Abstract
Purpose:
The epidermal growth factor (EGF) pathway is important in esophageal adenocarcinoma (EAC) tumorigenesis. We hypothesized that the EGF A61G homozygous variant genotype (GG) (i) is both a risk and poor prognostic factor for EAC; and (ii) is associated with higher EGF serum levels in individuals with gastroesophageal reflux disease (GERD).
Experimental Design:
Using unconditional logistic regression, we compared EGF A61G in 312 EAC cases and 447 GERD-free controls, adjusting for age, gender, smoking history and healthy adult body mass index. Using the method of Kaplan&Meier, log rank tests, and Cox proportional hazard models, we correlated EGF A61G with overall and failure-free survival in the EAC cases. Serum EGF levels and EGF genotype (G/G vs. others) were correlated in 144 GERD patients using Wilcoxon rank sum tests.
Results:
The EGF A61G G/G genotype conferred increased EAC risk with an adjusted odds ratio (AOR) of 1.81 (95% CI, 1.2-2.7), and was even higher in the subgroup of EAC patients with concurrent Barrett's esophagus (BE): AOR 2.18 (95%CI 1.3-3.7). However, EGF A61G was not associated with a more aggressive phenotype or prognosis in EAC patients. Higher serum EGF levels were found in GERD patients carrying G/G compared with A/A or A/G (p=0.03, Wilcoxon rank test).
Conclusion:
The EGF A61G G/G genotype is associated with a near 2-fold greater risk of EAC. The G/G allele was also associated with higher EGF levels in tumor free patients with GERD. EGF genotyping can potentially identify high risk patients with GERD and Barrett's metaplasia who might benefit from increased surveillance.
INTRODUCTION
Esophageal adenocarcinoma (EAC) is an aggressive tumor with a dismal 5-year overall survival rate of under 15%.[1,2] Over the past 30 years, EAC incidence has surpassed that of esophageal squamous cell carcinoma in the Western World.[3] The majority of patients have unresectable tumors or detectable metastases at the time of diagnosis.[4] Many clinical risk factors have been established for EAC including gastroesophageal reflux disease (GERD), Barrett's esophagus (BE) [5], tobacco use, obesity, and male gender.[2,4,6] In the era of targeted therapy, evidence for molecular risk factors for EAC are mounting[7], though the prognostic value of these predictors are largely unknown.
Examples of molecular targets evaluated in EAC therapy include epidermal growth factor receptor (EGFR), matrix metalloproteinases (MMPs), and cyclooxygenase-2 (COX-2). Epidermal growth factor (EGF) is an important gene for proliferation and differentiation of epithelial cells. Its over-expression in esophageal cancer has been implicated in tumor invasion and prognosis.[8,9,10] Moreover, a single nucleotide A→G polymorphism at position +61 in the 5′UTR of the EGF gene has been associated with an elevated risk of glioma [11,14], malignant melanoma [12,15], gastric cancer [13], lung cancer[16] and transformation of cirrhosis to hepatocellular carcinoma [17].
In the present study, we evaluated the role of the EGF A61G polymorphism in EAC development and risk, using a case-control design. To further study the potential functional impact of EGF A61G in EAC risk, we measured serum EGF levels in a GERD population with endoscopic evidence of BE. Because several studies suggested malignant progression and worse prognosis in other types of cancer patients carrying the G variant of EGF A61G [13,14], we also evaluated this polymorphism for EAC survival.
EXPERIMENTAL DESIGN
Study Populations
All participants provided informed, written consent. Approval for this study was granted by the institutional review boards at the Massachusetts General Hospital, Harvard School of Public Health, and Princess Margaret Hospital.
The case-control study enrolled 313 incident EAC patients, all of which had DNA available, where 312 had successful DNA genotyping. Patients were recruited at the Massachusetts General Hospital between November, 1999 and December, 2005. The 447 controls with successful genotyping (454 screened and 454 with DNA) consisted of healthy friends and non-blood related family members who were visiting hospital patients; these hospital patients were admitted with either upper aerodigestive cancers or for cardiothoracic surgery. No controls were actual hospital patients, and none reported ever having a diagnosis of cancer. We selected controls with no self-reported history of GERD. Cases and controls were frequency-matched for age and gender distributions. Recruitment rate was over 85% for cases and for controls.
All participants were interviewed by a trained research nurse, who collected important covariate information such as age, gender, height, weight, ethnicity, gastroesophageal reflux symptoms, presence of BE, tobacco and alcohol exposure. Healthy adult body mass index (BMI) was estimated using the self-reported average heights and weights of participants during their third decade of life.
