies 791 kb upstream of Trps1. The hair abnormalities that were detected in the Koa mutants, in addition to the profound decreases in the expression of both Trps1 transcript and protein levels at sites of wild-type Trps1 expression, indicate that the hypertrichosis phenotype of Koa mice is likely due to a position effect on Trps1, similar to AS in humans.
Interestingly, Koa mice share a number of phenotypic traits with the Trps1tm1.1Shiv mutant mouse model. As reported here, the vibrissae follicles of Koa mice are reduced in number and more widely spaced than in wild-type mice. Trps1tm1.1Shiv/tm1.1Shiv mice were reported to completely lack vibrissae follicles at birth (16). To investigate the point at which this phenotype develops, we discovered that Trps1tm1.1Shiv/tm1.1Shiv mice have reduced and irregularly spaced vibrissae follicles during embryogenesis, which subsequently degenerate by the time of birth (data not shown). Furthermore, the vibrissa phenotype of Koa/+ mice is intermediate between that of wild-type and homozygous mutant Koa mice, typical of a semidominant mutation. While the vibrissa phenotype of Trps1tm1.1Shiv mice only applies to homozygous mutants, the pelage hair defects appear to be dose-dependent. Neonatal Trps1tm1.1Shiv/tm1.1Shiv mice have an approximately 50% reduction in pelage follicle density compared with their wild-type counterparts, whereas heterozygous Trps1tm1.1Shiv/+ mice have an intermediate phenotype. By adulthood, however, this pelage follicle defect is no longer observed in Trps1tm1.1Shiv/+ mice. Trps1tm1.1Shiv/tm1.1Shiv mice die six hours after birth and are therefore not informative for postnatal pelage hair follicle development (16). In addition, both homozygous Koa/Koa and Trps1tm1.1Shiv/tm1.1Shiv mice (16) exhibit spinal curvature resulting in hunched posture. Combined, the overlapping phenotypes of Koa and Trps1 mutant mice indicate that the observed defects are likely due to changes in spatiotemporal regulation of the same gene, Trps1.
Supporting evidence that the Koa phenotype is due to a position effect stemming from the location of the proximal inversion breakpoint near Trps1 comes from the hairy ears (Eh) mutant mouse. Like Koa, the Eh mutation is associated with a paracentric inversion of the distal half of chromosome 15 (33,34). Heterozygous Eh/+ mice have small, hairy pinnae, while homozygous Eh/Eh mice die as neonates (33–36). Eh +/+ Koa compound heterozygous mice display characteristics of both mutations, including the small pinna of Eh mice, the bushy muzzle of Koa mice and the hairy ears common to both mutant phenotypes (33,67). Crossing of the compound heterozygous mice to wild-type mice generated two recombinant chromosome products: one carrying the Eh proximal inversion breakpoint and the Koa distal inversion breakpoint, and the second with the Koa proximal inversion breakpoint and the Eh distal inversion breakpoint. The former product resulted in mice that were indistinguishable from Eh animals, while the latter produced mice that were phenotypically identical to Koa animals, indicating that the Koa proximal inversion breakpoint is responsible for the dominant hair phenotype observed in these mice (33,67).
As is the case for other position effect genes, the phenotype generated by mutations in the coding region of a gene may be distinct from the phenotype caused by disruption of a regulatory element for the same gene. The roles of SHH in holoprosencephaly 3 (HPE3) (OMIM 142945) and preaxial polydactyly II (PPD2) (OMIM 174500) illustrate this point. HPE is characterized by defects in the development of the brain and face. One of the loci for HPE, HPE3, was mapped to 7q36 (37). Point mutations in SHH were shown to cause HPE in cases of autosomal dominant inheritance of the condition (38,39). In addition to the brain and face, the role of SHH in PPD2 demonstrates that the gene is also involved in establishing the anterior–posterior axis of the limb. PPD2 is a congenital limb malformation resulting in a mirror-image duplication of one or more digits. Shh, which is normally expressed in the posterior of the limb bud in the zone of polarizing activity (ZPA), is ectopically expressed in the anterior region of the bud in mouse models of PPD (40). This ectopic expression is caused by translocation breakpoints in humans (41), transgene insertions in mice (41), or point mutations in both species in a SHH regulatory element (the ZPA regulatory sequence, ZPS) that lies in intron 5 of the LMBR1 gene approximately 1 Mb from SHH (12). Similar to the situation for SHH, intragenic mutations in TRPS1 and the position effect described in this study result in different phenotypes, although within the same organs. Unlike SHH, in the case of TRPS1, the sites of pathology in these phenotypes are precisely colocalized, underscoring the importance of TRPS1 in hair development.
