Abstract
Human saliva contains thousands of mRNAs, some of which have translational value as diagnostic markers for human diseases. We have found that more than 30% of the mRNAs detected in human saliva contain AU-rich elements (ARE) in their 3′ untranslated regions (3′UTR). Since AREs are known to contribute to RNA turnover by forming complexes with ARE-binding proteins, we hypothesized that salivary mRNA stability is mediated by ARE-binding proteins in human saliva. To test this hypothesis, we monitored the in vitro degradation of a radiolabeled ARE-containing salivary mRNA (IL-8) in salivary protein extracts. The degradation of IL-8 mRNA was accelerated by competition for saliva ARE-binding proteins through the addition of excess unlabeled IL-8 mRNA fragments containing 4 tandem AREs. UV cross-linking and immunoprecipitation experiments revealed 2 ARE-binding proteins, AUF1 and HuR, associated with IL-8 mRNA in saliva. These results demonstrate that ARE-binding proteins contribute to the stability of ARE mRNAs in human saliva.
Keywords: Human saliva, mRNA stability, AU-rich elements, AUF1, HuR
INTRODUCTION
Human saliva mRNA biomarker discovery is an emerging field for noninvasive diagnostic applications. Discoveries resulting from the mRNA profiling of saliva from healthy humans and persons with oral cancer have opened up a new area of research for the study of salivary gene expression (Li et al., 2004a,b). In addition, the mRNA profiling of saliva by real-time PCR analysis has been used in forensic applications (Juusola and Ballantyne, 2005, 2007; Setzer et al., 2008). Most recently, salivary amylase mRNA was identified as a biomarker for sleep disorders (Seugnet et al., 2006). Of interest is that salivary amylase mRNA is more informative than amylase activity as a biomarker for sleep deprivation. At present, the mechanisms that regulate salivary RNA stability are not clear. We have shown that mRNA in the saliva is associated with certain macromolecules and is protected against ribonucleases (Park et al., 2006). However, detailed studies on the stability of salivary RNA are required before we can understand the molecular mechanisms underlying protection of RNA from RNases in human saliva.
There are several RNA-binding proteins that are either directly or indirectly involved in mRNA stability in higher eukaryotes. Cis-acting adenylate-uridylate-rich elements (AREs) present in the 3′ untranslated region (UTR) of certain mRNAs are known to be involved in the regulation of gene expression by affecting the mRNA stability through interaction with specific AU-rich RNA-binding proteins (Khabar, 2005). Of these, HuR and AUF1 are the best-characterized. HuR mediates stabilization of AU-rich mRNAs against exonucleases (Ma et al., 1996; Keene, 1999; Li et al., 2007). AUF1 is principally associated with destabilization of target mRNAs (Dreyfuss et al., 2002), but several lines of evidence suggest that this protein is also implicated in mRNA stabilization through interaction with other protein macromolecules (Kiledjian et al., 1997). Therefore, the level of RNA-protein interaction determines the stability of the mRNAs.
The goal of this study was to identify the molecular factors responsible for the stabilization of mRNAs in human saliva. Based on our observation that ~30% of salivary mRNAs contain an ARE motif in their 3′UTR, we hypothesize that these ARE motifs are involved in salivary mRNA stability. In this paper, we demonstrate that two ARE-binding proteins, AUF1 and HuR, are present in human saliva and are involved in the stability of AU-rich-containing mRNAs, providing the first evidence that the ARE-binding proteins can modulate mRNA stability in human saliva.
MATERIALS & METHODS
Participants
Saliva samples were obtained from healthy volunteers from the Division of Otolaryngology, Head and Neck Surgery, at the Medical Center, University of California, Los Angeles (UCLA), CA, in accordance with a protocol approved by the UCLA Institutional Review Board. All participants gave written informed consent, and the ethics committee of UCLA approved the study. The mean age of the volunteers was 31 yrs (range, 26–43 yrs). The volunteers had no history of malignancy, immune deficiencies, autoimmune disorders, hepatitis, or HIV infection.
Preparation of Salivary Protein Extracts
The unstimulated supernatant of the whole saliva was collected as described previously (Li et al., 2004a,b). Further, the supernatant saliva samples were concentrated 10X by means of Amicon ultracentrifugal filters (Millipore, Bedford, MA, USA) with a molecular-weight cut-off of 5 kDa. The concentrated saliva was recovered and then stored at −80°C until further use.