Blood samples from a cancer-free population were collected on the day of esophagoscopy for 144 patients with a history of GERD and processed within two hours. Thus, 82 GERD patients with histologically-confirmed BE and 62 GERD patients with endoscopically normal esophagus were analyzed.
DNA Extraction and Genotyping of EGF gene
DNA was extracted from whole blood for cases and controls, using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN). Commercial primers were used to perform polymerase chain reactions (PCR) using Taqman (Applied Biosystems, Foster City, CA) to identify a SNP at position 61 in the 5′ untranslated region of the EGF gene (rs4444903). The primers and conditions are available upon request, and were obtained from Assays-by-Demand® (www.appliedbiosystems.com). Cases and controls were genotyped blinded to all clinical information and case-control status, in mixed batches. Two co-authors checked all results independently and a third arbitrated discrepancies. A random 15 percent of the samples and all equivocal or failed genotyping results were repeated.
EGF ELISA
For serum analysis, 5-6 ml of blood was collected in an uncoated plastic blood tube at the time of esophagoscopy, and processed within two hours. EGF levels were measured in serum as opposed to plasma because the major source of EGF in the blood is platelets.[18] The serum was allowed to clot for 30 min at 4°C before centrifugation at 2,000 rpm for 10 min at 4°C. Serum was isolated and stored at −80°C prior to use. EGF protein was subsequently quantified in a blinded fashion using an ELISA (PeproTech, Rocky Hill, NJ). Each ELISA plate well was incubated overnight with 100μl capture antibody (1 μg/ml) before blocking with 1% BSA in PBS for 1h. Serum samples (diluted 1:20 in PBS containing 25% FBS) were analyzed in triplicate and incubated in the plate for 2h, followed by addition of 100μl of detection antibody (0.25 μg/ml) for 2h and 100μl of avidin peroxidase (1:2000) for 30 min. Wells were washed 4X with PBS containing 0.05% Tween-20 between each step. Color development was monitored after the addition of 100μl of tetramethylbenzidine (TMB; R & D Systems) using a spectrophotometric plate reader (Emax; Molecular Devices). An equal number of samples representing each genotype were run on every ELISA plate and the average concentration of each sample was determined based on a plate-specific EGF standard curve.
Outcomes Data Collection
Of 312 cases, 262 had follow-up outcomes data available. Notes and results from Massachusetts General Hospital, referring and primary care physicians and the social security death index were all utilized to determine overall (OS) and failure-free survival (FFS). FFS is defined as the time from the date of first histologic diagnosis through to the first date of either (i) cancer progression or recurrence, per RECIST (Response Evaluation Criteria in Solid Tumors) [19]; or (ii) death from any cause. Individuals, who at their last radiologic, physical, or pathologic evaluation had no evidence of recurrence or progression by RECIST, were censored at the date of their last evaluation for FFS. The last date known to be alive was used for censoring in OS analyses.
STATISTICAL METHODS
Hardy Weinberg equilibrium was assessed in cases and controls separately using Chi-squared tests. Unconditional logistic regression was used to analyze the case-control study. Although cases and controls were frequency-matched on age and gender, we adjusted for age, adult BMI, and pack-years of smoking using continuous variables, and gender and smoking status (current, exor never smoker) using indicator variables. Crude odds ratios (OR) and adjusted ORs with 95% confidence intervals were calculated for homozygous (G/G) and heterozygous (A/G) genotypes, using the wild type genotype (A/A) as the reference. Survival curves for overall and failure-free survival were constructed using the Kaplan-Meier method, and compared using the log-rank test. Cox proportional hazards models for both overall and failure-free survival were used to adjust for clinical covariates identified as being significant in univariate analyses (e.g., stage, performance status, age, gender, and smoking history).
For the serum analysis, we first assessed assay variability by comparing the percent differences in absolute serum levels between each of the triplicate samples and the serum level corresponding with the arithmetic mean of the raw data obtained from the triplicate serum analysis (termed the average serum level). The average of these percent differences (assay variability) was compared with the percent differences in absolute serum levels across different individuals (inter-individual variability). The average serum level was used for all subsequent analyses. EGF serum levels were first analyzed using the Wilcoxon rank sum test. Then we compared G/G to A/G+A/A using t-tests. Because serum EGF levels were not normally distributed, we log-transformed the serum levels first prior to parametric testing. The statistical packages, SAS 9.1 9 (Cary, NC) and R, were used for all analyses.
RESULTS
DEMOGRAPHICS
Basic demographic data are presented in Table 1. Cases and controls had similar demographic distributions of age, gender, race and pack-years. Of the known risk factors for EAC, healthy adult BMIs were significantly higher and there were substantially greater numbers of ever-smokers in the cases.
Table 1.