While there are a number of common characteristics shared between the three AS patients described in this study, some subtle phenotypic differences exist. The first AS patient discussed, patient ME-1, was originally reported to have facial hypertrichosis in which the forehead, nose, cheeks, eyelids and preauricular region were evenly covered with hair, with additional excessive hair on the external ears (1). The second patient, SS-1, had a slightly different hair distribution, with no hypertrichosis of the nose and only mild hypertrichosis of the ears. Furthermore, the distribution of hair on the patient's face was not uniform (2,7,8). Finally, patient BN-1 was also reported to have facial hypertrichosis, especially pertaining to the eyebrows. However, excessive hair on the nose, cheeks and ears was absent. In addition, this patient possessed multiple exostoses and skeletal defects in both the spine and hands (3), which were not observed in the two aforementioned patients. As phenotypic heterogeneity can result from the exact same mutation in different individuals (42), it is perhaps not surprising that different chromosomal aberrations within the same vicinity could result in slight variations in phenotype. The subtle differences in phenotype between the three AS patients discussed here may result from the disruption of different regulatory elements in each case. Depending on the nature of the regulatory mutation, TRPS1 expression may be affected at different times in various tissues during development, resulting in the observed phenotypic heterogeneity.
Phenotypic differences also exist between AS and TRPS I patients. In addition to disparities in hair density and distribution, TRPS I patients exhibit skeletal abnormalities that are not characteristic of AS, including cone-shaped epiphyses at the phalanges, hip malformations and short stature (19–21). As the position effect described in this study likely disrupts a TRPS1 regulatory element(s), expression of the protein may only be affected in a subset of tissues at very distinct developmental timepoints, providing an explanation for why a bone phenotype like that found in TRPS I is not observed in all of the AS patients. Furthermore, mutations in TRPS1 found in families with TRPS I have previously been shown to encode proteins that dominantly antagonize the wild-type protein (14). In these cases, essentially no functional, wild-type protein remained. As we have demonstrated for AS, however, patient ME-1 still expresses TRPS1 transcripts, albeit at a very low level (2.65 ± 0.16% of wild-type levels).
There are therefore many possible explanations for the differences in hair phenotypes between patients with TRPS I and those with AS. One explanation is that TRPS1 is a transcription factor capable of acting as both a transcriptional repressor and/or activator, depending on the context and amount of protein present. A second possibility is that TRPS1 is capable of binding to numerous cofactors with various functions. Still a third possibility is that TRPS1 transcriptionally regulates both genes that activate and repress hair growth, to varying degrees. In the case of AS, where low levels of TRPS1 transcript are present, these balances of transcriptional regulation may be skewed, resulting in an excessive hair phenotype. Additional studies are necessary to define the precise function of TRPS1 in the context of hair follicle growth.
Similarly, while both Koa and Trps1tm1.1Shiv mice have abnormal muzzle hair (hypertrichosis in the case of Koa and hypotrichosis in the case of Trps1tm1.1Shiv) Koa mice have an additional hypertrichosis phenotype of the ear, while Trps1tm1.1Shiv mice also exhibit hypotrichosis of the pelage hair. Furthermore, Trps1tm1.1Shiv mice have a number of skeletal defects reminiscent of TRPS I in humans that are not found in Koa mice (16). In order to establish whether regulatory elements that control Trps1 transcription are disrupted in Koa mutant mice, we determined the location of transcription factor binding sites in a highly-conserved region spanning the proximal inversion breakpoint in wild-type and Koa mice. Interestingly, the inversion creates a new Sp1 binding site and places three additional Sp1 sites within this 100 bp region. Sp1 is a zinc finger transcription factor that regulates expression by binding to GC-box and GT-box sequences in the promoter region of target genes (43–45). Whereas Sp1 expression is ubiquitous in the mouse embryo, higher expression levels are detected in specific cell types during development, such as developing hematopoietic cells, fetal cells and spermatids (46). While most commonly functioning as a transcriptional activator (47,48), Sp1 has also been shown to act as a transcriptional repressor (49), with the ability to recruit additional factors that fine-tune transcriptional activity (50–52). Interestingly, when numerous Sp1 binding sites are present in the promoter of a gene, Sp1 has the capacity to form multimers. These protein–protein interactions loop the intervening DNA, and have been shown to join Sp1 sites over long distances (53,54). In the context of the Koa inversion, we postulate that the additional Sp1 sites created by the inversion may act to repress Trps1 transcription during development, possibly in conjunction with the multiple Sp1 sites present in the promoter of the gene. The potential regulation of Trps1 by Sp1 provides insight into a possible mechanism of transcriptional control of Trps1 expression during development.