RNA Substrates and Synthesis
The pCMV6 vector containing full-length IL-8 ORF under control of the T7 promoter was obtained from OriGene Technologies, Inc. (Rockville, MD, USA). We prepared the 1700-nt full-length IL-8 mRNA by linearizing the vector using Bgl II. We prepared β-actin mRNA (approximately 1300 nt of the transcript) by linearizing the pGEM3 vector containing the full-length cDNA under the control of the T7 promoter (1.8 kb inserted using a Pst I site) with Hind III. The remaining IL-8 ARE-containing short transcripts (see Appendix for RNA sequences) were a kind gift from Helmut Holtmann (Winzen et al., 2004). Labeled RNA (107 to 108 cpm/μg) was synthesized with the use of a Maxiscript in vitro transcription kit (Ambion, Austin, TX, USA), and prepared according to the manufacturer’s protocol.
RNA Decay Assay
The mRNA decay assay was conducted as described previously (Ford and Wilusz, 1999; Viswanathan et al., 2003), with slight modifications (for details, see the Appendix).
UV Cross-linking Assay
The UV cross-linking assay was performed with 10 μg of saliva protein extracts as described previously (Moraes et al., 2006).
RNA-Protein Co-immunoprecipitation
Following UV cross-linking, samples were incubated with RNase A (30 U/sample) for 20 min at 37°C. Next, a 200-μL quantity of Net2 buffer (50 mM Tris, pH 7.6, 75 mM NaCl, 0.05% NP-40) was added to the samples, which were then incubated for 1 hr at 4°C with anti-AUF1 (Upstate Biotechnologies, Lake Placid, NY, USA) or anti-HuR (Zymed Laboratories, San Francisco, CA, USA) antibodies. Antigen-antibody complexes were collected on protein-A sepharose beads and washed 5 times with Net2 buffer. The immunoprecipitated cross-linked proteins were separated on a 10% SDS polyacrylamide gel. The gels were dried and scanned with a phosphor imager.
Preparation of Recombinant HuR
Recombinant GST-HuR was prepared according to a previously described method (Abdelmohsen et al., 2007).
RESULTS
Enriched AU-rich mRNAs in Human Saliva
We have previously shown, through microarray analysis, that saliva from healthy individuals contains more than 3000 transcripts (Li et al., 2004b). Of these transcripts, 185 (core transcriptome) are commonly present in all persons’ saliva. Here, we found that 30% (56/185) of these mRNAs contain the nanomeric “WWAUUUAWW” ARE motif (Appendix Table 1) at their 3′ UTR, and that 60% (110/180) contain the pentameric “AUUUA” motif. These ARE motifs were confirmed by means of the ARED database (Bakheet et al., 2006). Next, we determined whether the previously identified 7 salivary oral cancer mRNA biomarkers contained ARE motifs (Li et al., 2004a). Interestingly, 5 of the 7 salivary oral cancer mRNA biomarkers (71%) contained ARE at the 3′ UTR, i.e., DUSP1, H3F3A, IL1β, IL-8, OAZ1, S100P, and SAT (Appendix Table 2). It should be noted that approximately 5–8% of mRNAs in the human transcriptome contain putative ARE sequences (Bakheet et al., 2006). The salivary transcriptome therefore contains a five-fold greater content of ARE-containing mRNAs, suggesting that these elements may contribute to a common feature of salivary mRNA that may account for their collective presence in the saliva.
Excess IL-8 AU-rich mRNA Leads to Rapid Degradation of Full-length IL-8 RNA in vitro
To evaluate the significance of AREs in salivary mRNA stability, we used human saliva extracts to examine decay of ARE-containing salivary mRNAs. IL-8 mRNA was selected, since it is an ARE-containing salivary mRNA present in the normal salivary transcriptome core. Radiolabeled full-length endogenous IL-8 mRNA slowly degrades in the presence of human salivary extract, with more than 60% of transcript remaining after 5 min of incubation with salivary extracts (Fig. 1B). To determine whether ARE motifs and potential ARE-binding proteins are important for IL-8 mRNA stability, we performed competition assays with a 60-nucleotide (nt) stretch of IL-8 mRNA containing 4 AREs. Competition for potential ARE-binding proteins by the addition of an excess cold 60-nt ARE fragment led to increased and more rapid degradation of the full-length transcript. After 1 min in the presence of the 60-nt ARE fragment, IL-8 mRNA degradation had already exceeded that seen at the five-minute timepoint in the absence of competitor (Fig. 1C). The addition of equimolar concentrations of β-actin mRNA or unlabeled 60-nt ARE mutant fragment (60-nt IL-8 AREmt), in which point mutations had been introduced into the AREs, did not decrease IL-8 mRNA stability (Figs. 1B, 1C). To confirm that ARE motifs contribute to IL-8 mRNA stability, we tested the effect of ARE mutation on the degradation of the 60-nt ARE fragment. As expected, mutation of ARE motifs in IL-8 mRNA (Figs. 1D, 1E) accelerated degradation. Interestingly, the degradation rate of an IL-8 mRNA fragment harboring mutation in only 1 ARE (IL-8 ARE 3g-mt) was comparable with that of the un-mutated fragment, suggesting that the 3 AUUUA motifs are needed for IL-8 mRNA stability in saliva. These results suggest that the ARE motif is important for RNA stability in saliva.