Demographics of patients with esophageal adenocarcinoma (cases) compared to patients who clinically and historically have no GERD symptoms (controls).
| Cases (n=312) |
Controls (n=447) |
P value comparing cases and controls |
Cases with survival outcomes (n=262) |
|
|---|---|---|---|---|
| Median Age (Range) | 64.0 (21-91) | 64.4 (19-96) | 0.57* | 64.2 (21-91) |
| Gender | ||||
| Male | 89% | 86% | 0.46** | 90% |
| Female | 11% | 14% | 10% | |
| Race | ||||
| Caucasian | 98% | 98% | 0.65** | 98% |
| Median BMI in 20's (Range) | 23.0 (15-37) | 22.5 (14-36) | 0.01* | 23.1 (15-37) |
| Smoking status | ||||
| Never Smoker | 20% | 32% | <0.001** | 19% |
| Ex-smoker | 55% | 51% | 55% | |
| Current smoker | 25% | 17% | 26% | |
|
Median pack-years of ever smokers (Range) |
34 (0.2-212) | 30 (0.1-218) | 0.21* | 35 (0.2-212) |
| Tumor Stage | ||||
| I | 7% | -- | 8% | |
| IIA | 22% | -- | 21% | |
| IIB | 18% | -- | 19% | |
| III | 25% | -- | 25% | |
| IVA | 9% | -- | 9% | |
| IVB | 18% | -- | 18% | |
| EGF genotype | ||||
| A/A | 32% (100) | 39% (174) | 0.01** | 32% |
| A/G | 43% (134) | 45% (201) | 44% | |
| G/G | 25% (78) | 16% (72) | 24% | |
| ECOG Performance Status | ||||
| 0, 0-1, and 1 (Good) | 80% | -- | 80% | |
| 1-2 and 2 (Fair) | 19% | -- | 19% | |
| 2-3 (Poor) | 1% | -- | 1% | |
Wilcoxon rank sum test
Chi-squared test
EGF GENOTYPE AND EAC RISK (Table 2)
Table 2.
Adjusted odds ratio of EGF A61G polymorphism overall, and in subsets defined by the presence of absence of concurrent BE
| Category | Crude Analysis | Adjusted Analysis * | ||||||
|---|---|---|---|---|---|---|---|---|
| G/G vs A/A | A/G vs A/A | G/G vs A/A | A/G vs A/A | |||||
| OR (95% CI) | P-value (cases/ controls) |
OR (95% CI) | P-value (cases/ controls) |
OR (95% CI) | P-value (cases/ controls) |
OR (95% CI) | P-value (cases/ controls) |
|
| All Adenocarcinomas | 1.83 (1.2-2.7) | P=0.003 (312/447) |
1.17 (0.8-1.6) | P=0.35 (312/447) |
1.81 (1.2-2.7) | p=0.005 (307/443) |
1.20 (0.9-1.7) | p=0.55 (307/443) |
|
Subset with histologic Barrett's esophagus on endoscopy/resection |
2.29 (1.4-3.8) | P=0.001 (145/447) |
1.25 (0.8-1.9) | P=0.32 (145/447) |
2.18 (1.3-3.7) | 0.004 (143/443) |
1.31 (0.8-2.1) | 0.24 (143/443) |
|
Subset of resected patients without histologic Barrett's esophagus |
1.38 (0.8-2.6) | P=0.31 (86/447) |
0.91 (0.5-1.5) | P=0.71 (86/447) |
1.37 (0.7-2.6) | 0.65 (84/443) |
0.88 (0.5-1.5) | 0.33 (84/443) |
adjusted for age, gender, smoking status, pack-years, healthy adult BMI
The concordance rate for the repeated genotyping samples was 99%. The chi-square test for Hardy Weinberg equilibrium was not significant for the 447 controls (p>0.5) but was significant for the 312 cases (p=0.04). There were no statistical differences between cases with and without survival outcome data for the following variables: age, gender, smoking status, performance status and stage (p>0.10 for each comparison).
The G/G genotype was significantly more common in the cases than controls (chi-squared test, p=0.01; Table 1). The risk of EAC in patients carrying the EGF A61G homozygous (G/G) genotype was significantly increased (Table 2) in both crude analyses (OR of 1.83 (95%CI: 1.2-2.7; p=0.003)) and after adjusting for age, gender, smoking status, cumulative tobacco exposure, and adult BMI: AOR, 1.81 (95% CI, 1.2-2.7; p=0.005). No increased risk of EAC was observed with the heterozygote (A/G) when compared to the wildtype A/A genotype (Table 2).