In a broader context, syndromes involving hypertrichosis are believed to represent examples of atavism in humans, defined as the recurrence of an ancestral phenotype (55,56). The amount of hair covering the human body is noticeably reduced when compared with other primates (57). Furthermore, the human primary hair follicle is unique in its ability to generate diverse hair shaft types of different caliber and length during different stages of life, such as scalp hair, eyebrows, eyelashes and secondary sexual hairs (58,59). It may be the case that genes promoting a fuller coat of hair in other mammals—genes that have been silenced during evolution, possibly through repression by TRPS1—have been unmasked in Ambras hypertrichosis. While the molecular basis of hair follicle induction is not fully defined, the work presented here indicates that TRPS1 may play a crucial role in determining the extent of hair distribution and growth, while disruption of its expression can result in a unique excessive hair phenotype across species.
MATERIALS AND METHODS
Human cytogenetic analysis
Transformed lymphoblast cell lines were generated for patients ME-1, SS-1 and their unaffected parents by Epstein-Barr virus transformation using standard procedures (60). FISH analysis was performed as previously described (5).
Human deletion (LOH) analysis
Genomic DNA was isolated from lymphoblastoid cell lines using the QIAGEN Blood and Cell Culture DNA Maxi Kit (Qiagen Inc., Valencia, CA, USA). For PCR-based genotyping with polymorphic microsatellite markers, 100 ng of genomic DNA was amplified using Invitrogen Platinum Taq PCR Supermix (Invitrogen, Carlsbad, CA, USA) in a cocktail containing 10 pmol primers in a 26 µL reaction volume. The following protocol was used for PCR amplification of genomic DNA: step 1: 95°C for 5 min; step 2: 94°C for 30 s; step 3: 55°C for 30 s; step 4: 72°C for 30 s; repeat steps 2–4 for 35 cycles; step 5: 72°C for 10 min. PCR products were resolved on non-denaturing 6% polyacrylamide sequencing gels (0.75 mm thick) and visualized by ethidium bromide staining. The sequences of commercially available microsatellite markers can be found in the Genome Database website (http://www.gdb.org). TAGA is an intragenic marker for TRPS1 (61). 46F10CA is an intragenic marker for EXT (62).
Human southern blot analysis
A TRPS1 cDNA probe was labeled with [α32P]dCTP in a random priming reaction using Stratagene Prime-It II Random Primer Labeling Kit (Stratagene, La Jolla, CA, USA). For Southern analysis, 10 µg of genomic DNA was treated with the appropriate restriction enzyme and resolved on a 0.7% agarose gel. Southern transfer was performed according to standard laboratory procedures (63).
Mouse cytogenetic analysis
Several BAC clones, including RPCI23–304J12 and RPCI24–161E24, were used as FISH probes on metaphase chromosome spreads prepared from Koa/+ mouse spleen cells. FISH analysis was performed as previously described (64).
Mouse Southern blot analysis and inversion breakpoint sequences determination
The Koa-P14 probe was amplified by PCR using BAC clone RPCI23–304J12 as a template. The following primers were used—F: 5′-GATTCAGTGTCTGCAATATCCAAG-3′ R: 5′-CAGTAGTCTCGGTTTTGTCCTTTT-3′.
The probe was labeled with Fl-dUTP in a random priming reaction using Gene Images Random Prime Labeling Kit (Amersham Biosciences, Piscataway, NJ, USA) and detected using the CDP-Star Detection System (Amersham Biosciences). For Southern analysis, genomic DNA was isolated from spleens snap-frozen in liquid nitrogen, digested with the appropriate restriction enzymes and resolved on a 0.8% agarose gel. Southern transfer was performed according to standard laboratory procedures (63).
After HindIII and XhoI digestion, probe Koa-P14 identified a 2.1 kb band of altered size which was cloned into a pBlueScript SK+ plasmid (Stratagene). Both ends of the insert were sequenced using T7 and T3 primers. Genomic sequence alignment indicated that 87 bp from the T3 primer sequence were located in the middle of chromosome 15, confirming that the clone contained the distal breakpoint (GenBank accession no. AY757365, http://www.ncbi.nlm.nih.gov/Genbank/index.html) from the inversion-bearing chromosome. The sequence of the proximal breakpoint (GenBank accession no. AY757366) was determined by PCR amplification of the Koa proximal breakpoint fragment using the following primers—KoaProx-4F: 5′-GAAATGACAGTGAGCTTGCTAAAA-3′; KoaProx-4R: 5′-CCTAAGAATGACCTTGTAGGGACT-3′.
The PCR product was electrophoresed on an agarose gel and the 1.8 kb band was cut and purified using the QIAquick® Gel Extraction Kit (Qiagen). Direct sequencing of the purified PCR product was performed using primers KoaProx-4F and KoaProx-4R.