Figure 1.
Addition of AUUUA-containing competitor accelerates IL-8 RNA degradation in vitro. (A) Sequences of RNA used for the study. (B) Representative autoradiograms showing in vitro decay of full-length, radiolabeled IL-8 mRNA in the absence of competitor or in the presence of unlabeled 60-nt IL-8 ARE competitor (100 ng), unlabeled mutated IL-8 ARE competitor (100 ng), or a control 950-nt β-actin mRNA (100 ng). (C) Kinetics of transcript degradation. (D) Representative autoradiographs showing decay of 60-nt IL-8 ARE-containing fragments in which all 4 AREs were intact (IL-8 ARE), fragments in which a single ARE was mutated (3g mt), or fragments in which all AREs were mutated (All mt). (E) Kinetics of transcript degradation. The data are from three independent experiments; values are means ± SD.
AUF1 and HuR Bind and Stabilize IL-8 Transcripts
To identify the potential proteins that bind to and stabilize salivary IL-8 mRNA, we performed UV cross-linking experiments. Analysis of cross-linked RNA-protein revealed that 2 proteins, at around 45 kDa and 36 kDa, were associated with IL-8 mRNA, but not with IL-8 mRNA harboring ARE mutations (Fig. 2). We performed immunoprecipitation of the cross-linked RNA-protein complexes with AUF1 or HuR antibodies, to determine whether these species could be positively identified. Both AUF1 and HuR antibodies specifically precipitated the 45- and 36-kDa species, respectively, from samples containing IL-8 AREs, but not from samples containing IL-8 ARE mutants or β-actin mRNAs (Fig. 2, lanes 5 and 8). Non-specific antisera did not precipitate any cross-linked material (lanes 7 and 10). Thus, analysis of our data indicates that AUF1 and HuR proteins bind to IL-8 AREs in saliva lysates. Further, immunodepletion of AUF1 and HuR proteins accelerated the degradation of IL-8 mRNA in an in vitro decay assay (Appendix Figs. 2A, 2B, 2C). The results suggest that both HuR and AUF1 are important for stabilization of AU-rich mRNAs in human saliva, and that HuR has a more significant role in stabilizing IL-8 mRNA (Appendix Fig. 2).
Figure 2.
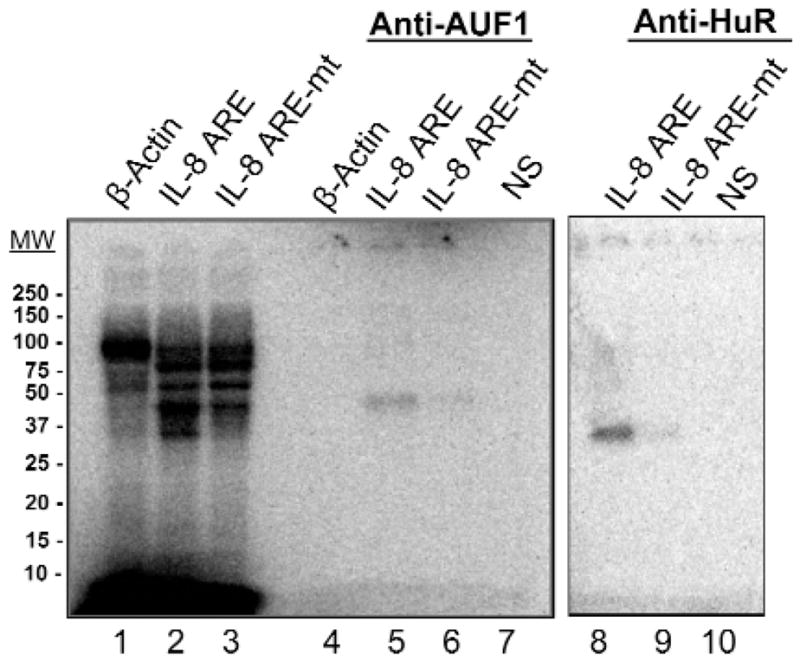
IL-8 mRNA associates with AUF1 and HuR in human saliva protein lysates. Salivary lysate was UV-cross-linked with IL-8 ARE, IL-8 ARE mutant, or β-actin. Cross-linked RNA-protein complexes were then immunoprecipitated with anti-AUF1 (lanes 4–6) or anti-HuR (lanes 8 and 9) and analyzed by SDS-PAGE. Lanes 1–3 show the cross-linked extract prior to immunoprecipitation. NS, non-specific serum.