Since BE is a known independent risk factor for the development of EAC [5,6], we explored two subsets of EACs, each compared with the entire set of controls (Table 2). In the first subset analysis, we evaluate patients with EAC that had pathologic or endoscopic evidence of BE. In this subgroup, the G/G genotype was significantly associated with EAC risk (AOR 2.18 (95%CI, 1.3-3.7; p=0.004). In the second subset of patients that had been resected and found not to have BE, the adjusted odds ratios were lower (AOR 1.37 (95%CI, 0.7-2.6; p=0.65.
We also performed exploratory stratified risk analyses (Table 3). For continuous variables such as age and pack-years, we stratified on the basis of median pack-years and quartiles for age in the controls. We stratified BMI using the standard obesity definition cut-off of 25. Deviation from Hardy-Weinberg equilibrium was not found in any control subset (p>0.20 for each subgroup), or in most of the case subsets, except for female cases (p<0.01). The risk conferred by EGF A61G appears to be limited to younger patients, earlier stages of disease, ever-smokers particularly light smokers, and individuals who were not obese at a younger age. Given these findings, we also re-analyzed the data adjusting for healthy adult BMI six months prior to diagnosis (BMI-6M) instead of in the third decade of life (BMI). We found similar results for both groups of patients using this new definition: the AOR for the G/G genotype was 2.23 (95%CI, 1.1-3.6; p=0.001) for BMI-6M<25, and 1.74 (95%CI, 1.0-3.0; p=0.94) for BMI-6M>25. Although the G/G associated risk of EAC was conferred to both genders, a substantially greater risk was found among females.
Table 3.
Adjusted odds ratio of EGF polymorphism in various subgroups
| Subset Categories | Cases/ Controls |
G/G vs A/A | |
|---|---|---|---|
| Adjusted OR (95% CI)* | P-value | ||
| Lowest of first quartile age of onset (<55.2 years) | 75/111 | 2.04 (0.8-5.3) | 0.14 |
| Second quartile age of onset (55.2-64.5 years) | 87/111 | 2.30 (1.0-5.0) | 0.04 |
| Third quartile age of onset (64.5-72.4 years) | 86/110 | 1.24 (0.6-2.7) | 0.59 |
| Fourth quartile age of onset (>72.4 years) | 59/111 | 1.49 (0.5-4.1) | 0.44 |
| Node-negative cancer (Stages I-IIA) | 94/443 | 2.26 (1.2-4.3) | 0.01 |
| Node-positive cancer (Stages IIB-III) | 157/443 | 1.64 (1.0-2.7) | 0.05 |
| Metastatic cancer (Stage IV) | 82/443 | 1.41 (0.7-2.8) | 0.31 |
| Women | 33/55 | 16.5 (1.5-220) | 0.005 |
| Men | 274/388 | 1.6 (1.0-2.4) | 0.05 |
| Never-smoker | 62/142 | 1.09 (0.5-2.5) | 0.85 |
| Ex-smoker | 168/228 | 2.07 (1.2-3.6) | 0.01 |
| Current smoker | 77/73 | 2.80 (1.1-7.6) | 0.04 |
| Ever smoker, pack-years (0.1-30) | 111/151 | 3.07 (1.5-6.2) | 0.002 |
| Ever smoker, pack-years > 30 | 134/150 | 1.69 (0.9-3.3) | 0.13 |
| Healthy adult BMI < 25 | 212/364 | 2.23 (1.4-3.6) | 0.001 |
| Healthy adult BMI > 25 | 95/79 | 1.00 (0.4-2.3) | 0.94 |
adjusted for age, gender, smoking status, pack-years, adult BMI , and the heterozygous variant, A/G; note that in some models, specific variables will not contribute to the model (e.g. smoking status in the never smoking subset of patients)
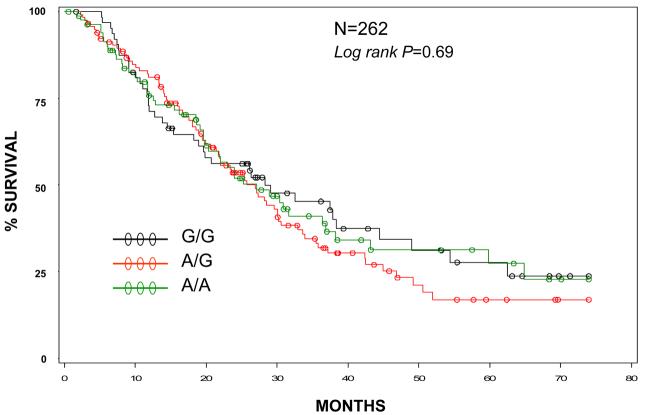
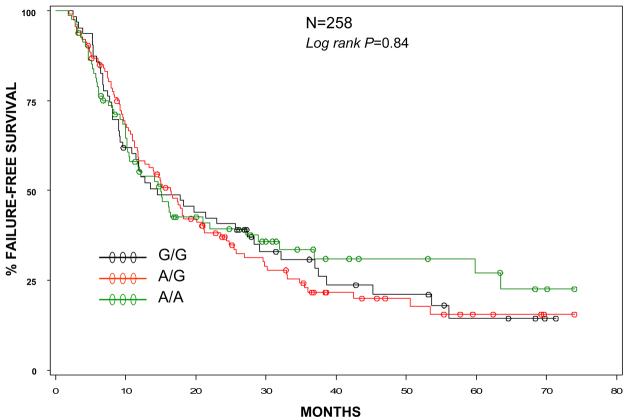
EGF GENOTYPE AND EAC PROGNOSIS (Figures 1 and 2)
Figure 1.