Quantitative RT-PCR
First-strand cDNA was synthesized from human lymphoblast total RNA, whole dorsal mouse skin total RNA or whole mouse muzzle skin total RNA using Oligo (dT) primer and SuperScript™ III RT (Invitrogen) according to the manufacturer's instructions. Real-time PCR (RT-PCR) was performed on an ABI 7300 machine and analyzed with ABI Relative Quantification Study software (Applied Biosystems, Foster City, CA, USA). Primers were designed according to ABI guidelines and all reactions were performed using Power SYBR® Green PCR Master Mix (Applied Biosystems), 250 nm primers (Invitrogen) and 100 ng cDNA in a 20 µL reaction volume. The following protocol was used—step 1: 50°C for 2 min; step 2: 95°C for 10 min; step 3: 95°C for 15 s; step 4: 60°C for 1 min; repeat steps 3 and 4 for 40 cycles. All samples were run in quadruplicate for three independent runs and normalized against an endogenous internal control, β-2-microglobulin. PCR products were electrophoresed on a 1% agarose/TBE gel containing ethidium bromide and photographed on a Kodak Electrophoresis Documentation and Analysis System 120 Camera (Kodak, Rochester, NY, USA) to confirm amplicon size. The following primers were used—hB2M: F: 5′-CACAGCCCAAGATAGTTAAGTG-3′ R: 5′-GCATAAAGTGTAAGTGTATAAGCATA-3′; hOXR1: F: 5′-TTCTCCCGAGCAGTTGTTAC-3′; R: 5′-CTGTTGGATGGACATCAGGA-3′; hEBAG9: F: 5′-TGGAACAACTGGAACCTGAC-3′; R: 5′-TGCCAGGTATCTAAGTCACC-3′; hTRPS1: F: 5′-TCTACCAGAAGCTTCACTCG-3′; R: 5′-CTCTCTAACGGGCTTCCATT-3′; hRAD21: F: 5′-CCACTGCCTGACTTAGATGA-3′; R: 5′-CGTCATCCTCAAAAGCACTG-3′; mB2 m: F: 5′-ACTGACCGGCCTGTATGCTA-3′; R: 5′-TGAAGGACATATCTGACATCTCTA-3′; mTrps1: F: 5′-GAGATCTCGAGACACTACAG-3′; R: 5′-CTCTTCGCCATTAGCAGTAG-3′.
Scanning electron microscopy
Scanning electron microscopy was performed as described previously (65). Briefly, whole ears of 27-day-old male mice were fixed overnight at 4°C by immersion in 2.5% glutaraldehyde in 0.1 m phosphate buffer (pH 7.2). After multiple rinses in phosphate buffer, samples were post-fixed in 1% osmium tetroxide in phosphate buffer for 48 h at 4°C. Samples were dehydrated in an ethanol series, critical point dried, mounted and sputter-coated with a 4 nm layer of gold. Inner ear samples were analyzed on a Hitachi S3000N VP Scanning Electron Microscope (Hitachi Science Systems, Japan).
Hair plucking and muzzle hair length determination
Hairs were manually plucked from the muzzle or forehead of +/+ and Koa/+ 8-week-old female littermates and photographed using a SPOT digital camera (Diagnostic Instruments, Sterling Heights, MI, USA) fitted onto an SV11 Stemi stereomicroscope (Carl Zeiss, Thornwood, NY, USA). The length of straight (n = 72) and vibrissae hairs (n = 24) from +/+ and Koa/+ muzzles were measured.
Whole mount in situ hybridization
A 700 bp cDNA in situ hybridization probe template in the first exon of Trps1 was amplified by PCR from an mTrps1/pCS vector (generous gift of Dr Ramesh Shivdasani, Dana-Farber Cancer Institute, Harvard Medical School, USA). The following primers were used—F: 5′-GCCAACAGCTACAGAGAGCA-3′ R: 5′-CCTGGTGCGATTATGCAGTC-3′.
The PCR product was cloned into the pCRII dual promoter (T7 and SP6) vector (Invitrogen) and standard procedures were followed for the preparation of DIG-labeled cRNA (Roche Applied Science, Indianapolis, IN, USA) anti-sense (AS) and control sense (S) probes. In situ hybridization was performed on E14.0 embryos as per detailed published protocols (66). The embryos were photographed using an HRC Axiocam fitted onto an SV Stemi stereomicroscope (Carl Zeiss).