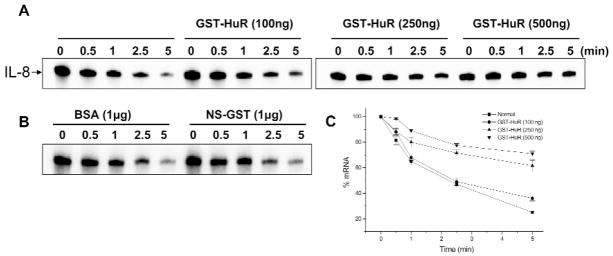
Recombinant HuR Protects IL-8 mRNA against Degradation by Salivary Proteins in vitro
Since HuR clearly binds to IL-8 salivary mRNA, we next determined the effect of recombinant HuR binding on IL-8 mRNA stability as measured by in vitro degradation assays. To test whether HuR is important for IL-8 mRNA stability, we performed in vitro decay assays with saliva proteins and increasing amounts of recombinant GST-HuR. The addition of increasing amounts of recombinant GST-HuR resulted in dose-dependent protection against IL-8 mRNA degradation (Figs. 3A, 3B). IL-8 mRNA stability was unaffected by 100 ng GST-HuR, while larger amounts of GST-HuR in the range of 250 to 500 ng greatly enhanced the stability of IL-8 transcripts. BSA and non-specific GST proteins did not alter degradation of IL-8 transcripts incubated in HuR-depleted saliva proteins. Analysis of these data clearly shows that HuR promotes IL-8 mRNA stability in saliva.
Figure 3.
Recombinant HuR protects IL-8 mRNA against in vitro degradation by human saliva lysates. (A) Representative autoradiographs showing decay of IL-8 mRNA in the presence of increasing concentrations of recombinant HuR. (B) Autoradiographs showing decay in the presence of equal amounts of BSA or non-specific recombinant GST fusion protein. (C) Effect of recombinant HuR on the kinetics of transcript degradation. The data are from three independent experiments; values are means ± SD.
Detection of ARE-binding Proteins in Human Salivary Glands and Saliva
The large concentration of ARE-containing mRNAs in saliva, coupled with the finding that ARE-binding proteins confer stability to ARE-containing mRNAs, prompted us to determine whether the major salivary glands produce ARE-binding proteins. Four AUF1 isoforms have been identified (Lu and Schneider, 2004), and all were present in human submandibular (SM) and parotid (PA) glands (Fig. 4) as well as in saliva (Appendix Fig. 1). HuR was also present in human salivary glands (Fig. 4) as well as in the saliva (Appendix Fig. 1). Interestingly, the ARE mRNA decay-promoting protein TTP (Tristetraprolin) was present in both salivary glands, but was completely absent from saliva (Appendix Fig. 1). These findings suggest that salivary glands are sources of saliva ARE-binding proteins.
Figure 4.
AUF1 and HuR protein levels in human salivary glands and saliva. Western blot analysis of AUF1 isoforms and HuR in the submandibular gland (SM), the parotid gland (PA), and the saliva (SL).