Overall survival by EGF genotype.
Figure 2.
Failure free survival by EGF genotype.
Median OS was 27.1 months and median FFS was 15.1 months. There were 161 deaths of 262 evaluable patients and184 recurrences or disease progression of 258 evaluable individuals. Median follow-up for OS was 29.4 months and 31.4 months for FFS.
Individuals with EAC and a G/G genotype had similar overall survival (logrank p=0.69, Figure 1) and failure free survival (logrank p=0.84, Figure 2) compared to either the A/A or A/G genotype. When adjusted for disease stage, performance status, age and pack-years (each of which was statistically significant in univariate analyses), the adjusted hazard ratios for the G/G and A/G genotypes (compared to reference group A/A) were 1.02 (95%CI 0.7-1.6) and 1.03 (0.7-1.5), respectively for OS. For FFS, the models were adjusted for stage and performance status only (the only statistically significant variables in univariate analyses) with adjusted hazard ratios of 1.05 (95%CI 0.7-1.5) for the G/G genotype and 1.12 (95%CI 0.8-1.7) for the A/G genotype. The G/G genotype did not confer a worse prognosis in any subgroup analyses, including stratified analyses by stage, smoking status, performance status, treatment or presence or absence of concurrent BE.
EGF SERUM LEVELS AND GERD (Table 4)
Table 4.
Serum EGF level in patients with gastroesophageal reflux disease (GERD)
| Analysis | Genotype | N | Mean (SD) log-EGF level (log (pg/ml)) |
Median (Range) EGF level (pg/ml) |
|---|---|---|---|---|
|
All GERD patients |
G/G | 34 | 5.79 (0.97)* | 242 (147-7092)** |
| A/- | 113 | 5.38 (0.37) | 216 (100-811) | |
|
Analyzing three genotypes separately |
G/G | 34 | 5.79 (0.97)*** | 242 (147-7092)**** |
| A/G | 68 | 5.41 (0.38) | 216 (100-811) | |
| A/A | 45 | 5.34 (0.34) | 215 (122-444) | |
|
Endoscopically normal |
G/G | 11 | 5.79 (1.10) | 214 (153-4030) |
| A/- | 54 | 5.36 (0.40) | 216 (105-810) | |
|
Barrett's Esophagus |
G/G | 23 | 5.80 (0.93)***** | 254 (148-7092)**** |
| A/- | 59 | 5.40 (0.34) | 216 (100-444) |
p<0.02 (t-test)
p=0.03 (Wilcoxon sum rank test)
p<0.0001 (ANOVA)
p=0.05 (sum rank test)
p=0.06 (t-test)
We assessed the serum EGF levels of an endoscopically evaluated population of GERD patients known to be free of esophageal cancer. This population had a median of four episodes of either acid reflux, regurgitation, or heartburn per week (range 2-20 episodes). Eighty two patients are known to have histologic evidence of BE, while the remaining 62 each have an endoscopically normal esophagus. Of the 82 patients with BE, 40 had moderate to severe dysplasia, while 42 had no or mild dysplasia.
Patients with a G/G genotype (N=34) had a statistically significant higher median serum EGF level compared with the individuals carrying at least one A allele (242 pg/ml vs. 216 pg/ml, p=0.03 Wilcoxon rank sum). In a subanalysis, serum EGF levels were also significantly elevated in the subset of GERD patients with BE carrying G/G vs other genotypes (median, 254 pg/ml vs 216 pg/ml); p=0.05 but not in endoscopically normal patients (median 214 pg/ml (G/G) vs 216 pg/ml (A/G + A/A); p>0.20). There was no correlation with the degree of Barrett's dysplasia and EGF level.
The mean variation in the average EGF concentrations across different individuals (inter-individual variation) was 6.0 fold greater than the mean variation in the repeated triplicate samples (test variability), based on assessment of the first 100 samples.