Immunofluorescence
A rabbit TRPS1 antibody (generous gift of Dr Ramesh Shivdasani, Dana-Farber Cancer Institute, Harvard Medical School) was used for immunofluorescence analysis. Frozen sections of whole mouse muzzle skin were fixed in 4% paraformaldehyde/0.1% Triton-X for 10 min and washed in phosphate-buffered saline (PBS). The sections were then blocked for 1 h in 10% heat-inactivated goat serum (HIGS)/PBS and incubated overnight at 4°C in primary antibody (TRPS1, 1:5000) diluted in 1% HIGS/PBS. After washing in PBS, the sections were incubated in an Alexa Fluor®488 goat anti-rabbit IgG (Molecular Probes®, Invitrogen) secondary antibody (1:500) diluted in 1% HIGS/PBS. Sections were mounted in VECTASHIELD® mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA) and photographed using an HRC Axiocam fitted onto an Axioplan2 fluorescence microscope (Carl Zeiss).
FUNDING
New York State Foundation for Science, Technology and Innovation (no. CO40072 to A.M.C.); the Kirsch Foundation (no. 2002-0359 to A.M.C.); Office of Biological and Environmental Research, United States Department of Energy (no. DE-AC05-00OR22725 to Y.Y.); North American Hair Research Society Mentorship Award (to Y.Y. and J.S.); National Institutes of Health (1T32HD055165-01 to K.A.F.).
ACKNOWLEDGEMENTS
We are deeply grateful to Dr Ramesh Shivdasani of the Dana-Farber Cancer Institute at Harvard Medical School for the generous gift of the Trps1 antibody and construct. We thank Dr Hermann-Josef Lüdecke for the marker TAGA and for stimulating discussions. We thank Mr Ming Zhang for his technical assistance and Mr Jason Miller for his mouse husbandry work. We thank Mr Donald Carpenter and Ms Patricia Hunsicker for their maintenance of Koa mutant mouse stocks at Oak Ridge National Laboratory. We also thank members of the Christiano Laboratory and Dr Colin Jahoda for their helpful discussions.
Conflict of Interest statement. All authors have no conflict of interest in this work.
REFERENCES
- 1.Baumeister F.A., Egger J., Schildhauer M.T., Stengel-Rutkowski S. Ambras syndrome: delineation of a unique Hypertrichosis Universalis congenital and association with a balanced pericentric inversion (8) (p11.2; q22) Clin. Genet. 1993;44:121–128. doi: 10.1111/j.1399-0004.1993.tb03862.x. [DOI] [PubMed] [Google Scholar]
- 2.Balducci R., Toscano V., Tedeschi B., Mangiantini A., Toscano R., Galasso C., Cianfarani S., Boscherini B. A new case of Ambras syndrome associated with a paracentric inversion (8) (q12; q22) Clin. Genet. 1998;53:466–468. doi: 10.1111/j.1399-0004.1998.tb02596.x. [DOI] [PubMed] [Google Scholar]
- 3.Wuyts W., Roland D., Lüdecke H.J., Wauters J., Foulon M., Van Hul W., Van Maldergem L. Multiple exostoses, mental retardation, hypertrichosis, and brain abnormalities in a boy with a de novo 8q24 submicroscopic interstitial deletion. Am. J. Med. Genet. 2002;113:326–332. doi: 10.1002/ajmg.10845. [DOI] [PubMed] [Google Scholar]
- 4.Belengeanu V., Rozsnyai K., Gug C., Bănăteanu M., Farcas S., Belengeanu A. Ambras syndrome: report on two affected siblings with no prior family history. Clin. Dysmorphol. 2004;13:265–267. [PubMed] [Google Scholar]
- 5.Tadin M., Braverman E., Cianfarani S., Sobrino A.J., Levy B., Christiano A.M., Warburton D. Complex cytogenetic rearrangement of chromosome 8q in a case of Ambras syndrome. Med. Genet. 2001;102:100–104. doi: 10.1002/1096-8628(20010722)102:1<100::aid-ajmg1396>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 6.Tadin-Strapps M., Warburton D., Baumeister F.A.M., Fischer S.G., Yonan J., Gilliam T.C., Christiano A.M. Cloning of the breakpoints of a de novo inversion of chromosome 8, inv (8) (p11.2q23.1) in a patient with Ambras syndrome. Cytogenet. Genome Res. 2004;107:68–76. doi: 10.1159/000079573. [DOI] [PubMed] [Google Scholar]
- 7.Baumeister F.A. Differentiation of Ambras syndrome from Hypertrichosis Universalis. Clin. Genet. 2000;57:157–158. doi: 10.1034/j.1399-0004.2000.570213.x. [DOI] [PubMed] [Google Scholar]
- 8.Cianfarani S. Ambras syndrome or Hypertrichosis Universalis, does it really matter? Clin. Genet. 2000;57:158. doi: 10.1034/j.1399-0004.2000.570213.x. [DOI] [PubMed] [Google Scholar]