DISCUSSION
Saliva is a non-invasive body fluid that can be used for the identification of biomarkers for disease detection. Our laboratory (Li et al., 2004b) and other groups (Juusola and Ballantyne, 2005; Gomes et al., 2006) have shown that human salivary secretions contain mRNAs that can be used as biomarkers for oral cancer, periodontal disease, and forensic applications. We have previously shown that the association of RNAs with certain macromolecules confers stability against nucleases present in the saliva (Park et al., 2006). Here, we provide evidence that a mechanism of salivary mRNA stability in the majority of salivary mRNA is mediated by AU-rich elements. Using IL-8 mRNA as an example, we have shown that ARE-containing transcripts are stabilized in the presence of human salivary protein lysates and are associated with HuR and AUF1, as shown by UV cross-linking and co-immunoprecipitation experiments. Importantly, immuno-depletion experiments revealed that both HuR and AUF1 promoted the stability of ARE mRNAs in the saliva. The former appeared to be particularly important for IL-8 mRNA stability, as shown by in vitro reconstitution experiments.
ARE-containing genes comprise 5–8% of the human transcriptome (Bakheet et al., 2006) and encode proteins that are involved in diverse biological processes. We have found that more than 30% of the normal saliva RNA transcripts contain AUUUA motifs, making up the majority of the salivary transcriptome. Analysis of oral cancer transcriptomic biomarkers revealed that 70% (5/7) of the ARE-containing mRNAs, suggesting that the ARE sequence contributes to their stability and function. It is tempting to speculate that the half-life of these salivary mRNAs is determined by cooperative interaction with ARE-binding proteins regulating their stabilization and destabilization. Findings demonstrating enhanced mRNA stability in tumor cells suggest that altered recognition of ARE sequences in neoplasia may lead to improper function of mRNAs (Schuler and Cole, 1988; Xu et al., 1997). Here we show that binding of AUF1 and HuR to the IL-8 mRNA enhanced stability of the ARE transcripts in saliva.
In humans, salivary nucleic acid has been viewed as unstable due to the presence of strong salivary RNases (Eichel et al., 1964; Bobek et al., 1993). Nevertheless, the protein-associated RNAs are protected against ribonucleases (Moallem et al., 1998), with decreased protein-binding rendering RNAs susceptible to RNase activity. Thus, protein-RNA interactions are vital for RNA stability. We have previously shown that secreted salivary RNA is protected from degradation, whereas mRNA that is exogenously (in vitro-transcribed) added to saliva is rapidly degraded (Park et al., 2006). The current study extends these findings by identifying the salivary proteins (AUF1 and HuR) that bind to and stabilize salivary ARE-containing RNAs, regulating salivary mRNA stability. HuR has been shown to bind to and stabilize many different ARE-containing mRNAs in different cell systems (Ma et al., 1996; Keene, 1999; Dixon et al., 2001). In salivary gland carcinomas, HuR was overexpressed and stabilized Cox-2 mRNA (Cho et al., 2007). We showed that recombinant HuR promoted stabilization of IL-8 mRNA in salivary lysates, suggesting that it regulates ARE mRNA stability in the saliva. AUF1 is a cytosolic protein that reportedly associates with other proteins to stabilize mRNAs such as α-globin mRNA (Kiledjian et al., 1997; Chkheidze et al., 1999). The additive function of AUF1 and HuR may possibly determine the rate of AU-rich mRNA decay in human saliva, although it is currently unknown whether different mRNA species may have different stabilities in saliva. Although HuR and AUF1 stabilize salivary ARE mRNAs, it is unclear whether these proteins are associated with mRNAs prior to their passage from the salivary ductal fluid into the oral cavity. Future study of protein-RNA interactions in secretions from the three major glands will circumvent buccal cell and other contamination and will enable us to obtain a more complete understanding of the results from these salivary RNA studies. In conclusion, the results presented here provide a novel link between the mRNA stability and ARE-binding proteins in human saliva.
Supplementary Material
A supplemental appendix to this article is published electronically only at http://jdr.iadrjournals.org/cgi/content/full/87/8/772/DC1.
Acknowledgments
We thank Myriam Gorospe and Lynn Wu for supplying pGST-HuR plasmid and protein preparations. Technical guidance regarding the ARE transcriptome was provided by Khalid Khabar and is gratefully acknowledged. This work is supported by NIH grants R01DE017170 (to D.T.W.), U01DE16275 (D.T.W.), and K99DE018165-01A1 (V.P.).