DISCUSSION
This study provides the first report of a relationship between the EGF A61G homozygous variant genotype and risk of EAC. More specifically, patients with the G/G genotype were found to have almost a 2-fold greater risk of EAC compared to the wildtype genotype. In contrast, the risk associated with the G/A heterozygote appears to be similar to the wildtype genotype. We also detected elevated EGF levels in tumor-free individuals with GERD and G/G genotype when compared with individuals with all other genotypes, suggesting that EGF and its signaling pathways may potentiate esophageal tumorigenesis in patients with GERD, a well known clinical risk factor of EAC. Since EGF/EGFR has been implicated in the progression of metaplasia to dysplasia to adenocarcinoma in patients with Barrett's epithelium[20], EGF genotype may serve as a novel biomarker to assess the risk of malignant transformation in endoscopic surveillance programs for patients with BE.
Caution must be made about over-interpreting interesting results from exploratory analyses. For example, EGF A61G appears to confer a substantially greater risk in women, but the confidence interval is wide and women formed a small proportion of study subjects. Nonetheless these exploratory results do suggest that EGF A61G may confer risk to important subsets of patients that previously had no known risk factor – namely females, early-onset EAC, and non-obese individuals. The greater association in light smokers may represent confounding by gender and age of onset of EAC, since both factors are associated with lower cumulative smoking exposures. Alternatively, heavier smokers may result in the activation of alternative carcinogenic pathways that bypass the role of the EGF A61G polymorphism. The association between EGF A61G G/G and EAC risk in earlier stage patients might suggest that the EGF A61G is associated with a less aggressive phenotype, paralleling lung cancer where individuals with EGF pathway driven tumors appear to have fewer molecular alterations [21,22]; such a conclusion is premature. A greater association of this polymorphism and EAC risk in non-obese individuals is only hypothesis generating, since using a different definition of BMI strata (i.e., BMI based on patient six months prior to diagnosis in cases and six months ago for controls) found similar results across strata. We did not have established non-GERD healthy controls to assess EGF levels, and thus, could not exclude the possibility that serum EGF levels are higher in individuals independent of GERD.
Since none of our controls had GERD symptoms, we were not able to analyze GERD-polymorphism relationships. Because we shared a common pool of controls with other aerodigestive cancers (from which the present age and gender frequency-matched control sample is derived), we had only limited information on GERD in the controls. Thus, we opted to utilize a homogeneous set of controls all lacking a history of GERD symptoms, as we have previous done.[23]
Although our EAC cases were not in Hardy Weinberg equilibrium (HWE), we have also genotyped an additional 2500 cancer cases and 1500 controls for other studies using the identical technique; each group of these other samples were in Hardy-Weinberg equilibrium (P>0.10). This suggests that our cases were properly genotyped, and that the cases out of HWE are reflective of a strong risk association for the G/G genotype.
We selected a set of endoscopically evaluated GERD patients for comparing constitutive serum EGF levels and EGF genotype. A little over half of this set of patients had BE with GERD, and the remainder had GERD without BE. GERD is the most clinically assessable risk factor for EAC risk, and we wanted to determine if EGF A61G G/G is linked with serum levels of EGF in this population. Previous studies [12] have already shown that the G allele was linked to elevated EGF production in lymphocytes in a general population; we are able to confirm an association with serum EGF levels in vivo in a more specific and clinically relevant population at risk of developing EAC. Though we would have wanted to evaluate serum EGF levels in EAC patients prior to the onset of cancer, this was clearly not feasible in a case-control setting. Finally, serum EGF levels may not entirely reflect the endoluminal milieu of the esophagus, since EGF is abundant in saliva and esophageal mucosa. Our overall results, nonetheless, are consistent with a genetically recessive genotype-phenotype correlation in at-risk GERD patients.
In contrast to investigations in gastric cancer [13] and glioma [11], the presence of an A61G single nucleotide polymorphism in the EGF gene did not impart a more aggressive phenotype or affect prognosis in patients with EAC. There are several potential explanations. First, esophageal cancer is among the most lethal of cancers. Thus, even in the earliest clinical stages, multiple pathways are already deregulated and the cancer is unlikely to be driven by the EGF pathway alone. Second, EGF A61G may not be prognostic, but predictive in the setting of clinical inhibition of the EGF pathway. However, fewer than one percent of our population received an EGFR pathway inhibitor such as an EGFR tyrosine kinase inhibitor or targeted antibody. Third, the entire EGF pathway may be affected by racial differences. In lung cancer patients, being of Asian descent (where the prevalence of a GG allele is ∼ 40%) significantly increases the chance of responding to an inhibitor of this pathway. Of note, the distribution of EGF genotype in our control population (predominantly Caucasian) was strikingly similar to previously reported studies in Caucasians [14,24].