- 9.Baumeister F.A. Diagnosis of Ambras syndrome: comments on complex cytogenetic rearrangement of chromosome 8q in a case of Ambras syndrome. Am. J. Med. Genet. 2002;109:77–78. [PubMed] [Google Scholar]
- 10.Kleinjan D.J., van Heyningen V. Position effect in human genetic disease. Hum. Mol. Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 11.Kleinjan D.A., van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 13.Momeni P., Glockner G., Schmidt O., von Holtum D., Albrecht B., Gillessen-Kaesbach G., Hennekam R., Meinecke P., Zabel B., Rosenthal A., et al. Mutations in a new gene, encoding a zinc finger protein, cause tricho-rhino-phalangeal syndrome type I. Nat. Genet. 2000;24:71–74. doi: 10.1038/71717. [DOI] [PubMed] [Google Scholar]
- 14.Malik T.H., Shoichet S.A., Latham P., Kroll T.G., Peters L.L., Shivdasani R.A. Transcriptional repression and developmental functions of the atypical vertebrate GATA protein TRPS1. EMBO J. 2001;20:1715–1725. doi: 10.1093/emboj/20.7.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunath M., Lüdecke H.J., Vortkamp A. Expression of Trps1 during mouse embryonic development. Mech. Dev. 2002;119(Suppl. 1):S117–S120. doi: 10.1016/s0925-4773(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 16.Malik T.H., von Stechow D., Bronson R.T., Shivdasani R.A. Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder of inherited tricho-rhino-phalangeal syndromes. Mol. Cell. Biol. 2002;22:8592–8600. doi: 10.1128/MCB.22.24.8592-8600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fantauzzo K.A., Bazzi H., Jahoda C.A., Christiano A.M. Dynamic expression of the zinc-finger transcription factor Trps1 during hair follicle morphogenesis and cycling. Gene Expr. Patterns. 2008;8:51–57. doi: 10.1016/j.modgep.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Lüdecke H.J., Schaper J., Meinecke P., Momeni P., Groß S., von Holtum D., Hirche H., Abramowicz M.J., Albrecht B., Apacik C., et al. Genotypic and phenotypic spectrum in tricho-rhino-phalangeal syndrome types I and III. Am. J. Med. Genet. 2001;68:81–91. doi: 10.1086/316926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giedion A. Das tricho-rhino-phalangeale syndrome. Helv. Paediatr. Acta. 1966;21:475–485. [PubMed] [Google Scholar]
- 20.Giedion A., Burdea M., Fruchter Z., Meloni T., Trosc V. Autosomal dominant transmission of the tricho-rhino-phalangeal syndrome. Report of 4 unrelated families, review of 60 cases. Helv. Paediatr. Acta. 1973;28:249–259. [PubMed] [Google Scholar]
- 21.Felman A.H., Frias J.L. The trichorhinophalangeal syndrome: study of 16 patients in one family. Am. J. Roentgenol. 1977;129:631–638. doi: 10.2214/ajr.129.4.631. [DOI] [PubMed] [Google Scholar]
- 22.Suemoto H., Muragaki Y., Nishioka K., Sato M., Ooshima A., Itoh S., Hatamura I., Ozaki M., Braun A., Gustafsson E., et al. Trps1 regulates proliferation and apoptosis of chondrocytes through Stat3 signaling. Dev. Biol. 2007;312:572–581. doi: 10.1016/j.ydbio.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Ball S.T., Peters J. Koala, a dominant mutation. Mouse News Lett. 1989;83:163–164. [Google Scholar]
- 24.Peters J., Tease C., Ball S.T. Koala, Koa, is associated with an inversion on mouse chromosome 15. Genet. Res. 1992;59:237–238. [Google Scholar]
- 25.Katayama K., Furuno A., Miyamoto S., Nakamura M., Ojika I., Shinkai Y., Akiyama K., Tsuji T., Kunieda T. Suppressed recombination on mouse chromosome 15 defined regions of chromosomal inversions associated with koala (koa) and hairy ears (eh) mutations. Exp. Anim. 2008;57:73–77. doi: 10.1538/expanim.57.73. [DOI] [PubMed] [Google Scholar]
- 26.Broman K.W., Murray J.C., Sheffield V.C., White R.L., Weber J.L. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies A.F., Mirza G., Flinter F., Raqoussis J. An interstitial deletion of 6p24-p25 proximal to the FKHL7 locus and including AP-2alpha that affects anterior eye chamber development. J. Med. Genet. 1999;36:708–710. [PMC free article] [PubMed] [Google Scholar]