References
- Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobek LA, Tsai H, Biesbrock AR, Levine MJ. Molecular cloning, sequence, and specificity of expression of the gene encoding the low molecular weight human salivary mucin (MUC7) J Biol Chem. 1993;268:20563–20569. [PubMed] [Google Scholar]
- Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alphaCP. Mol Cell Biol. 1999;19:4572–4581. doi: 10.1128/mcb.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NP, Han HS, Soh Y, Son HJ. Overexpression of cyclooxygenase-2 correlates with cytoplasmic HuR expression in salivary mucoepidermoid carcinoma but not in pleomorphic adenoma. J Oral Pathol Med. 2007;36:297–303. doi: 10.1111/j.1600-0714.2007.00526.x. [DOI] [PubMed] [Google Scholar]
- Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Eichel HJ, Conger N, Chernick WS. Acid and alkaline ribonucleases of human parotid, submaxillary, and whole saliva. Arch Biochem Biophys. 1964;107:197–208. doi: 10.1016/0003-9861(64)90322-4. [DOI] [PubMed] [Google Scholar]
- Ford LP, Wilusz J. An in vitro system using HeLa cytoplasmic extracts that reproduces regulated mRNA stability. Methods. 1999;17:21–27. doi: 10.1006/meth.1998.0703. [DOI] [PubMed] [Google Scholar]
- Gomes MA, Rodrigues FH, Afonso-Cardoso SR, Buso AM, Silva AG, Favoreto S, Jr, et al. Levels of immunoglobulin A1 and messenger RNA for interferon gamma and tumor necrosis factor alpha in total saliva from patients with diabetes mellitus type 2 with chronic periodontal disease. J Periodontal Res. 2006;41:177–183. doi: 10.1111/j.1600-0765.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- Juusola J, Ballantyne J. Multiplex mRNA profiling for the identification of body fluids. Forensic Sci Int. 2005;152:1–12. doi: 10.1016/j.forsciint.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Juusola J, Ballantyne J. mRNA profiling for body fluid identification by multiplex quantitative RT-PCR. J Forensic Sci. 2007;52:1252–1262. doi: 10.1111/j.1556-4029.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar KS. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J Interferon Cytokine Res. 2005;25:1–10. doi: 10.1089/jir.2005.25.1. [DOI] [PubMed] [Google Scholar]
- Kiledjian M, DeMaria CT, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the alpha-globin mRNA stability complex. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. erratum in Mol Cell Biol 17:6202, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XL, Andersen JB, Ezelle HJ, Wilson GM, Hassel BA. Post-transcriptional regulation of RNase-L expression is mediated by the 3′-untranslated region of its mRNA. J Biol Chem. 2007;282:7950–7960. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004a;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhou X, St John MA, Wong DT. RNA profiling of cell-free saliva using microarray technology. J Dent Res. 2004b;83:199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]
- Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- Moallem E, Kilav R, Silver J, Naveh-Many T. RNA-protein binding and post-transcriptional regulation of parathyroid hormone gene expression by calcium and phosphate. J Biol Chem. 1998;273:5253–5259. doi: 10.1074/jbc.273.9.5253. [DOI] [PubMed] [Google Scholar]
- Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NJ, Li Y, Yu T, Brinkman BM, Wong DT. Characterization of RNA in saliva. Clin Chem. 2006;52:988–994. doi: 10.1373/clinchem.2005.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD, Cole MD. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell. 1988;55:1115–1122. doi: 10.1016/0092-8674(88)90256-5. [DOI] [PubMed] [Google Scholar]
- Setzer M, Juusola J, Ballantyne J. Recovery and stability of RNA in vaginal swabs and blood, semen, and saliva stains. J Forensic Sci. 2008;53:296–305. doi: 10.1111/j.1556-4029.2007.00652.x. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Boero J, Gottschalk L, Duntley SP, Shaw PJ. Identification of a biomarker for sleep drive in flies and humans. Proc Natl Acad Sci USA. 2006;103:19913–19918. doi: 10.1073/pnas.0609463104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan P, Chen J, Chiang YC, Denis CL. Identification of multiple RNA features that influence CCR4 deadenylation activity. J Biol Chem. 2003;278:14949–14955. doi: 10.1074/jbc.M211794200. [DOI] [PubMed] [Google Scholar]
- Winzen R, Gowrishankar G, Bollig F, Redich N, Resch K, Holtmann H. Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol Cell Biol. 2004;24:4835–4847. doi: 10.1128/MCB.24.11.4835-4847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A supplemental appendix to this article is published electronically only at http://jdr.iadrjournals.org/cgi/content/full/87/8/772/DC1.