There are studies examining populations with melanoma [24,25], gastric cancer [26], glioma [27] and cervical cancer [28] that have not shown a consistent correlation between EGF A61G polymorphism and risk of disease. This discordance can be partially attributed to ethnic variation and population stratification.
In summary, the potential future role of EGF A61G may lie in its ability to identify patients at risk for developing EAC. This novel correlation requires verification in other patient populations with EAC, particularly in other races, since our population was overwhelmingly Caucasian. EGF genotyping may thus eventually form one of a panel of molecular markers that includes p53 expression status [29] and cyclin D1 genotyping [30,31] that is clinically useful in predicting which GERD patients should have increased surveillance endoscopy. Although these molecular markers are not yet applicable in clinical practice, the EGF A61G polymorphism may be among the first markers that may identify EAC risk, with particular interest in women, non-obese individuals, and early-onset EAC, where few risk factors (either clinical or molecular) have been identified.
ACKNOWLEDGMENTS
The authors wish to thank Peggy Suen, William Puricelli, Richard Rivera Massa, Andrea Shafer, Matthew Kulke.
Grant support : Alan B. Brown Chair in Molecular Genomics, Kevin Jackson Memorial Fund, Doris Duke Charitable Foundation, Flight Attendant Medical Research Institute; NIH R01 CA109193, R01 CA074386, R03 CA110822, R01 CA090578, R01 CA 092824.
REFERENCES
- 1.Aklilu M. Ilson DH: Targeted Agents and Esophageal Cancer-The Next Step? Semin Radiat Oncol. 2006;17:62–9. doi: 10.1016/j.semradonc.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Holmes RS, Vaughan TL. Epidemiology and Pathogenesis of Esophageal Cancer. Semin Radiat Oncol. 2006;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.de Jonge P, Steyerberg EW, Kuipers EJ, et al. Risk Factors for the Development of Esophageal Adenocarcinoma in BE. Am J Gastroenterol. 2006;101:1421–29. doi: 10.1111/j.1572-0241.2006.00626.x. [DOI] [PubMed] [Google Scholar]
- 4.Enzinger PC, Mayer RJ. Esophageal Cancer. N Engl J Med. 2003;349:2241–52. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald RC. Molecular Basis of Barrett's Oesophagus and Oesophageal Adenocarcinoma. Gut. 2006;55:1810–18. doi: 10.1136/gut.2005.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaheen N, Ransohoff DF. Gastroesophageal reflux, Barrett esophagus, and esophageal cancer: scientific review. JAMA. 2002;287:1972–81. doi: 10.1001/jama.287.15.1972. [DOI] [PubMed] [Google Scholar]
- 7.Lagarde SM, ten Kate FJW, Richel DJ, Offerhaus GJA, van Lanschot JJB. Molecular prognostic factors in adenocarcinoma of the esophagus and gastroesophageal Junction. Ann Surg Oncol. 2007;14:977–91. doi: 10.1245/s10434-006-9262-y. [DOI] [PubMed] [Google Scholar]
- 8.Lu SH, Hsieh LL, Luo FC, Weinstein IB. Amplification of the EGF receptor and cmyc genes in human esophageal cancer. Int J Cancer. 1988;42:502–5. doi: 10.1002/ijc.2910420406. [DOI] [PubMed] [Google Scholar]
- 9.Hollstein MC, Smits AM, Galiana C, et al. Amplification of epidermal growth gene but no evidence for ras mutations in primary human esophageal cancers. Cancer Res. 1988;48:5119–5123. [PubMed] [Google Scholar]
- 10.Stoscheck CM, King LE. Role of Epidermal growth factor in carcinogenesis. Cancer Research. 1986;46:1030–1037. [PubMed] [Google Scholar]
- 11.Costa BM, Ferreira P, Costa S, et al. Association between functional EGF +61 polymorphism and glioma risk. Clin Cancer Res. 2007;13:2621–26. doi: 10.1158/1078-0432.CCR-06-2606. [DOI] [PubMed] [Google Scholar]
- 12.Shahbazi M, Pravica V, Nasreen N, et al. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet. 2002;359:397–401. doi: 10.1016/S0140-6736(02)07600-6. [DOI] [PubMed] [Google Scholar]
- 13.Hamai Y, Matsumura S, Matsusaki K, et al. A single nucleotide polymorphism in the 5′ untranslated region of the EGF gene is associated with occurrence and malignant progression of gastric cancer. Pathobiology. 2005;72:133–138. doi: 10.1159/000084116. [DOI] [PubMed] [Google Scholar]