- 28.Jamieson R.V., Perveen R., Kerr B., Carette M., Yardley J., Heon E., Wirth M.G., van Heyningen V., Donnai D., Munier F., et al. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum. Mol. Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- 29.Velagaleti G.V., Bien-Willner G.A., Northup J.K., Lockhart L.H., Hawkins J.C., Jalal S.M., Withers M., Lupski J.R., Stankiewicz P. Position effects due to chromosome breakpoints that map approximately 900 kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am. J. Hum. Genet. 2005;76:652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McElreavy K., Vilain E., Abbas N., Costa J.M., Souleyreau N., Kucheria K., Boucekkine C., Thibaud E., Brauner R., Flamant F., et al. XY sex reversal associated with a deletion 5′ to the SRY ‘HMG box’ in the testis-determining region. Proc. Natl Acad. Sci. USA. 1992;89:11016–11020. doi: 10.1073/pnas.89.22.11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McElreavey K., Vilain E., Barbaux S., Fugua J.S., Fechner P.Y., Souleyreau N., Doco-Fenzy M., Gabriel R., Quereux C., Fellous M., et al. Loss of sequences 3′ to the testis-determining gene, SRY, including the Y pseudoautosomal boundary associated with partial testicular determination. Proc. Natl Acad. Sci. USA. 1996;93:8590–8594. doi: 10.1073/pnas.93.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crackower M.A., Scherer S.W., Rommens J.M., Hui C.C., Poorkaj P., Soder S., Cobben J.M., Hudgins L., Evans J.P., Tsui L.C. Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum. Mol. Genet. 1996;5:571–579. doi: 10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- 33.Katayama K., Furuno A., Akiyama K., Tsuji T., Kunieda T. Characterization of chromosomal inversion of the mouse hairy ears (Eh) mutation associated with cleft palate. Mamm. Genome. 2007;18:246–254. doi: 10.1007/s00335-007-9015-7. [DOI] [PubMed] [Google Scholar]
- 34.Davisson M.T., Roderick T.H., Akeson E.C., Hawes N.L., Sweet H.O. The hairy ears (Eh) mutation is closely associated with a chromosomal rearrangement in mouse chromosome 15. Genet. Res. 1990;56:167–178. doi: 10.1017/s0016672300035266. [DOI] [PubMed] [Google Scholar]
- 35.Bangham J.W. Hairy ears, Eh. Mouse News Lett. 1965;33:68. [Google Scholar]
- 36.Bangham J.W. Hairy ears, Eh. Mouse News Lett. 1968;38:32. [Google Scholar]
- 37.Lurie I.W., Ilyina H.G., Podleschuk L.V., Gorelik L.B., Zaletajev D.V. Chromosome 7 abnormalities in parents of children with holoprosencephaly and hydronephrosis. Am. J. Med. Genet. 1990;35:286–288. doi: 10.1002/ajmg.1320350229. [DOI] [PubMed] [Google Scholar]
- 38.Belloni E., Muenke M., Roessler E., Traverso G., Siegel-Bartelt J., Frumkin A., Mitchell H.F., Donis-Keller H., Helms C., Hing A.V., et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat. Genet. 1996;14:353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 39.Roessler E., Belloni E., Guadenz K., Jay P., Berta P., Scherer S.W., Tsui L.C., Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat. Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- 40.Sharpe J., Lettice L., Hecksher-Sorensen J., Fox M., Hill R., Krumlauf R. Identification of sonic hedgehog as a candidate gene responsible for the polydactylous mouse mutant Sasquatch. Curr. Biol. 1999;9:97–100. doi: 10.1016/s0960-9822(99)80022-0. [DOI] [PubMed] [Google Scholar]
- 41.Lettice L.A., Horikoshi T., Heaney S.J., van Baren M.J., van der Linde H.C., Breedveld G.J., Joosse M., Akarsu N., Oostra B.A., Endo N., et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl Acad. Sci. USA. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf U. Identical mutations and phenotypic variation. Hum. Genet. 1997;100:305–321. doi: 10.1007/s004390050509. [DOI] [PubMed] [Google Scholar]
- 43.Dynan W.S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983;32:669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- 44.Dynan W.S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 45.Kadonaga J.T., Carner K.R., Masiarx F.R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 46.Saffer J.D., Jackson S.P., Annarella M.B. Developmental expression of Sp1 in the mouse. Mol. Cell. Biol. 1991;11:2189–2199. doi: 10.1128/mcb.11.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Courey A.J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 48.Courey A.J., Holtzman D.A., Jackson S.P., Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 49.Shou Y., Baron S., Poncz M. An Sp1-binding silencer element is a critical negative regulator of the megakaryocyte-specific alphaIIb gene. J. Biol. Chem. 1998;273:5716–5726. doi: 10.1074/jbc.273.10.5716. [DOI] [PubMed] [Google Scholar]