- 14.Bhowmick DA, Zhuang Z, Wait SD, Weil RJ. A functional polymorphism in the EGF gene is found with increased frequency in gliobastoma multiforme patients and is associated with more aggressive disease. Cancer Res. 2004;64:1220–23. doi: 10.1158/0008-5472.can-03-3137. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto I, Roka F, Krogler J, et al. The EGF A61G polymorphism is associated with disease free period and survival in malignant melanoma. J Invest Dermatol. 2007;127:969–70. [Google Scholar]
- 16.Lim YJ, Kim JW, Song JY, et al. Epidermal growth factor gene polymorphism is different between schizophrenia and lung cancer patients in Korean population. Neurosci Lett. 2005;374:157–60. doi: 10.1016/j.neulet.2004.10.055. [DOI] [PubMed] [Google Scholar]
- 17.Tanabe KK, Lemoine A, Finkelstein DM, et al. A functional polymorphism in the EGF gene increases the risk of hepatocellular carcinoma in patients with cirrhosis. JAMA. 2008;299:53–60. doi: 10.1001/jama.2007.65. [DOI] [PubMed] [Google Scholar]
- 18.Oka Y, Orth DN. Human plasma epidermal growth factor/beta-urogastrone is associated with blood platelets. J Clin Invest. 1983;72:249–59. doi: 10.1172/JCI110964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, NCI US and Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Wo JM, Ray MB, et al. Cyclooxygenase 2 and epithelial growth factor up-regulation during progression of BE to adenocarcinoma. World J. Gastroenterol. 2006;12(6):928–34. doi: 10.3748/wjg.v12.i6.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niklinski J, Niklinska W, Chyczewski L, Becker HD, Pluygers E. Molecular genetic abnormalities in pre malignant lung lesions: biological and clinical implications. Eur J Cancer Prev. 2001;10:213–26. doi: 10.1097/00008469-200106000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Shibata T, Hanada S, Kokubu A, et al. Gene expression profiling of epidermal growth factor receptor/KRAS pathway activation in lung adenocarcinoma. Cancer Sci. 2007;98:985–91. doi: 10.1111/j.1349-7006.2007.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu G, Zhou W, Yeap BY, et al. XRCC1 and XPD polymorphisms and esophageal adenocarcinoma risk. Carcinogenesis. 2007;28:1254–58. doi: 10.1093/carcin/bgm020. [DOI] [PubMed] [Google Scholar]
- 24.Amend KL, Elder JT, Tomsho LP, et al. EGF gene polymorphism and the risk of incident primary melanoma. Cancer Res. 2004;64:2668–72. doi: 10.1158/0008-5472.can-03-3855. [DOI] [PubMed] [Google Scholar]
- 25.McCarron SL, Bateman AC, Theaker JM, Howell WM. EGF +61 gene polymorphism and susceptibility to and prognostic markers in cutaneous maliganant melanoma. Int J Cancer. 2003;107:673–75. doi: 10.1002/ijc.11448. [DOI] [PubMed] [Google Scholar]
- 26.Goto Y, Ando T, Goto H, Hamajima N. No association between EGF gene polymorphism and gastric cancer. Cancer Epidemiol Biomarkers Prev. 2005;(14):2454–6. doi: 10.1158/1055-9965.EPI-05-0401. [DOI] [PubMed] [Google Scholar]
- 27.Vauleon E, Auger N, Benouaich-Amiel A, et al. The 61A/G EGF polymorphism is functional but is neither a prognostic marker nor a risk factor for glioblastoma. Cancer Genet Cytogenet. 2007;(1):33–7. doi: 10.1016/j.cancergencyto.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Kang S, Kim JW, Park NH, et al. Epidermal growth factor 61 A/G polymorphism and uterine cervical cancer. Int J Gynecol Cancer. 2007;17:492–6. doi: 10.1111/j.1525-1438.2007.00870.x. [DOI] [PubMed] [Google Scholar]
- 29.Casson AG, Tammemagi M, Eskandarian S, Redston M, McLaughling J, Ozcelik H. P53 alterations in esophageal cancer : association with clinicopathological features, risk factors, and survival. Mol Pathol. 1998;51:71–79. doi: 10.1136/mp.51.2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casson AG, Zheng Z, Evans SC, et al. Cyclin D1 polymorphism (G870A) and risk for esophageal adenocarcinoma. Cancer. 2005;104:730–39. doi: 10.1002/cncr.21229. [DOI] [PubMed] [Google Scholar]
- 31.Izzo jg, Wu TT, Wu X, et al. Cyclin D1 Guanine/Adenine 870 polymorphism with altered protein expression is associated with genomic instability and aggressive clinical biology of esophageal adenocarcinoma. J Clin Onc. 2007;25:698–706. doi: 10.1200/JCO.2006.08.0283. [DOI] [PubMed] [Google Scholar]