- 50.Murata Y., Kim H.G., Rogers K.T., Udvadia A.J., Horowitz J.M. Negative regulation of Sp1 trans-activation is correlated with the binding of cellular proteins to the amino terminus of the Sp1 trans-activation domain. J. Biol. Chem. 1994;269:20674–20681. [PubMed] [Google Scholar]
- 51.Sun J.M., Chen H.Y., Moniwa H., Litchfield D.W., Seto E., Davie J.R. The transcriptional repressor Sp3 is associated with CK2- phosphorylated histone deacetylase 2. J. Biol. Chem. 2002;277:35783–35786. doi: 10.1074/jbc.C200378200. [DOI] [PubMed] [Google Scholar]
- 52.Jang S.I., Steinert P.M. Loricrin expression in cultured human keratinocytes is controlled by a complex interplay between transcription factors of the Sp1, CREB, AP1 and AP2 families. J. Biol. Chem. 2002;277:42268–42279. doi: 10.1074/jbc.M205593200. [DOI] [PubMed] [Google Scholar]
- 53.Su W., Jackson S., Tjian R., Echols H. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 54.Mastrangelo I.A., Courey A.J., Wall J.S., Jackson S.P., Hough P.V. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc. Natl Acad. Sci. USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall B.K. Developmental mechanisms underlying the formation of atavisms. Biol. Rev. 1984;59:89–124. doi: 10.1111/j.1469-185x.1984.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 56.Cantú J.M., Ruiz C. On atavisms and atavistic genes. Ann. Genet. 1985;28:141–142. [PubMed] [Google Scholar]
- 57.Figuera L.E., Pandolfo M., Dunne P.W., Cantú J.M., Patel P.I. Mapping of the congenital generalized hypertrichosis locus to chromosome Xq24-q27.1. Nat. Genet. 1995;10:202–207. doi: 10.1038/ng0695-202. [DOI] [PubMed] [Google Scholar]
- 58.Miner R.W., editor. The Growth, Replacement, and Types of Hair. New York: Annals of New York Academic Science; 1951. [Google Scholar]
- 59.Montagna W., Ellis R.A. The Biology of Hair Growth. New York: Academic Press; 1958. [Google Scholar]
- 60.Speck S.H., Strominger J.L. Epstein-Barr virus transformation. Prog. Nucleic Acid Res. Mol. Biol. 1987;34:189–207. [PubMed] [Google Scholar]
- 61.Riedl S., Giedion A., Schweitzer K., Müllner-Eidenböck A., Grill F., Frisch H., Lüdecke H.J. Pronounced short stature in a girl with tricho-rhino-phalangeal syndrome II (TRPS II, Langer-Giedion syndrome) and growth hormone deficiency. Am. J. Med. Genet. A. 2004;131:200–203. doi: 10.1002/ajmg.a.30374. [DOI] [PubMed] [Google Scholar]
- 62.Bernard M.A., Hall C.E., Hogue D.A., Cole W.G., Scott A., Snuggs M.B., Clines G.A., Lüdecke H.J., Lovett M., Van Winkle W.B., et al. Diminished levels of the putative tumor suppressor proteins EXT1 and EXT2 in exostosis chondrocytes. Cell Motil. Cytoskeleton. 2001;48:149–162. doi: 10.1002/1097-0169(200102)48:2<149::AID-CM1005>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 63.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Analysis of genomic DNA by Southern hybridization; pp. 9.31–9.62. [Google Scholar]
- 64.Chick W.S.H., Mentzer S.E., Carpenter D.A., Rinchik E.M., You Y. Modification of an existing chromosomal inversion to engineer a balancer for mouse chromosome 15. Genetics. 2004;167:889–895. doi: 10.1534/genetics.104.026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mecklenburg L., Paus R., Halata Z., Bechtold L.S., Fleckman P., Sundberg J.P. FOXN1 is critical for onycholemmal terminal differentiation in nude (Foxn1) mice. J. Invest. Dermatol. 2004;123:1001–1011. doi: 10.1111/j.0022-202X.2004.23442.x. [DOI] [PubMed] [Google Scholar]
- 66.Wilkinson D. In situ Hybridization: A Practical Approach. 2nd edn. Oxford: Oxford University Press; 1998. [Google Scholar]
- 67.Mentzer S.E., Sundberg J.P., Awgulewitsch A., Chao H.H.J., Carpenter D.A., Zhang W.-D., Rinchik E.M., You Y. The mouse hairy ears mutation exhibits an extended growth (anagen) phase in hair follicles and altered Hoxc gene expression in the ears. Veterinary Dermatology. 2008 doi: 10.1111/j.1365-3164.2008.00709.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]